Abstract
The premature aging disease Hutchinson–Gilford Progeria Syndrome (HGPS) is caused by a mutant lamin A (LAΔ50). Nuclei in cells expressing LAΔ50 are abnormally shaped and display a loss of heterochromatin. To determine the mechanisms responsible for the loss of heterochromatin, epigenetic marks regulating either facultative or constitutive heterochromatin were examined. In cells from a female HGPS patient, histone H3 trimethylated on lysine 27 (H3K27me3), a mark for facultative heterochromatin, is lost on the inactive X chromosome (Xi). The methyltransferase responsible for this mark, EZH2, is also down-regulated. These alterations are detectable before the changes in nuclear shape that are considered to be the pathological hallmarks of HGPS cells. The results also show a down-regulation of the pericentric constitutive heterochromatin mark, histone H3 trimethylated on lysine 9, and an altered association of this mark with heterochromatin protein 1α (Hp1α) and the CREST antigen. This loss of constitutive heterochromatin is accompanied by an up-regulation of pericentric satellite III repeat transcripts. In contrast to these decreases in histone H3 methylation states, there is an increase in the trimethylation of histone H4K20, an epigenetic mark for constitutive heterochromatin. Expression of LAΔ50 in normal cells induces changes in histone methylation patterns similar to those seen in HGPS cells. The epigenetic changes described most likely represent molecular mechanisms responsible for the rapid progression of premature aging in HGPS patients.
Keywords: histone methylation, heterochromatin, progeria
Hutchinson–Gilford Progeria Syndrome (HGPS) is a premature aging disease usually diagnosed in the first 12–18 months of life (1). HGPS is characterized by a rapid progression of disorders including hair loss, growth retardation, lack of s.c. fat, aged-looking skin, osteoporosis, and arteriosclerosis (2, 3). Patients with HGPS usually die from heart attacks or strokes at ≈13 years (4). The common form of HGPS is caused by a conservative heterozygous mutation (1824 C>T) in the human nuclear lamin A (LA) gene (LMNA), which introduces a splice site resulting in the synthesis of LA with 50 amino acids deleted near its C terminus [mutant LA (LAΔ50)] (5, 6).
Nuclear lamins are divided into A and B types. Lamins A and C (LA/C) are derived from LMNA by alternative splicing, whereas lamins B1 and B2 are derived from different genes. The B type lamins are expressed in every cell, whereas the expression of A type lamins is developmentally regulated. Lamins have a common tripartite structure with an α-helical central rod domain flanked by globular N- and C-terminal domains (7). The basic structural unit of lamins is a dimer consisting of two parallel and in-register protein chains that form a coiled coil through the association of their rod domains (8). Lamin dimers assemble in a head-to-tail fashion forming protofilaments that interact laterally to form numerous higher-order structures (9).
Lamins are the major components of the nuclear lamina, a protein network located adjacent to the inner nuclear membrane. The lamina maintains the mechanical properties and shape of nuclei, and it has been proposed that it provides a molecular docking site for peripheral heterochromatin (7, 10, 11). Lamins are also distributed throughout the nucleoplasm, where they appear to be essential for DNA replication and RNA polymerase II transcription (7). Interest in the lamins has increased because of recent reports of ≈200 mutations in LMNA causing >15 distinct diseases, collectively known as the “laminopathies” (12).
HGPS fibroblasts accumulate LAΔ50 as a function of their age in culture and coincidentally display changes in nuclear shape and architecture, most notably a loss of heterochromatin (1). In this study, we examine changes in the epigenetic histone marks, H3K27me3 for facultative heterochromatin, histone H3 trimethylated on lysine 9 (H3K9me3), and H4 trimethylated on lysine 20 (H4K20me3) for constitutive heterochromatin (13), which take place in HGPS cells as they age in culture. The data define alterations in repressive histone lysine methylation (14) as early events in disease manifestation and suggest that HGPS-specific LMNA mutations induce perturbed epigenetic control of chromatin structure.
