Abstract
Nuclear hormone receptors (NRs) function as ligand dependent DNA binding proteins that translate physiological/nutritional signals into gene regulation. Dysfunctional NR signaling leads to many disorders in reproduction, inflammation, and metabolism. The opportunity to identify novel regulatory pathways in the context of human health and disease drives the challenge to unravel the biological function of the “orphan nuclear hormone receptors”. For example, the Rev-erb (NR1D) subgroup (Rev-erbα/NR1D1 and Rev-erbβ/NR1D2) of orphan NRs are transcriptional silencers and negative regulators of RORα mediated trans-activation. The NR1D subgroup is highly enriched in peripheral tissues with onerous energy demands including skeletal muscle, brown and white adipose, brain, liver and kidney. This alludes to the involvement of this subgroup in metabolism. In this context, Rev-erbα-/- mice have a dyslipidemic phenotype. Recent studies in vascular smooth and skeletal muscle cells also suggest that the NR1D subgroup modulates inflammation by regulating IκBα/NFκB dependent gene expression. Rev-erbα has been identified as a critical regulator (and target) of circadian rhythm, a factor in blood pressure control and inflammation. Finally, two recent reports have demonstrated: (i) lithium mediated regulation of Rev-erbα stability and (ii) E75 (the Drosophila orthologue of human Rev-erbα) is tightly bound by heme, and functions as a “gas sensor” through interaction with CO/NO and interferes with the repression of DHR3 (the Drosophila orthologue of human RORα). In conclusion, the role of these receptors at the cross-roads of metabolism, inflammation, and circadian cycling underscores the importance of understanding the organ-specific function of the NR1D subgroup in homeostasis.
The NR1D subgroup of orphan NRs, Rev-erbα (NR1D1) and Rev-erbβ (NR1D2)
All members of the NR superfamily display a highly conserved structural organisation with an amino terminal AB region (that encodes activation function 1, AF-1); followed by the C-region which encodes the DNA binding domain (DBD); a linker region D and the C-terminal E region. The DE region encodes the ligand binding domain (LBD), and a transcriptional domain (in helix 12), denoted as activation function 2 (AF-2) [Gronemeyer et al., 2004].
The Rev-erb/NR1D subgroup contains two members, Rev-erbα (NR1D1;[Lazar et al., 1989]) and Rev-erbβ (NR1D2;[Dumas et al., 1994; Retnakaran et al., 1994]). The Rev-erbs, and retinoid related orphan receptors (RORs; [Giguere et al., 1994; Laitinen and Staels, 2003] bind as monomers to the consensus NR half-site motif, flanked by an 6bp AT-rich sequence (A/T)6 PuGGTCA, or as a homodimer to a direct repeat of the core motif separated by 2 bp. Although these receptors are closely related, and bind to the same motif, they function in an opposing manner. The NR1D subgroup function as dominant transcriptional silencers, and they repress trans-activation mediated by RORα [Forman et al., 1994]. Interestingly, RORα activates Rev-erbα transcription [Delerive et al., 2002; Raspe et al., 2002b], and Rev-erbα represses its own transcription [Adelmant et al., 1996].
Homology modeling of the putative LBDs of the NR1D subgroup suggested that the pocket is occupied by bulky side chains, and the small residual cavity could not “accommodate” a classical ligand. Concordantly, NR1D1 and 2 are the only members of the superfamily that do not encode helix 12 and an AF-2 domain [Renaud et al., 2000]. However, the existence of a novel small molecular regulator cannot be precluded. Molecular modeling also revealed a very hydrophobic surface due to the absence of H12, exposing residues from H3, loop 3-4, H4, and H11 that mediate corepressor recruitment.
