Abstract
Cytosine DNA methylation protects eukaryotic genomes by silencing transposons and harmful DNAs, but also regulates gene expression during normal development. Loss of CG methylation in the Arabidopsis thaliana met1 and ddm1 mutants causes varied and stochastic developmental defects that are often inherited independently of the original met1 or ddm1 mutation. Loss of non-CG methylation in plants with combined mutations in the DRM and CMT3 genes also causes a suite of developmental defects. We show here that the pleiotropic developmental defects of drm1 drm2 cmt3 triple mutant plants are fully recessive, and unlike phenotypes caused by met1 and ddm1, are not inherited independently of the drm and cmt3 mutations. Developmental phenotypes are also reversed when drm1 drm2 cmt3 plants are transformed with DRM2 or CMT3, implying that non-CG DNA methylation is efficiently re-established by sequence-specific signals. We provide evidence that these signals include RNA silencing though the 24-nucleotide short interfering RNA (siRNA) pathway as well as histone H3K9 methylation, both of which converge on the putative chromatin-remodeling protein DRD1. These signals act in at least three partially intersecting pathways that control the locus-specific patterning of non-CG methylation by the DRM2 and CMT3 methyltransferases. Our results suggest that non-CG DNA methylation that is inherited via a network of persistent targeting signals has been co-opted to regulate developmentally important genes.
Synopsis
The majority of DNA in large eukaryotic genomes (such as the human genome) consists of transposons, sequences that can reproduce at the expense of their host. Plants and animals mark transposon DNA with a chemical modification called DNA methylation. DNA methylation prevents the functional information in transposons from being copied into RNA and utilized—this process is termed “gene silencing.” Using a flowering plant called Arabidopsis, the authors created mutants lacking a particular type of DNA methylation, and found that these plants had defects in leaf shape, plant height, and fertility. This shows that a gene-silencing mechanism used to defend the genome from transposons is also important for normal plant development. When the mutated genes are restored, plant development returns to normal, showing that one type of DNA methylation can be efficiently re-established (other gene-silencing marks can be lost irreversibly). Small RNA molecules are important for targeting DNA methylation to transposons and harmful DNAs. Mutants in genes that are important for making small RNAs have similar developmental defects to those lacking DNA methylation. This implies that normal plant development requires DNA methylation that is targeted by small RNAs.
Introduction
The met1 and ddm1 mutations that affect maintenance of CG DNA methylation cause severe and variable developmental defects, suggesting that DNA methylation can affect many developmental genes [1–4]. MET1 encodes a maintenance DNA methyltransferase orthologous to mouse Dnmt1, and DDM1 encodes a SNF2-like chromatin-remodeling ATPase [5,6]. Some of the developmental phenotypes in met1 and ddm1 clearly result from loss of CG DNA methylation at particular genes. In the case of the FWA gene, loss of CG DNA methylation in met1 causes overexpression of the FWA transcription factor, resulting in a dominant and heritable late-flowering phenotype [3,4,7]. Unmethylated fwa segregates as an independent trait because CG DNA methylation is not regained when met1 is crossed to wild type. An independently segregating developmental phenotype caused by loss of CG DNA methylation has also been observed at the BAL locus, which encodes a pathogen-resistance gene within a repetitive gene cluster [8].
DNA methylation is found at cytosines in three different sequence contexts, CG, CNG (where N is any base), and asymmetric CHH (where H = A, T, or C). The maintenance activity of MET1 can replicate CG DNA methylation even when the initial trigger for DNA methylation is genetically removed [9,10]. This may be explained in part by the fact that Dnmt1-type DNA methyltransferases have a strong preference for hemimethylated substrates, such as those left by DNA replication of a CG dinucleotide that is methylated on both strands [11]. However, non-CG DNA methylation is inherited differently and appears to require active signals to continually target regions of DNA for methylation [12]. In the case of CNG methylation, this signal seems to come from histones that are associated with the DNA. CNG methylation is mostly controlled by the methyltransferase CMT3, and also often requires histone H3 lysine 9 dimethylation (H3K9me2) by the SET domain protein SUVH4/KRYPTONITE (KYP) [13,14].