Results
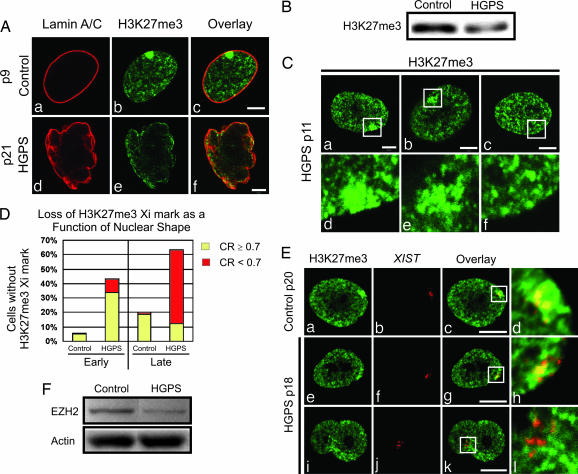
To initiate these studies, we examined the inactive X chromosome (Xi) of female HGPS patient fibroblasts and controls. The Xi is identifiable as a heterochromatic domain usually associated with the nuclear lamina. Silencing of the Xi is regulated by trimethylation of lysine 27 in histone H3 (H3K27me3) and X-inactive specific transcript (XIST) RNA (15–17). Fibroblasts from an age-matched normal female sibling of an HGPS patient were used as controls. In early passages [passages 9–13 (p9–13)], ≈94% of control cells (n = 193) contained an Xi closely associated with the lamina, as determined by immunofluorescence with anti-H3K27me3 (Fig. 1A a–c and D). This number decreased slightly to ≈80% (n = 103) at later passages (p20–25) (Fig. 1D). In addition, punctate H3K27me3 staining was present throughout the nucleoplasm of control cells at all passages (Fig. 1A a–c and data not shown). In early-passage HGPS cells, ≈57% (n = 200) of the cells possessed an Xi that reacted with anti-H3K27me3, which decreased to ≈36% by p21 (n = 186; see Fig. 1D). Interestingly, ≈58% (n = 200) of the H3K27me3-positive Xi consisted of loose arrays of discrete fluorescent granules in p9–13 cells (Fig. 1C b and e). These loose arrays suggest the existence of transition states during the loss of the H3K27me3 mark (Fig. 1C a–f). Late-passage HGPS cells also showed a reduction in overall staining compared with controls, which was confirmed by immunoblotting (Fig. 1 A d–f and B). Significantly, these observations revealed that many normally shaped early-passage HGPS cell nuclei lost the H3K27me3 mark on their Xi (Fig. 1 C c and f and D). To quantify this, we correlated the H3K27me3 mark on Xi with the nuclear contour ratios of control and HGPS cells at different passages. In controls, the contour ratio remained 0.9 ± 0.2 from early to late passages, whereas in HGPS cells, there was a significant decrease from early (0.9 ± 0.2) to late (0.5 ± 0.2) passages. In early-passage HGPS cells, where the nuclear contour ratio was ≈0.9, ≈34% did not have a detectable Xi by H3K27me3 staining (Fig. 1D). These observations demonstrate that significant alterations in chromatin regulation precede the nuclear shape changes that have been considered a pathological hallmark of HGPS cells (1, 18, 19).
Fig. 1.
H3K27me3 decreases and XIST RNA remains with Xi at all passages in HGPS cells. (A) Cells were processed for immunofluorescence with antibodies against LA/C (a and d) and H3K27me3 (b and e; overlay, c and f). Control cell nuclei at p9 displayed a distinct LA/C nuclear rim (a) associated with a compact Xi (b). The HGPS cells showed a change in nuclear morphology by p21 by using anti-LA/C (d), and there was no obvious Xi revealed by anti-H3K27me3 (e). (Scale bars, 5 μm.) (B) There was a decrease in H3K27me3 by Western blot analysis of HGPS compared with control total cell extracts at p20. (C) In early-passage HGPS cells stained with anti-H3K27me3, the Xi appeared either as a uniformly stained compact domain (a and d) or a loose array of closely spaced granules (b and e), or it could not be detected (a typical lamina region in a nucleus with no Xi staining is depicted in the white box; c and f). d–f are enlargements (×4) of the regions in the white boxes in a–c. (Scale bars, 5 μm.) (D) Nuclear contour ratios (CR; see also ref. 1) were determined for control and HGPS cells at early and late passages (nuclei with a CR ≥ 0.7 were considered nonlobulated, and those with a CR < 0.7 were considered lobulated). In early-passage HGPS cells, ≈43% of the nuclei lost their H3K27me3 Xi mark, and ≈80% of these were normally shaped. In late-passage cells, ≈63% lost this mark, whereas ≈20% of these nuclei were normally shaped. In early-passage controls, ≈6% of the nuclei did not contain an obvious Xi, as distinguished by anti-H3K27me3. At late passages, this number increased to ≈20%. (E) Control (a–d) and HGPS cells (e–l) were prepared for XIST RNA FISH and stained with anti-H3K27me3. (a–d) In controls at early and late passages (p13–20), XIST RNA was associated with the Xi, and ≈93% (n = 102) also contained the H3K27me3 mark. (e–l) In HGPS cells, XIST FISH revealed an Xi at all passages, whereas by p17–18, the H3K27me3 staining on Xi was lost (≈53%; n = 82). (i–l) In lobulated HGPS nuclei lacking the H3K27me3 mark on Xi, the XIST RNA staining was more dispersed. d, h, and l are enlargements (×5.6) of the overlay regions in the white boxes (c, g, and k). (Scale bars, 10 μm.) (F) Western blotting shows a decrease in EZH2 in HGPS cells compared with controls at p14. Actin was used as a loading control.
We also determined whether there were changes in the association between XIST RNA and the Xi. All HGPS and control cells at early and late passages contained an Xi, as determined by XIST RNA FISH (Fig. 1E). When the Xi also stained with anti-H3K27me3, the signals colocalized (Fig. 1E d and h). When the Xi did not stain with anti-H3K27me3, the FISH signal was more dispersed (Fig. 1E i–l).