Rev-erbs and lipid metabolism
Rev-erbα has been shown to be involved in repressing rat apoA1 gene expression [Vu-Dac et al., 1998]. in vivo studies have shown that apoA1 reduces free cholesterol accumulation in atherosclerotic lesions of ApoE-deficient mice. Moreover, human Rev-erbα and β specifically repress apoCIII gene expression in rats [Coste and Rodriguez, 2002] and humans [Raspe et al., 2002a]. ApoCIII is a component of HDL and VLDL that regulates triglyceride levels and lipoprotein lipase activity. In this context Rev-erbα null mutant mice [Chomez et al., 2000] have elevated VLDL triglyceride and apoCIII levels [Raspe et al., 2002a]. It is interesting to note that plasma triglyceride and apoC-III protein concentrations in staggerer (sg/sg) mice, homozygous for a deletion in the RORα gene, were significantly lower than in wild type littermates [Laitinen and Staels, 2003]. This is in agreement with the repression and activation of apoCIII gene expression by Rev-erbα and RORα, respectively. Moreover, it highlights the regulatory crosstalk between Rev-erbα and RORα in the context of the blood lipid profile.
Hypolipidemic fibrate drugs increase Rev-erbα expression in human liver cells [Gervois et al., 1999]. Interestingly, the fibrate mediated activation occurs at the transcriptional level in a PPARα dependent manner. The mechanism involves a previously characterized Rev-erbα response element; a unique DR2 element whose 5’ region is flanked by AT-rich sequence. Rev-erbα and PPARα compete for binding to this response element in the Rev-erbα promoter. Moreover, Rev-erbα has been shown to modulate hydratase-dehydrogenase expression, a key gene involved in peroxisomal β-oxidation pathway through repression of PPARα-dependent transactivation [Kassam et al., 1999]. Similarly, the mechanisms involved direct binding to a PPRE, however, the Rev-erbα and PPARα binding characteristics to the response element were distinct. Furthermore, fibrates regulate apoA1 gene expression in a species specific manner. For example, fibrates increase and decrease apoA1 expression in humans and rats respectively. Firstly, the repression of apoA1 expression in fibrates is due to a 3 nucleotide difference in the rat promoter that produces a non-functional PPARα response element. Secondly, the rat but not the human promoter harbors a Rev-erbα response element that mediates Rev-erbα dependent repression in the presence of fibrates. These studies underscore the crosstalk between Rev-erbα and PPARα in the control of lipid homeostasis.
Rev-erbα and β are regulated in a biphasic manner during DMI (Dexamethasone/IBMX/Insulin) and rosiglitazone induced adipocyte differentiation [Chawla and Lazar, 1993; Fu et al., 2005; Laitinen et al., 2005]. Rosiglitazone induced adipogenesis involves PPAR-γ mediated induction of Rev-erbα mRNA expression. In agreement, exogenous expression of Rev-erbα in 3T3-L1 cells promoted adipogenic differentiation [Fontaine et al., 2003]. In contrast, Rev-erbα and β antagonistically regulate mammalian muscle differentiation and their expression levels decrease during myogenesis [Burke et al., 1996; Downes et al., 1995].
In vivo studies in mice suggest that Rev-erbα is preferentially expressed in fast-twitch type IIB and intermediate typeIIA fibers. Moreover, in slow twitch (type I) soleus muscle of Rev-erbα homozygous and heterozygous null mutant mice, there is a significant increase in the expression of the β/slow myosin heavy chain (MyHC) isoform [Pircher et al., 2005]. Type IIB fast fibers utilize glycolysis, in contrast, type IIA combine fast twitch glycolytic capacity with aerobic metabolism. Type I fibers are dependent on the aerobic metabolism of fatty acids. The expression of Rev-erbα in energy demanding tissues, the fiber specific expression characteristics, and the effect of the null mutation on the muscle phenotype suggest a role in metabolism.