On the other hand, asymmetric methylation (which lacks an adjacent methylcytosine to provide epigenetic information after DNA replication) is mostly controlled by the DNA methyltransferase DRM2, which is targeted by 24-nucleotide short interfering RNAs (siRNAs) produced though RNA interference pathways [15–18]. DRM2 and the closely linked gene DRM1 encode proteins that are homologous to the mammalian de novo DNA methyltransferase Dnmt3 [19]. Notably, the drm2 single mutant is identical to the drm1 drm2 double mutant for all phenotypes tested [18]. siRNAs and DRM2 are also important for the initial establishment of DNA methylation in all sequence contexts, since a suite of siRNA metabolism mutants in the genes encoding nuclear RNA POLYMERASE IV (NRPD1a), RNA-DEPENDENT RNA POLYMERASE2 (RDR2), DICER-LIKE3 (DCL3), and ARGONAUTE4 (AGO4) fail to establish DNA methylation at the direct repeats of the FWA locus when a new copy of FWA is transformed into plants [15].
The maintenance of non-CG DNA methylation at endogenous sequences shows very locus-specific differences in its requirement for different DNA methyltransferases [17]. For example, the direct repeat loci FWA and MEA-ISR require RNA interference (RNAi) and DRM2 for both the CNG and asymmetric DNA methylation, whereas non-CG methylation at the centromeric retrotransposon Ta3 solely depends on CMT3. At other loci such as the small euchromatic transposon AtSN1 and at silent alleles of the SUPERMAN gene, DRM2 and CMT3 act redundantly to maintain non-CG DNA methylation [17,20]. Another example of this redundancy between DRM2 and CMT3 is the fact that neither drm1 drm2 nor cmt3 mutants show any morphological defects, but the drm1 drm2 cmt3 triple mutant shows a pleiotropic suite of developmental abnormalities [17]. Thus, like CG DNA methylation, non-CG DNA methylation can affect developmentally important gene expression.
We sought to understand the mechanisms that underlie propagation of non-CG DNA methylation during plant development. Here we show that non-CG methylation that controls developmental genes can be readily restored after it is lost, implying the existence of persistent targeting signals that remain in the absence of DNA methyltransferase function. We provide evidence that these signals include input from RNA silencing pathways, from the chromatin remodeling protein DRD1, and from histone methylation. We further show that DRD1 works along with the 24-nucleotide siRNA pathway in the establishment of DNA methylation, and works through both the DRM2 and CMT3 methyltransferases in the maintenance of DNA methylation. These results help to define the different mechanisms that control non-CG DNA methylation and its involvement in developmental gene regulation.
Results/Discussion
Inheritance of drm1 drm2 cmt3 Developmental Phenotypes Is Strongly Correlated with the drm2 cmt3 Genotype
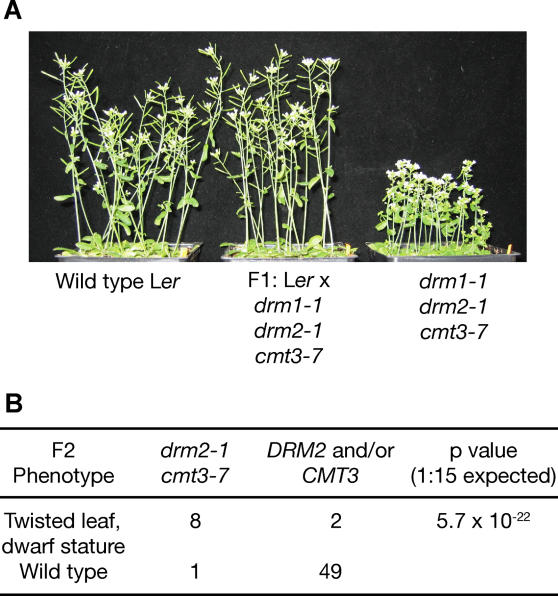