We also examined the expression level of EZH2, the methyltransferase responsible for the trimethylation of H3K27 (20). Immunoblotting demonstrated a significant reduction in EZH2 in HGPS cells compared with controls at p14 (Fig. 1F). There was also a reduction in the level of the mRNA encoding EZH2 in HGPS relative to control cells, as determined by RT-PCR (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site). The EZH2 mRNA level was decreased 10-fold in p25 HGPS cells relative to p14 control cells (see Fig. 6). In agreement with the FISH data, RT-PCR revealed no differences in the amounts of XIST RNA between early- and late-passage HGPS cells and late-passage controls (see Fig. 6 and data not shown).
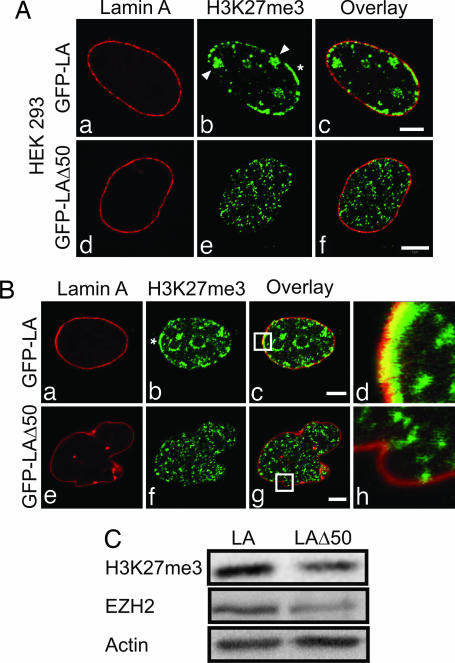
We next studied the effects of transient expression of LAΔ50 in human embryonic kidney 293 (HEK293) cells. In the nuclei of nontransfected or GFP-LA-expressing cells, ≈80% (n = 100) contained multiple Xi that stained with anti-H3K27me3. Prominent staining not associated with the Xis was also observed in the lamina region and in a few nucleoplasmic foci (Fig. 2A a–c). In contrast, ≈20% (n = 100) of the cells expressing GFP-LAΔ50 contained a distinct Xi with a significant reduction in H3K27me3 staining in the lamina region (Fig. 2A d–f). This H3K27me3 staining was restricted to small foci distributed throughout the nucleoplasm (Fig. 2A d–f). Furthermore, in all untransfected and transfected HEK293 cells examined, Xis were observed by XIST RNA FISH (data not shown).
Fig. 2.
Expression of GFP-LAΔ50 in HEK293 and HeLa cells. (A) HEK293 cells transiently expressing either GFP-LA or GFP-LAΔ50 were stained with anti-H3K27me3. In controls (GFP-LA), the lamina appeared normal (a), and H3K27me3 staining revealed several Xi (arrowheads, b) and arrays of peripheral heterochromatin (∗, b) colocalizing with the lamina (c). Most cells expressing GFP-LAΔ50 showed a dramatic loss of Xi and peripheral heterochromatin staining with anti-H3K27me3, even in those retaining normal nuclear shapes (d–f). (B) HeLa cells expressing either GFP-LA (a–d) or GFP-LAΔ50 (e–h) were stained with anti-H3K27me3. GFP-LA-expressing cells displayed a normally shaped nucleus (a) and punctate H3K27me3 staining throughout the nucleus, with prominent arrays at the nuclear periphery (∗, b), where it overlaps with the lamina (c and d). Expression of GFP-LAΔ50 caused lobulations in the nuclear envelopes of the majority of HeLa cells (e), and there was an overall loss of H3K27me3 staining, especially in the region of the lamina (f–h). Enlargements (×7.4) of the overlay areas indicated by white boxes (c and g) are shown (d and h). (C) Whole-cell extracts derived from HeLa cells expressing GFP-LA or GFP-LAΔ50 for 24–48 h were analyzed by immunoblotting. Note the reductions in H3K27me3 and EZH2 in cells expressing the mutant protein. Actin was used as a loading control. (Scale bars, 5 μm.)
The reduction in H3K27me3-positive chromatin domains related to the expression of LAΔ50 was further investigated by using HeLa cell lines induced to express GFP-LA or GFP-LAΔ50. In noninduced control and GFP-LA-expressing cells, small and large foci were present throughout the nucleoplasm, and there was a significant colocalization between LA and chromatin regions marked by H3K27me3 at the lamina (Fig. 2B a–d). Most nuclei of cells expressing GFP-LAΔ50 were convoluted, and there was an overall reduction in H3K27me3 fluorescence in ≈83% of the cells (n = 100; Fig. 2Bf). In addition, the large nucleoplasmic foci were not detected, and there was a substantial reduction of H3K27me3 staining in the lamina region (Fig. 2B e–h). Immunoblotting revealed a reduction in both H3K27me3 and EZH2 in cells expressing LAΔ50 (Fig. 2C).