In this context, it has been shown that ectopic Rev-erbβΔE (that lacks the LBD) expression in skeletal muscle cells attenuated the expression of many genes involved in fatty acid/lipid absorption (including FAT/CD36, FABP-3 and -4), and an expected increase in RORα expression [Ramakrishnan et al., 2005]. Interestingly, these authors reported a dramatic decrease in the expression of the mRNA encoding myostatin, a TGF-β family member that plays a crucial role in regulating muscle and adipose mass [Gonzalez-Cadavid and Bhasin, 2004]. Myostatin -/- mice have increased skeletal muscle mass (from muscle hypertrophy and hyperplasia) accompanied by decreased adiposity. McPherron and Lee reported that the introduction of the myostatin mutation in mouse models of obesity and diabetes, suppressed fat accumulation and hyperglycemia [McPherron and Lee, 2002]. The Rev-erbα and Rev-erbβ target genes and tissues are schematically depicted in Figure 1.
Figure 1. Target genes of the NR1D subgroup.
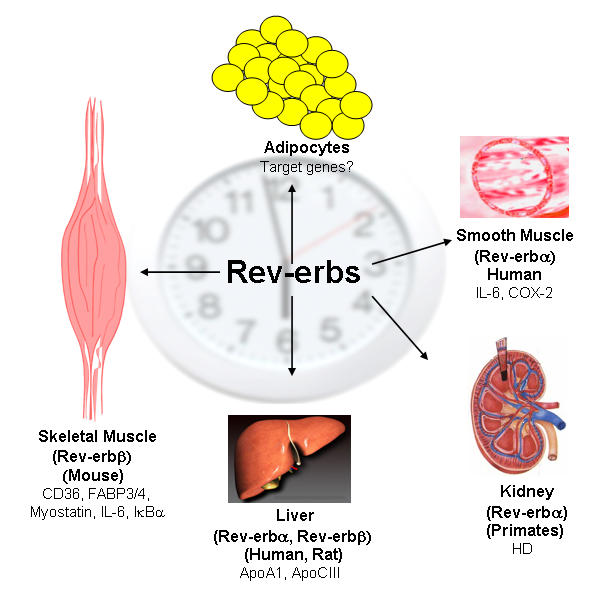
“Rev-erbs” regulate gene expression events in various species and tissues. Genes listed include primary and secondary targets that have been documented in the literature.
Rev-erbs and circadian rhythm
Rev-erbα is involved in circadian timing in brain and liver tissue, and regulates BMAL1, and to a lesser extent CLOCK expression (see Figure 2) [Balsalobre et al., 1998; Torra et al., 2000]. Rev-Erbα itself is driven directly by the molecular oscillator, it is activated by the BMAL1-CLOCK heterodimer, and repressed by PERIOD1/2 and CRYPTOCHROME1/2 proteins [Preitner et al., 2002; Triqueneaux et al., 2004]. It is interesting to note that Rev-erbα and RORα opposingly regulate Bmal1 expression and are an integral component of a complex transcriptional feedback loop (see Figure 2) [Balsalobre et al., 1998; Emery and Reppert, 2004; Sato et al., 2004; Torra et al., 2000]. The amplitude of Bmal1, Clock, and Cry1 mRNA oscillations are significantly attenuated in Rev-erbα -/- mice. Rev-erbα has been shown to influence period length and phase shifting. Schibler and colleagues demonstrated that Rev-erbα -/- mice kept under constant darkness and constant light conditions had a shorter period length compared to wild-type animals [Preitner et al., 2002]. Curiously, the maintenance of circadian pacemaker by Rev-erbα is also conserved in other organisms such as zebrafish [Delaunay et al., 2000]. Laudet and colleagues have hypothesized that Rev-erbα can function as integrator of circadian and physiological events [Triqueneaux et al., 2004]. The literature provides copious examples of circadian regulation of blood pressure, and hypertensive fluctuations associated with cardiovascular disease [Giles, 2005; Ikeda et al., 2004]. Furthermore, aberrant sleep cycles in humans lead to increased secretion of proinflammatory cytokines (for example TNFα and IL-6) [Vgontzas et al., 2004] and reduced cortisol secretion [Takahashi et al., 2001]. In this context, mice treated with prednisolone repressed Rev-erbα and Bmal1 expression [Koyanagi et al., 2006]. Very recently, Yin et al. demonstrated that glycogen synthase kinase 3 (GSK3)-mediated phosphorylation of Rev-erbα and stabilizes the expression of the nuclear receptor [Yin et al., 2006]. Excitingly, they went on to demonstrate that lithium treatment (commonly used to treat bipolar disorder) leads to proteosomal degradation of Rev-erbα and induction of Bmal 1. This process is mediated by lithium induced inhibition of GSK3. Finally, a report from Cermakian and colleagues also highlights that Rev-erbβ can regulate circadian transcription of the Bmal1 gene [Guillaumond et al., 2005].