It was previously shown that drm1 drm2 cmt3 triple mutants display a pleiotropic set of developmental abnormalities [17]. Interestingly, we found that these developmental phenotypes are strongly penetrant and largely homogeneous in a population of drm1 drm2 cmt3 plants, unlike the stochastic nature of the developmental phenotypes seen in ddm1 and met1 mutants. Furthermore, successive generations of inbreeding did not exacerbate the developmental phenotype of drm1 drm2 cmt3. drm1 drm2 cmt3 plants in the Landsberg erecta (Ler) ecotype show three major defects: a twisted leaf shape, shorter stature, and partial sterility which is evidenced by short siliques that produce fewer seeds than wild type (100% penetrance of the sterility phenotype is shown in Table S1). Flowering time in drm1 drm2 cmt3 is similar to wild type. We found that, unlike met1 phenotypes, all of these defects were entirely recessive when drm1–1 drm2–1 cmt3–7 was crossed to wild type Ler (Figure 1A). To further characterize inheritance of the drm1 drm2 cmt3 phenotype, we selected ten plants from the F2 generation of this cross that showed a twisted leaf and short stature phenotype, and an additional 50 plants with wild-type morphology, and all were genotyped for the drm2–1 and cmt3–7 mutations. We did not genotype drm1–1, because the drm2–1 single mutant has all of the phenotypes of drm1–1 drm2–1, and because the DRM1 and DRM2 genes are tightly linked at a distance of approximately 1 cM [18]. Twisted leaf and dwarf stature phenotypes in the F2 segregated strongly with the drm2–1 cmt3–7 genotype (Figure 1B). Only one plant out of 50 scored with a wild-type morphology had the drm2–1 cmt3–7 genotype, and this observation may have resulted from incomplete penetrance of the developmental phenotype. Only two plants with a twisted leaf and dwarf phenotype were heterozygous for the cmt3–7 mutant, and thus contained a wild-type CMT3 gene. These plants may have indeed inherited these developmental defects epigenetically, or might have been scored as dwarf due to developmental variability caused by growth conditions. Thus, in 57/60 cases tested, the predicted phenotype of the F2 plants correlated with their genotype. Overall, these F2 segregation data demonstrate a fundamental difference between developmental defects in drm1 drm2 cmt3 and those seen in ddm1 and met1—the former are generally not inherited independent of the drm1 drm2 cmt3 genotype.
Figure 1. Inheritance of drm1 drm2 cmt3 Developmental Phenotypes.
(A) Developmental phenotypes of drm1–1 drm2–1 cmt3–7 are fully recessive when crossed to wild-type Ler.
(B) The developmental phenotypes of the original drm1–1 drm2–1 cmt3–7 plants are inherited with the drm2–1 cmt3–7 genotype in the F2 of a backcross. Phenotypes were scored blindly before the plants were genotyped. The p-value was calculated from a chi square test based on the null hypothesis that the developmental phenotypes were segregating independently of the drm2–1 and cmt3–7 genotypes, such that one would only expect 1/16th of the twisted leaf dwarf plants to be drm2–1 cmt3–7 double mutants. The two plants scored as having a mutant phenotype that were not homozygous for drm2–1 cmt3–7 were drm2–1 cmt3–7/CMT3.