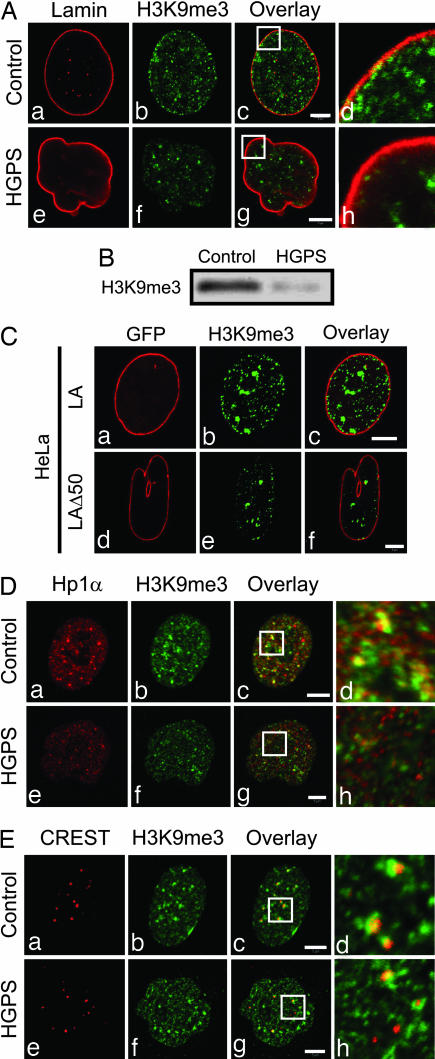
The above data indicate there are major aberrations in the facultative heterochromatin of HGPS cells, as detected by anti-H3K27me3. To extend our analyses, we investigated possible changes in constitutive heterochromatin by examining H3K9me3, a known mark for pericentric heterochromatin (21). In early- and late-passage control cells, the staining pattern consisted mainly of small foci interspersed with a few larger foci both in the nucleoplasm and in the lamina region (Fig. 3A a–d and data not shown). Identical patterns were seen in early-passage HGPS cells (data not shown). In late-passage HGPS cells, there was an overall reduction of H3K9me3-positive foci in highly lobulated nuclei and a dramatic reduction in the lamina region (Fig. 3A e–h). An overall decrease of H3K9me3 in late-passage HGPS cells was also observed by immunoblotting (Fig. 3B).
Fig. 3.
LAΔ50 expression results in changes in constitutive heterochromatin. (A) HGPS and control cells were stained by using antibodies directed against LA/C and H3K9me3. (a–d) Early- and late-passage control cell nuclei appeared normal. The H3K9me3 staining pattern consisted of small foci dispersed throughout the nucleoplasm and in the lamina region. (e–h) In late-passage (p21) HGPS cells, the H3K9me3 immunofluorescence pattern was altered in lobulated nuclei. These nuclei showed an overall decrease in the amount of H3K9me3 staining, which was most obvious in the lamina region. Enlargements (×4.5) of the overlay areas indicated by white boxes (c and g) are shown (d and h). (B) There is a decrease in the amount of H3K9me3 in HGPS cells (p23) compared with control cells (p24), as indicated by Western blotting. (C) HeLa cells expressing either GFP-LA or GFP-LAΔ50 were stained with anti-H3K9me3. (a–c) In controls, the H3K9me3 staining pattern consisted of a few large foci interspersed among smaller foci throughout the nucleoplasm and in close proximity to the lamina. When GFP-LAΔ50 was expressed, the majority of HeLa cell nuclei were lobulated (d–f), and there were fewer H3K9me3 foci both in the nucleoplasm and the lamina (d–f). (D) Midpassage (p16) control and HGPS cells were stained with antibodies to H3K9me3 and Hp1α. Both antibodies stained large and small foci throughout the nucleoplasm. (a–d) In controls, many of the large foci stained with both antibodies, as seen in the overlay. (e–h) In HGPS cells, this colocalization is reduced. Enlargements (×4) of the overlay areas indicated by white boxes (c and g) are shown (d and h). (E) Control and HGPS cells were also double-labeled with CREST antiserum and anti-H3K9me3 (p16). (a–d) In controls, CREST revealed a typical pattern of kinetochores, and the majority of these were associated with H3K9me3. (e–h) In HGPS cells, many kinetochores were not associated with H3K9me3. Enlargements (×4.4) of the overlay areas indicated by white boxes (c and g) are shown (d and h). (Scale bars, 5 μm.)
We further investigated the relationship between constitutive heterochromatin and the expression of LAΔ50 by analyzing the patterns of H3K9me3 in the HeLa cell line expressing GFP-LAΔ50 (Fig. 3C d–f). Nuclei in control cells expressing GFP-LA show H3K9me3 foci distributed throughout the nucleoplasm and in linear arrays in the lamina region (Fig. 3C a–c). Expression of GFP-LAΔ50 resulted in a decrease in the number of nucleoplasmic and lamina-associated foci, especially in highly lobulated nuclei (Fig. 3C d–f).
Based on the changes in H3K9me3, we examined one of its binding partners, heterochromatin protein 1α (Hp1α; see ref. 22). In control nuclei, immunofluorescence observations revealed many bright foci containing both Hp1α and H3K9me3 (Fig. 3D a–d). In midpassage HGPS cells (p16), in addition to an overall reduction in fluorescence intensity for both Hp1α and H3K9me3, their association decreased. This loss of association appeared in many nuclei before extensive nuclear lobulation (Fig. 3D e–h). We also examined the relationship between H3K9me3 and kinetochores using CREST autoimmune serum (23). In controls, foci stained with H3K9me3 are closely associated with kinetochores (Fig. 3E a–d), whereas in midpassage HGPS cells, the number of such associations was reduced (Fig. 3E e–h). However, the overall H3K9me3 pattern in HGPS cells did not appear significantly altered at this passage number compared with p22 (compare Fig. 3 A b and f with E b and f). In addition, transcript levels for SUV39H1 and SUV39H2 (24), the histone methyltransferases generating H3K9me3, were reduced in late-passage HGPS and control cells by RT-PCR (data not shown).