Figure 2. Cross-talk between Rev-erbα and RORα signaling in circadian gene regulation.
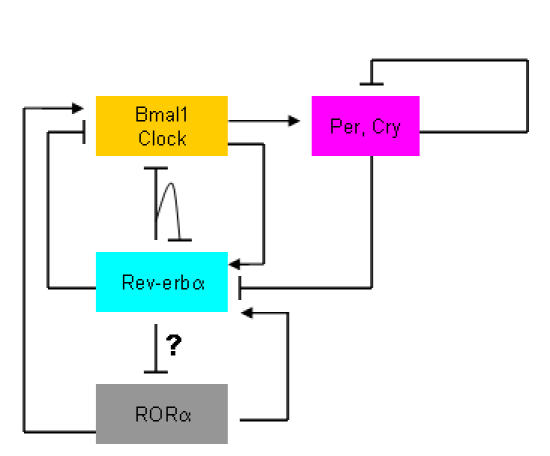
Rev-erbα and RORα mutually regulate circadian Clock genes. Dysregulation in these pathways leads to chronic disease state such as metabolic syndrome.
Rev-erbs and inflammation
Lipopolysaccharide induced activation of bone marrow derived macrophages induces biphasic regulation of Rev-erbβ [Barish et al., 2005]. Recent studies have shown that inflammatory cytokines play crucial roles in endothelial and smooth muscle cell responses in diseases associated with the vasculature. Vascular smooth muscle cells over-expressing Rev-erbα induced NF-κB mediated trans-activation (and p65 translocation). In addition, increased expression of the pro-inflammatory cytokines IL-6 and COX-2 was observed [Migita et al., 2004]. Concordantly, ectopic expression of RORαin human smooth muscle cells inhibits TNFα mediated induction of IL-6, IL-8, and COX-2 expression through induction of IκBα [Delerive et al., 2001]. The attenuation of Rev-erbβ expression/function in skeletal muscle cells has been reported to induce IL-6 mRNA expression. Paradoxically, this was associated with increased expression of RORα and IκBα[Ramakrishnan et al., 2005]. IL-6 is an exercise induced myokine that induces lipolysis in adipose tissue, and suppresses TNFα production. The increased expression of IL-6 is consistent with decreased myostatin expression. For example, it has been reported that IL-6 deficiency leads to late-onset obesity [Wallenius et al., 2002], whereas myostatin deficiency leads to the reduced accumulation of body fat [McPherron and Lee, 2002]. Therefore, the inverse correlation between IL-6 and myostatin expression after attenuation of Rev-erbβ expression is in concordance with the reported phenotypic effects of these proteins. The studies above highlight the emerging role of Rev-erbs in the inflammation cascade.
Concluding remarks and future directions
Molecular modeling suggests the cavities in the LBDs of the NR1D subgroup are occupied by bulky side chains. This indicates that pharmacological manipulation of this orphan NR subgroup by agonists and antagonists in the near future may present technical challenges. However, a recent report from Reinking et al [Reinking et al., 2005] has shown that the ligand binding pocket of E75, a Drosophila orthologue of human Rev-erbα is tightly bound by heme. In recent years, various heme containing molecules have been identified to regulate lipid homeostasis. For example, heme containing neuronal nitric oxide synthase regulates genes involved in the lipogenesis pathway [Schild et al., 2006]. Curiously, Reinking and colleagues also identified E75 as a potential gas sensor and that CO or NO bind to E75 to interfere with E75 mediated repression of DHR3, a Drosophila orthologue of human RORα, see Figure 3). Moreover, heme, NO and CO are important regulatory molecules in mammalian circadian clocks (reviewed in [Pardee et al., 2004]). Recently, cholesterol/cholesterol sulphate and all-trans retinoic acid have been identified as RORα and RORβ agonists, respectively [Kallen et al., 2004; Kallen et al., 2002; Stehlin-Gaon et al., 2003]. In summary, these studies suggest that identification of novel synthetic small molecule regulators for the NR1D subgroup by contemporary chemistry in the near future is possible.