Transforming drm1 drm2 cmt3 with DRM2 or CMT3 Restores Normal Development
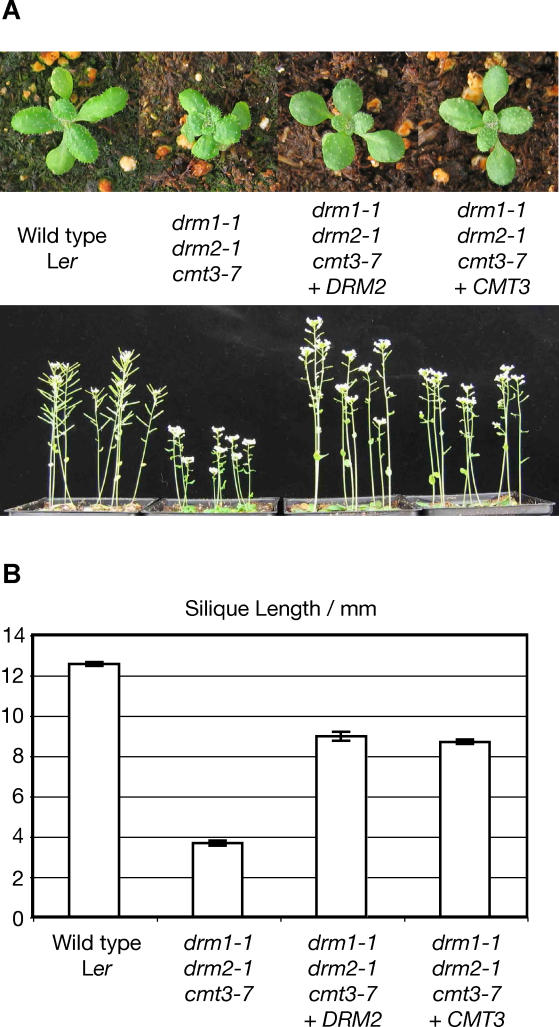
In the backcross experiment shown in Figure 1A, a correctly expressed and methylated parental genome is introduced along with the wild-type DRM2 and CMT3 genes. This complicates interpretation of the experiment, because the genome from the wild-type parent may bring in signals that confer correct developmental regulation to chromosomes derived from the drm1 drm2 cmt3 parent. Therefore, as a further test of whether normal development could be restored to drm1 drm2 cmt3 mutants, we introduced either DRM2 or CMT3 by Agrobacterium-mediated plant transformation into these plants, and asked whether these transgenes could confer a wild-type morphological phenotype. Both DRM2 and CMT3 completely restored normal leaf shape and wild-type stature when transformed into drm1–1 drm2–1 cmt3–7 (Ler) (Figure 2A). Importantly, recovery of the normal phenotype in DRM2 and CMT3 transformants implies that the active signals that target non-CG DNA methylation are still present in the drm1 drm2 cmt3 triple mutant. This restoration is consistent with a model in which drm1 drm2 cmt3 developmental phenotypes result mostly from genes that are overexpressed when silencing-associated non-CG methylation is lost. In this scenario, these genes would be re-silenced when DRM2 or CMT3 are introduced by transformation. Alternatively, loss of non-CG DNA methylation might also result in inappropriately low expression of endogenous genes. For instance, the loss of DNA methylation on silencer elements could result in transcriptional suppression. The sterility defect seen in drm1–1 drm2–1 cmt3–7 plants was greatly reduced in drm1–1 drm2–1 cmt3–7 plants transformed with DRM2 or CMT3. However, this defect was not completely reversed because the silique length in transformed plants did not reach wild-type levels in the T2 generation (Figure 2B). Failure of DRM2 or CMT3 transgenes to fully reverse the sterility defect of drm1–1 drm2–1 cmt3–7 may reflect incomplete complementation. However, we did observe shorter siliques in multiple independent lines of both DRM2- and CMT3-transformed plants (unpublished data). These data therefore suggest that, although most of the developmental defects seen in drm1–1 drm2–1 cmt3–7 are completely restored when wild-type DRM2 and CMT3 genes are reintroduced, the sterility defect may be to some extent inherited epigenetically.
Figure 2. drm1 drm2 cmt3 Phenotypes Are Efficiently Restored to Wild Type by Transformed DRM2 or CMT3 .
(A) Normal rosette leaf shape and stature are restored in drm1–1 drm2–1 cmt3–7 transformed with either DRM2 or CMT3.
(B) The sterility of drm1–1 drm2–1 cmt3–7 plants is partially restored in the T2 generation of DRM2 or CMT3 transformants. The length of ten mature siliques on the primary stem was measured; analysis was started at the third silique of the stem. Between eight and 36 individual plants were measured. Error bars show standard error of the mean.