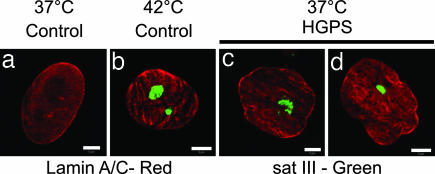
The loss of H3K9me3 associated with pericentric heterochromatin in HGPS cells suggested there may be an up-regulation of the transcriptional activity of pericentric regions. This possibility was investigated by examining the expression levels of two major pericentric DNA sequences, satellite III (sat III) and α satellite repeats (25, 26). There was no change in α satellite transcripts in HGPS cells even in late passages, as determined by RT-PCR (data not shown). In contrast, a significant increase in the level of chromosome 9 sat III transcripts was detected in mid- to late-passage HGPS cells by RT-PCR (see Supporting Text and Fig. 7, which are published as supporting information on the PNAS web site). This was confirmed by FISH using probes for chromosome 9 sat III RNA (Fig. 4c and d). No sat III signal was detected in controls (Fig. 4a) unless they were heat-shocked for 1 h at 42°C, a treatment that induces chromosome 9 sat III transcription in nuclear stress bodies (Fig. 4b; see ref. 25). These data demonstrate that the normal epigenetic silencing at DNA repeats associated with pericentric heterochromatin is altered in HGPS cells.
Fig. 4.
Up-regulation of sat III transcripts in HGPS cells. Control and HGPS cells were analyzed by sat III RNA FISH. (a) No signal was observed in control cells grown at 37°C. (b) After heat shock, prominent stress bodies (green) were observed. (c and d) In HGPS cells grown at 37°C, stress bodies were observed in nuclei regardless of the extent of their lobulation. (Scale bars, 5 μm.)
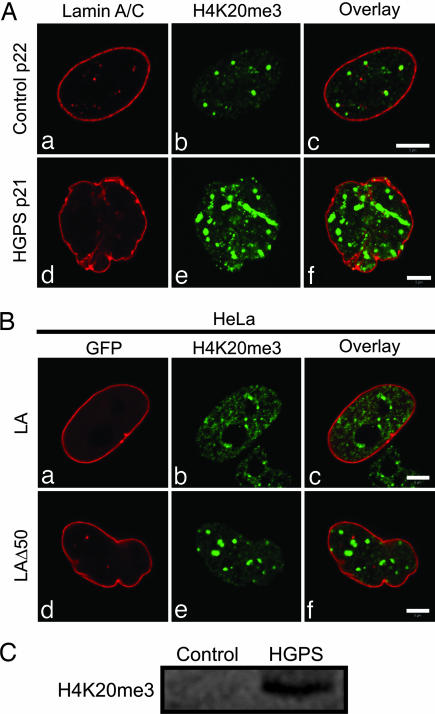
A second major epigenetic mark for pericentric heterochromatin is the presence of H4K20me3 (27). In late-passage HGPS cells prepared for immunofluorescence with anti-H4K20me3, there is an increase in the number and size of brightly stained nuclear foci compared with control cells (Fig. 5A). Similar results were obtained with HeLa cells expressing LAΔ50 (Fig. 5B). Immunoblotting of late-passage progeria cells supports the up-regulation of the H4K20me3 mark (Fig. 5C). These data provide additional evidence for the role of LA in the regulation of pericentric heterochromatin.
Fig. 5.
Increased staining of H4K20me3 in late-passage HGPS cells. (A) Late-passage HGPS and control cells were stained with antibodies directed against LA/LC and H4K20me3. The fluorescence pattern looked similar in early and late passages (a; data not shown). (a–c) Nuclei of control cells at p22 displayed foci of H4K20me3 throughout the nucleoplasm that were frequently adjacent to lamin foci. In late-passage HGPS cells (p21), the H4K20me3 staining pattern consisted of much larger intensely staining structures. (d–f) There was also a loss of the association with lamin foci. (B a and d) HeLa cells expressing either GFP-LA or GFP-LAΔ50 were prepared for immunofluorescence by using anti-H4K20me3. (b and c) The H4K20me3 staining pattern consisted of mainly small foci throughout the nucleoplasm. (d–f) When GFP-LAΔ50 was expressed in HeLa cells, the majority of their nuclei were lobulated, and the H4K20me3 antibody showed large bright foci throughout the nucleoplasm. (C) Immunoblotting of control (p23) and HGPS cells (p24) for H4K20me3 showed a significant increase in HGPS cells. (Scale bars, 5 μm.)