Figure 3. A speculative model for ligand-dependent Rev-erbα modulation in mammals.
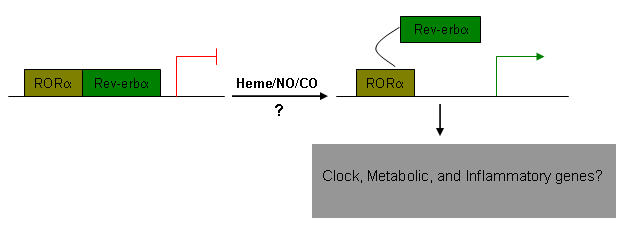
Like Drosophila, endogenous molecules such as heme and diatomic gases could possibly bind to Rev-erbα and activate/modulate its ability to trans-repress RORα and other gene expression events.
Further insights into the function of the NR1D subgroup in the short term will require targeted genetic analysis of both Rev-erbα and β in specific tissues and organs. Cell specific attenuation of the NR1D subgroup (by Cre-Lox and/or transgenic techniques) in the major mass tissues (including skeletal muscle, liver and adipose tissue) will provide novel insights into the physiological role of these orphan NRs in metabolism.
The NR1D subgroups are transcriptional silencers, and identification of target genes is certainly an area requiring additional study. Current studies indicate Rev-erb signaling impacts on RORα and PPARα-mediated control of inflammation and metabolism. Furthermore, it is increasingly apparent that Rev-erbα is an integral component of complex transcriptional feedback loops that are involved in circadian control. Complete elucidation of the pathophysiologically relevant targets of the Rev-erbs and the crosstalk with other NR signaling pathways in a cell/organ specific context will provide unique insights into the links between metabolism, inflammation and the circadian cycle in homeostasis.
Acknowledgments
We apologize to those authors whose work was not cited in the pursuit of brevity. We thank Michael Pearen for his help in preparing Figure 1. SR holds an University of Queensland Postgraduate Scholarship. GEOM is a Principal Research Fellow of the National Health and Medical Research Council of Australia (NHMRC), and this study was supported by an NHMRC project grant.
Abbreviations
- apoA1
apolipoprotein A1
- apoCIII
apolipoprotein CIII
- apoE
apolipoprotein E
- BMAL1
Brain and Muscle Arnt-like protein-1
- bp
base pair(s)
- CO
carbon-monoxide
- COX-2
cyclooxygenase 2
- DR2
direct repeat 2
- FABP
fatty acid binding protein
- FAT/CD36
fatty acid translocase
- HDL
high density lipoprotein
- IL-6
Interleukin-6
- IκBα
Inhibitor of NF;
- LBD
ligand binding domain
- NFκB
nuclear factor κB
- NO
nitric-oxide
- NR
nuclear receptor
- PPAR
peroxisome proliferators-activated receptor
- ROR
RAR-related orphan receptor
- TGF-β
transforming growth factor β
- TNF-α
tumor necrosis factor α
- VLDL
very low density lipoprotein
References
- Adelmant G., Begue A., Stehelin D., Laudet V. A functional Rev-erb α responsive element located in the human Rev-erb α promoter mediates a repressing activity. Proc Natl Acad Sci U S A. 1996;93:3553–8. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Barish G. D., Downes M., Alaynick W. A., Yu R. T., Ocampo C. B., Bookout A. L., Mangelsdorf D. J., Evans R. M. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol. 2005;19:2466–77. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- Burke L., Downes M., Carozzi A., Giguere V., Muscat G. E. Transcriptional repression by the orphan steroid receptor RVR/Rev-erb β is dependent on the signature motif and helix 5 in the E region: functional evidence for a biological role of RVR in myogenesis. Nucleic Acids Res. 1996;24:3481–9. doi: 10.1093/nar/24.18.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A., Lazar M. A. Induction of Rev-ErbA α, an orphan receptor encoded on the opposite strand of the α-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–9. [PubMed] [Google Scholar]
- Chomez P., Neveu I., Mansen A., Kiesler E., Larsson L., Vennstrom B., Arenas E. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(α) orphan receptor. Development. 2000;127:1489–98. doi: 10.1242/dev.127.7.1489. [DOI] [PubMed] [Google Scholar]
- Coste H., Rodriguez J. C. Orphan nuclear hormone receptor Rev-erbalpha regulates the human apolipoprotein CIII promoter. J Biol Chem. 2002;277:27120–9. doi: 10.1074/jbc.M203421200. [DOI] [PubMed] [Google Scholar]
- Delaunay F., Thisse C., Marchand O., Laudet V., Thisse B. An inherited functional circadian clock in zebrafish embryos. Science. 2000;289:297–300. doi: 10.1126/science.289.5477.297. [DOI] [PubMed] [Google Scholar]
- Delerive P., Chin W. W., Suen C. S. Identification of Reverb(α) as a novel ROR(α) target gene. J Biol Chem. 2002;277:35013–8. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- Delerive P., Monte D., Dubois G., Trottein F., Fruchart-Najib J., Mariani J., Fruchart J. C., Staels B. The orphan nuclear receptor ROR α is a negative regulator of the inflammatory response. EMBO Rep. 2001;2:42–8. doi: 10.1093/embo-reports/kve007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M., Carozzi A. J., Muscat G. E. Constitutive expression of the orphan receptor, Rev-erbA α, inhibits muscle differentiation and abrogates the expression of the myoD gene family. Mol Endocrinol. 1995;9:1666–78. doi: 10.1210/mend.9.12.8614403. [DOI] [PubMed] [Google Scholar]
- Dumas B., Harding H. P., Choi H. S., Lehmann K. A., Chung M., Lazar M. A., Moore D. D. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol Endocrinol. 1994;8:996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- Emery P., Reppert S. M. A rhythmic Ror. Neuron. 2004;43:443–6. doi: 10.1016/j.neuron.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Fontaine C., Dubois G., Duguay Y., Helledie T., Vu-Dac N., Gervois P., Soncin F., Mandrup S., Fruchart J. C., Fruchart-Najib J., Staels B. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) γ target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278:37672–80. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- Forman B. M., Chen J., Blumberg B., Kliewer S. A., Henshaw R., Ong E. S., Evans R. M. Cross-talk among ROR α 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol. 1994;8:1253–61. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- Fu M., Sun T., Bookout A. L., Downes M., Yu R. T., Evans R. M., Mangelsdorf D. J. A Nuclear Receptor Atlas: 3T3-L1 adipogenesis. Mol Endocrinol. 2005;19:2437–50. doi: 10.1210/me.2004-0539. [DOI] [PubMed] [Google Scholar]
- Gervois P., Chopin-Delannoy S., Fadel A., Dubois G., Kosykh V., Fruchart J. C., Najib J., Laudet V., Staels B. Fibrates increase human REV-ERBalpha expression in liver via a novel peroxisome proliferator-activated receptor response element. Mol Endocrinol. 1999;13:400–9. doi: 10.1210/mend.13.3.0248. [DOI] [PubMed] [Google Scholar]
- Giguere V., Tini M., Flock G., Ong E., Evans R. M., Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR α, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–53. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- Giles T. Relevance of blood pressure variation in the circadian onset of cardiovascular events. J Hypertens. 2005;23 Suppl 1:S35–9. doi: 10.1097/01.hjh.0000165626.57063.b1. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cadavid N. F., Bhasin S. Role of myostatin in metabolism. Curr Opin Clin Nutr Metab Care. 2004;7:451–7. doi: 10.1097/01.mco.0000134365.99523.7f. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H., Gustafsson J. A., Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–64. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- Guillaumond F., Dardente H., Giguere V., Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- Ikeda T., Gomi T., Shibuya Y., Matsuo K., Kosugi T., Oku N., Uetake Y., Kinugasa S., Furutera R. Morning rise in blood pressure is a predictor of left ventricular hypertrophy in treated hypertensive patients. Hypertens Res. 2004;27:939–46. doi: 10.1291/hypres.27.939. [DOI] [PubMed] [Google Scholar]