Discussion
We demonstrate that there are significant changes in the epigenetic control of both facultative and constitutive heterochromatin in cells from HGPS patients. These changes are a direct consequence of the expression of the HGPS lamin mutant, LAΔ50, which alters the state of histone methylation. At early passages of cultured female HGPS cells, we observed a loss of the H3K27me3 mark for facultative heterochromatin (13), especially in association with the Xi (17). The loss of this mark occurred before obvious changes in nuclear shape and was accompanied by a 9- to 10-fold down-regulation of EZH2. These results suggest that the low levels of LAΔ50 present in early passages of HGPS cells are sufficient to alter chromatin organization before the extreme changes in nuclear shape and architecture that typify later-passage HGPS cells (1, 28). These shape changes are coincident with an increased concentration of LAΔ50 in the lamina region (1), which may be related to the abnormal association of this mutant protein with the inner nuclear membrane due presumably to its permanent state of farnesylation (29).
To determine whether factors other than the H3K27me3 mark regulating Xi are also lost, we examined whether there were changes in XIST RNA. Transcription of XIST RNA is initiated early in development when it coats the Xi. This coating is required for targeting the Eed/EZH2 methyltransferase complex to the Xi, resulting in the trimethylation of H3K27 (30). Both XIST RNA and H3K27me3 are necessary for the transcriptional silencing of the Xi (15–17). Furthermore, XIST RNA has been shown to be required for the maintenance of H3K27me3 on the Xi and for its silencing (17). Our results demonstrate that the loss of H3K27me3 is not accompanied by the loss of XIST RNA on the Xi in HGPS cells or in female cells expressing LAΔ50. However, XIST RNA does not appear as condensed or as closely associated with the nuclear lamina when the H3K27me3 mark is lost from the Xi. This suggests that facultative heterochromatic regions become transcriptionally active in HGPS cells.
In support of this idea, we show there is transcriptional activation of normally silenced pericentric constitutive heterochromatin in HGPS cells. Specifically, the sat III DNA repeats of chromosome 9 are transcribed in the patients’ cells when the H3K9me3 mark is down-regulated. Normally, sat III DNA repeats are up-regulated only in response to environmental stresses such as heat shock (25, 31). These changes in constitutive heterochromatin were detected at later passages than the changes in facultative heterochromatin reflected by the loss of the H3K27me3 mark in HGPS cells.
Our data demonstrate that the HGPS mutant protein, LAΔ50, alters sites of histone methylation known to regulate heterochromatin. This implies that, under normal conditions, LA is linked to heterochromatin regulation. The precise mechanisms linking LA to the factors associated with histone methylation complexes are unknown. It is conceivable that these linkages may be facilitated by a lamin network that provides a 3D molecular scaffold throughout the nucleus. It has been hypothesized that such a scaffold could connect, coordinate, and act as an assembly platform for the complex molecular machines involved in a wide range of functions, including chromatin organization (7, 10).
This study also provides evidence that the rapid aging phenotype of HGPS reflects aspects of normal aging at the molecular level. For example, the up-regulation of H4K20me3 seen in late-passage HGPS cells is also observed in tissues of old rats (32). The loss of the Xi mark, H3K27me3, in ≈20% of p21 control cells also supports this possibility. Therefore, it is obvious that specific changes in the normal epigenetic control of chromatin and gene regulation shed new light on the most probable causes of this devastating disease as well as the functions of LA in the aging process of normal cells.
Materials and Methods
Cell Culture.
Cells from a female HGPS patient (AG11513) and an age-matched unaffected female (AG08470) (Coriell Cell Repositories, Camden, NJ) were grown as described (1). HEK293 cells were grown in MEM (Invitrogen) containing 10% FBS. A HeLa Tet-On cell line was maintained in DMEM high glucose/10% FBS (Tet free)/penicillin/streptomycin/200 μg/ml G418 (Clontech).
Immunofluorescence.
Cells grown on coverslips were fixed either in methanol for 10 min at −20°C or in 3% paraformaldehyde (PFA) in PBS and processed for immunofluorescence (1). Rabbit antibodies against H3K27me3, H3K9me3, and H4K20me3 (all 1:500; see ref. 17); mouse antibodies against LA/C (1:30) (JoL2; Chemicon) and Hp1α (1:1,000) (Euromedex, Souffelwyershein, France); rat anti-LA/C (1:200; see ref. 1); and CREST human antiserum (1:50; from B. Brinkley, Baylor College of Medicine, Houston) were used. Goat secondary antibodies were anti-rabbit IgG–Alexa Fluor 568 or 488, anti-rat IgG–Alexa Fluor 568, and anti-mouse IgG–Alexa Fluor 568 (Molecular Probes). Cells were observed with a Zeiss LSM 510 META (Zeiss), and nuclear contour ratios were computed (1).
Immunoblotting.
Cells were lysed in Laemmli buffer (33), and equal cell numbers were analyzed by Western blotting by using primary rabbit antibodies (1:1,000) against H3K27me3, H3K9me3, H4K20me3, actin (Sigma), and EZH2 (1:200; from M. Busslinger, Research Institute of Molecular Pathology). Secondary antibodies were donkey anti-rabbit and anti-mouse IgG conjugated to horseradish peroxidase (1:5,000; Kirkegaard & Perry Laboratories).