- Kallen J., Schlaeppi J. M., Bitsch F., Delhon I., Fournier B. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–8. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- Kallen J. A., Schlaeppi J. M., Bitsch F., Geisse S., Geiser M., Delhon I., Fournier B. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure (Camb) 2002;10:1697–707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- Kassam A., Capone J. P., Rachubinski R. A. Orphan nuclear hormone receptor RevErbalpha modulates expression from the promoter of the hydratase-dehydrogenase gene by inhibiting peroxisome proliferator-activated receptor α-dependent transactivation. J Biol Chem. 1999;274:22895–900. doi: 10.1074/jbc.274.32.22895. [DOI] [PubMed] [Google Scholar]
- Koyanagi S., Okazawa S., Kuramoto Y., Ushijima K., Shimeno H., Soeda S., Okamura H., Ohdo S. Chronic treatment with prednisolone represses the circadian oscillation of clock gene expression in mouse peripheral tissues. Mol Endocrinol. 2006;20:573–83. doi: 10.1210/me.2005-0165. [DOI] [PubMed] [Google Scholar]
- Laitinen S., Staels B. Nucl Recept Signal. 2003;4 doi: 10.1621/nrs.01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen S., Fontaine C., Fruchart J. C., Staels B. The role of the orphan nuclear receptor Rev-Erb α in adipocyte differentiation and function. Biochimie. 2005;87:21–5. doi: 10.1016/j.biochi.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Lazar M. A., Hodin R. A., Darling D. S., Chin W. W. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA α transcriptional unit. Mol Cell Biol. 1989;9:1128–36. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron A. C., Lee S. J. Suppression of body fat accumulation in myostatin-deficient mice. J Clin Invest. 2002;109:595–601. doi: 10.1172/JCI13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita H., Morser J., Kawai K. Rev-erbalpha upregulates NF-kappaB-responsive genes in vascular smooth muscle cells. FEBS Lett. 2004;561:69–74. doi: 10.1016/S0014-5793(04)00118-8. [DOI] [PubMed] [Google Scholar]
- Pardee K., Reinking J., Krause H. Nuclear hormone receptors, metabolism, and aging: what goes around comes around.Transcription factors link lipid metabolism and aging-related processes. Sci Aging Knowledge Environ. 2004;2004 doi: 10.1126/sageke.2004.47.re8. [DOI] [PubMed] [Google Scholar]
- Pircher P., Chomez P., Yu F., Vennstrom B., Larsson L. Aberrant expression of myosin isoforms in skeletal muscles from mice lacking the rev-erbAalpha orphan receptor gene. Am J Physiol Regul Integr Comp Physiol. 2005;288:R482–90. doi: 10.1152/ajpregu.00690.2003. [DOI] [PubMed] [Google Scholar]
- Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S. N., Lau P., Burke L. J., Muscat G. E. Rev-erbbeta regulates the expression of genes involved in lipid absorption in skeletal muscle cells: evidence for cross-talk between orphan nuclear receptors and myokines. J Biol Chem. 2005;280:8651–9. doi: 10.1074/jbc.M413949200. [DOI] [PubMed] [Google Scholar]