FISH.
XIST RNA.
Cells grown on Lab-Tek slides (Nunc) were fixed with 4% PFA in PBS at 22°C, permeabilized in 0.1% sodium citrate (pH 6.0)/0.5% Triton X-100 for 5 min at 4°C, washed in PBS, washed twice in PBS/0.1% Tween-20 (PBST), incubated in blocking solution (PBST/2.5% BSA/0.4 units of RNasin per microliter), and overlaid with anti-H3K27me3 in blocking solution for 1.5 h at 22°C. The slides were washed twice in PBST and twice in PBST with 0.25% BSA, overlaid with goat anti-rabbit IgG–Alexa 488 for 1 h in blocking solution at 22°C, washed twice in PBST and PBS, and postfixed in 4% PFA in PBS. The slides were placed in cytobuffer (100 mM NaCl/300 mM sucrose/3 mM MgCl2/10 mM Pipes, pH 6.8) for 30 sec, washed in cytobuffer with 0.5% Triton X-100 for 5 min, washed in cytobuffer for 30 sec, and fixed for 10 min in 4% PFA in PBS. The cells were dehydrated in 70%, 80%, 95%, and 100% ethanol; air-dried; and hybridized overnight at 37°C with XIST RNA probes in hybridization buffer [2× sodium saline citrate (SSC)/0.3 M NaCl/30 mM Na-citrate (pH 7.0)/50% formamide]. Slides were washed three times for 5 min in hybridization buffer at 40°C, 10 min in 2× SSC at 40°C, and three times for 5 min in 4× SSC at 22°C.
sat III RNA.
Cells grown on coverslips were prepared essentially as described (34). In some cases, cells were heat-shocked for 1 h at 42°C followed by recovery at 37°C for 3 h. Cells were washed in PBS (all steps were at 22°C unless otherwise specified), fixed in 4% PFA for 10 min, washed in PBS/100 mM Tris (pH 7.4), washed twice on ice in PBS/0.5% Triton X-100, washed on ice in PBS, dehydrated stepwise with ice-cold ethanol (70%, 80%, 90%, and 100%) for 5 min each, and then air-dried. The sat III probes (25) were hybridized overnight at 37°C. The cells were washed three times for 5 min in hybridization buffer, three times for 5 min in 2× SSC and for 15 min in 4× SSC/0.1% Tween-20; incubated for 60 min in 20 μg/ml FITC–avidin (Vector Laboratories) in PBS; washed three times for 3 min in PBST, then in PBS; incubated for 60 min in 10 μg/ml biotinylated anti-avidin D (Vector Laboratories) in PBS; washed three times for 3 min in PBST; and then washed for 3 min in PBS. After a 60-min incubation in 20 μg/ml FITC–avidin (Vector Laboratories) in PBS and two 3-min washes in PBST, then PBS, the cells were blocked for 30 min in PBST with 10% FCS. The cells were stained with anti-LA/C (JoL2; Chemicon; 1:200) for 60 min, washed twice for 3 min in PBST and for 3 min in PBS, then incubated for 60 min with goat anti-mouse IgG–Alexa 633 (1:300) (Molecular Probes) and washed in PBS. Cells were examined by confocal microscopy.
Preparation of FISH Probes.
The DNA probes for XIST RNA FISH were amplified by PCR from a genomic bacterial artificial chromosome clone containing the XIST locus using the primer pairs 1aF (acccgtcttttgttggacag)/1aR (ctcccaaagtgctgggatta), 1bF (aagacaattgccctggaatc)/1bR (gcacataacaggccaagaaaa), and 12F (ctctcagaccccttttgcag)/12R (ggccttgtctggtcaacatt). XIST RNA FISH probes were Cy3-labeled (see ref. 17), and hybridization was carried out as described (16). The probes used for sat III have been described (25). FISH probes were provided by Integrated DNA Technologies (Coralville, IA).
Expression of GFP-LA and GFP-LAΔ50.
Transfections were performed by electroporation with a GenePulser Xcell (Bio-Rad) at 200 V and 950 μF using ≈1 × 107 cells and 10–20 μg of DNA in 400 μl of DMEM in a 4-mm gap cuvette using either pEGFP-myc-LMNA or pEGFP-myc-hLMNAΔ150 (1). The cells were plated onto coverslips and processed for immunofluorescence after 24–48 h.
Stable HeLa Tet-On cell lines were prepared expressing either EGFP-myc-LA or EGFP-myc-LAΔ50. The coding regions from pEGFP-myc-hLMNA or -hLMNAΔ150 were cut out with NheI and XbaI and ligated into the pTRE vector (Clontech) cut with XbaI. The resulting plasmids (pTRE-EGFP-myc-hLMNA and -hLMNAΔ50) were dual-transfected into HeLa Tet-On cells with the selection vector pTKHyg (Clontech) at a 20:1 ratio. The transfected cells were plated into two 10-cm culture dishes, incubated for 48 h, released by trypsinization, and each distributed to 10 10-cm dishes maintained in medium containing 200 μg/ml each G418 and hygromycin until visible colonies emerged. Single colonies were transferred to individual wells of 24-well plates. After the expansion of each colony, cells were induced with 2 mg/ml doxycycline for 24 h, and GFP fusion protein expression was analyzed by fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank A. Wutz and D. Pullirsch (Institute of Molecular Pathology, Vienna) for the XIST bacterial artificial chromosome clone and for advice on RNA FISH. R.D.G. is supported by the National Cancer Institute, the National Institute on Aging, the Ellison Foundation, and the Progeria Research Foundation. T.D. is supported by a Schroedinger Fellowship of the Austrian Science Foundation. T.J. is supported by the Institute of Molecular Pathology, Boehringer Ingelheim, the European Union (FP6 Network of Excellence, “The Epigenome”), and the Austrian GEN-AU initiative.
Abbreviations
- Xi
inactive X chromosome
- H3K27me3
histone H3 trimethylated on lysine 27
- H3K9me3
histone H3 trimethylated on lysine 9
- H4K20me3
histone H4 trimethylated on lysine 20
- HGPS
Hutchinson–Gilford Progeria Syndrome
- LA/C
lamin A/C
- LAΔ50
mutant LA in HGPS cells
- PFA
paraformaldehyde
- sat III
satellite III
- pn
passage n
- HEK293
human embryonic kidney 293.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Goldman R. D., Shumaker D. K., Erdos M. R., Eriksson M., Goldman A. E., Gordon L. B., Gruenbaum Y., Khuon S., Mendez M., Varga R., Collins F. S. Proc. Natl. Acad. Sci. USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hutchinson J. Medicochir. Trans. 1886;69:473–477. doi: 10.1177/095952878606900127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilford M. Practitioner. 1904;73:188–217. [Google Scholar]
- 4.Stables G. I., Morley W. N. J. R. Soc. Med. 1994;87:243–244. [PMC free article] [PubMed] [Google Scholar]
- 5.de Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C. L., Munnich A., Le Merrer M., Levy N. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson M., Brown W. T., Gordon L. B., Glynn M. W., Singer J., Scott L., Erdos M. R., Robbins C. M., Moses T. Y., Berglund P., et al. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman R. D., Gruenbaum Y., Moir R. D., Shumaker D. K., Spann T. P. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 8.Strelkov S. V., Schumacher J., Burkhard P., Aebi U., Herrmann H. J. Mol. Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- 9.Stuurman N., Heins S., Aebi U. J. Struct. Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 10.Shumaker D. K., Kuczmarski E. R., Goldman R. D. Curr. Opin. Cell Biol. 2003;15:358–366. doi: 10.1016/s0955-0674(03)00050-4. [DOI] [PubMed] [Google Scholar]
- 11.Gruenbaum Y., Margalit A., Goldman R. D., Shumaker D. K., Wilson K. L. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 12.Hegele R. Clin. Genet. 2005;68:31–34. doi: 10.1111/j.1399-0004.2005.00447.x. [DOI] [PubMed] [Google Scholar]
- 13.Sarma K., Reinberg D. Nat. Rev. Mol. Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- 14.Sims R. J., 3rd, Nishioka K., Reinberg D. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Clemson C. M., Chow J. C., Brown C. J., Lawrence J. B. J. Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wutz A., Jaenisch R. Mol. Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 17.Kohlmaier A., Savarese F., Lachner M., Martens J., Jenuwein T., Wutz A. PloS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capell B. C., Erdos M. R., Madigan J. P., Fiordalisi J. J., Varga R., Conneely K. N., Gordon L. B., Der C. J., Cox A. D., Collins F. S. Proc. Natl. Acad. Sci. USA. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaffidi P., Misteli T. Nat. Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 21.Peters A. H., Kubicek S., Mechtler K., O’Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., et al. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 22.Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 23.Van Hooser A. A., Mancini M. A., Allis C. D., Sullivan K. F., Brinkley B. R. FASEB J. 1999;13(Suppl. 2):S216–S220. doi: 10.1096/fasebj.13.9002.s216. [DOI] [PubMed] [Google Scholar]
- 24.Peters A. H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., et al. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 25.Rizzi N., Denegri M., Chiodi I., Corioni M., Valgardsdottir R., Cobianchi F., Riva S., Biamonti G. Mol. Biol. Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grady D. L., Ratliff R. L., Robinson D. L., McCanlies E. C., Meyne J., Moyzis R. K. Proc. Natl. Acad. Sci. USA. 1992;89:1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClintock D., Gordon L. B., Djabali K. Proc. Natl. Acad. Sci. USA. 2006;103:2154–2159. doi: 10.1073/pnas.0511133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young S. G., Fong L. G., Michaelis S. J. Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Rougeulle C., Chaumeil J., Sarma K., Allis C. D., Reinberg D., Avner P., Heard E. Mol. Cell. Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jolly C., Metz A., Govin J., Vigneron M., Turner B. M., Khochbin S., Vourc’h C. J. Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarg B., Koutzamani E., Helliger W., Rundquist I., Lindner H. H. J. Biol. Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U. K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Jolly C., Mongelard F., Robert-Nicoud M., Vourc’h C. J. Histochem. Cytochem. 1997;45:1585–1592. doi: 10.1177/002215549704501201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.