- Raspe E., Duez H., Mansen A., Fontaine C., Fievet C., Fruchart J. C., Vennstrom B., Staels B. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002b;43:2172–9. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- Raspe E., Mautino G., Duval C., Fontaine C., Duez H., Barbier O., Monte D., Fruchart J., Fruchart J. C., Staels B. Transcriptional regulation of human Rev-erbalpha gene expression by the orphan nuclear receptor retinoic acid-related orphan receptor α. J Biol Chem. 2002a;277:49275–81. doi: 10.1074/jbc.M206215200. [DOI] [PubMed] [Google Scholar]
- Reinking J., Lam M. M., Pardee K., Sampson H. M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Krause H. M. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Renaud J. P., Harris J. M., Downes M., Burke L. J., Muscat G. E. Structure-function analysis of the Rev-erbA and RVR ligand-binding domains reveals a large hydrophobic surface that mediates corepressor binding and a ligand cavity occupied by side chains. Mol Endocrinol. 2000;14:700–17. doi: 10.1210/mend.14.5.0444. [DOI] [PubMed] [Google Scholar]
- Retnakaran R., Flock G., Giguere V. Identification of RVR, a novel orphan nuclear receptor that acts as a negative transcriptional regulator. Mol Endocrinol. 1994;8:1234–44. doi: 10.1210/mend.8.9.7838156. [DOI] [PubMed] [Google Scholar]
- Sato T. K., Panda S., Miraglia L. J., Reyes T. M., Rudic R. D., McNamara P., Naik K. A., FitzGerald G. A., Kay S. A., Hogenesch J. B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–37. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schild L., Jaroscakova I., Lendeckel U., Wolf G., Keilhoff G. Neuronal nitric oxide synthase controls enzyme activity pattern of mitochondria and lipid metabolism. Faseb J. 2006;20:145–7. doi: 10.1096/fj.05-3898fje. [DOI] [PubMed] [Google Scholar]
- Stehlin-Gaon C., Willmann D., Zeyer D., Sanglier S., Van Dorsselaer A., Renaud J. P., Moras D., Schule R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR β. Nat Struct Biol. 2003;10:820–5. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Yokota S., Hara R., Kobayashi T., Akiyama M., Moriya T., Shibata S. Physical and inflammatory stressors elevate circadian clock gene mPer1 mRNA levels in the paraventricular nucleus of the mouse. Endocrinology. 2001;142:4910–7. doi: 10.1210/endo.142.11.8487. [DOI] [PubMed] [Google Scholar]
- Torra I. P., Tsibulsky V., Delaunay F., Saladin R., Laudet V., Fruchart J. C., Kosykh V., Staels B. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology. 2000;141:3799–806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- Triqueneaux G., Thenot S., Kakizawa T., Antoch M. P., Safi R., Takahashi J. S., Delaunay F., Laudet V. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vgontzas A. N., Zoumakis E., Bixler E. O., Lin H. M., Follett H., Kales A., Chrousos G. P. Adverse effects of modest sleep restriction on sleepiness, performance, and inflammatory cytokines. J Clin Endocrinol Metab. 2004;89:2119–26. doi: 10.1210/jc.2003-031562. [DOI] [PubMed] [Google Scholar]
- Vu-Dac N., Chopin-Delannoy S., Gervois P., Bonnelye E., Martin G., Fruchart J. C., Laudet V., Staels B. The nuclear receptors peroxisome proliferator-activated receptor α and Rev-erbalpha mediate the species-specific regulation of apolipoprotein A-I expression by fibrates. J Biol Chem. 1998;273:25713–20. doi: 10.1074/jbc.273.40.25713. [DOI] [PubMed] [Google Scholar]
- Wallenius V., Wallenius K., Ahren B., Rudling M., Carlsten H., Dickson S. L., Ohlsson C., Jansson J. O. Interleukin-6-deficient mice develop mature-onset obesity. Nat Med. 2002;8:75–9. doi: 10.1038/nm0102-75. [DOI] [PubMed] [Google Scholar]
- Yin L., Wang J., Klein P. S., Lazar M. A. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–5. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]