DRD1 Is Required for Establishment of DNA Methylation (De Novo DNA Methylation) of Transformed FWA
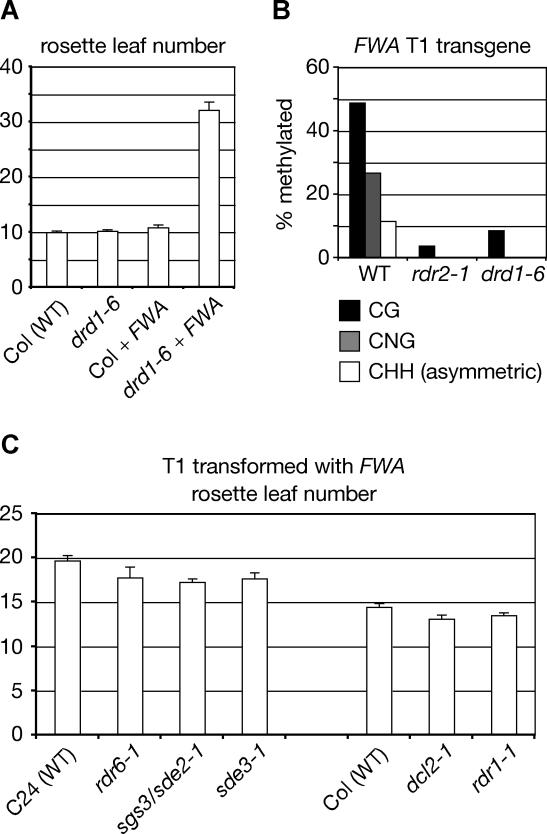
DRD1 is an SNF2-related ATPase and putative chromatin-remodeling protein that is required to establish and maintain RNA-directed non-CG DNA methylation triggered by a transcribed inverted repeat [21,22]. Tandem repeat sequences are probably recognized by a different mechanism than that used for inverted repeats, which have the capacity to generate double-stranded RNA by monodirectional transcription. We therefore sought to test whether DRD1 plays a role in de novo DNA methylation and silencing of transformed FWA, a tandem repeat–containing gene [18]. We found that, like drm2 and RNA-silencing mutants from the 24-nucleotide siRNA pathway, including rdr2, dcl3, and ago4 mutants [15], the drd1–6 mutant flowered late after FWA transformation, but had no defect in silencing of the endogenous FWA gene, as shown by early flowering prior to transformation (Figure 3A). As predicted from their inability to silence transformed FWA, drd1–6 plants also lacked de novo DNA methylation of the FWA transgene in the T1 generation (Figure 3B). These results indicate that DRD1 is essential for de novo DNA methylation of both transformed tandem repeats and targets of inverted repeat–generated siRNAs. They also suggest that DRD1 acts in concert with the RNA polymerase IV/RDR2/DCL3/AGO4 RNAi pathway to guide DRM2. We determined that de novo gene silencing was normal in the RNAi mutants rdr6–1/sde1, sgs3/sde2–1, sde3–1, dcl2–1, and rdr1–1 (Figure 3C). The fact that many RNAi proteins are not required for de novo DNA methylation is further confirmation that Arabidopsis RNAi pathways are functionally specialized [23].
Figure 3. DRD1 Is Required for De Novo DNA Methylation of Tandem Repeats.
(A) DRD1 is required for de novo silencing of transformed FWA. Flowering time in untransformed and transformed T1 plants is shown—overexpression of FWA causes late flowering. Col, Columbia ecotype; WT, wild type.
(B) DRD1 is required for de novo DNA methylation of transformed FWA. DNA methylation of the FWA transgene was measured by bisulfite genomic sequencing in T1 plants. Graph represents the percentage methylation in different sequence contexts.
(C) Several RNAi mutants are competent for de novo silencing of transformed FWA. Flowering time for each transformed mutant is shown adjacent to its corresponding wild-type ecotype.
DRD1 Acts through Both DRM2 and CMT3 to Maintain Endogenous Non-CG DNA Methylation
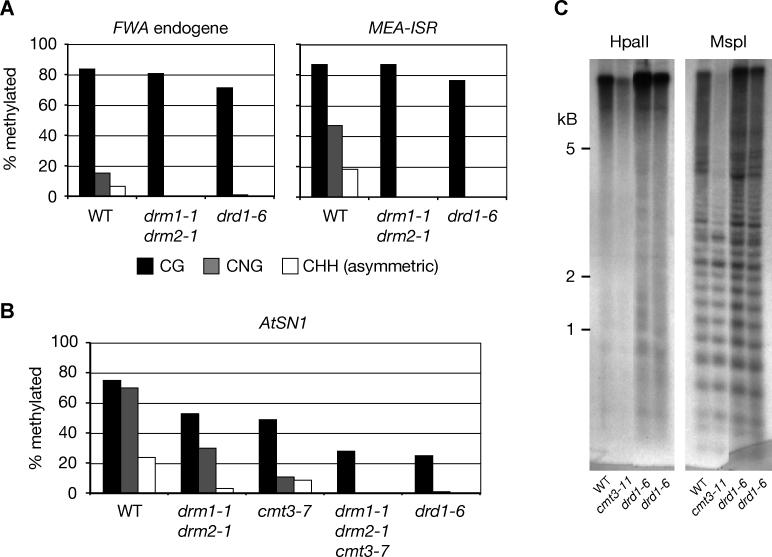
We tested whether DRD1 acts through the DRM2 and/or CMT3 methyltransferases in its control of non-CG methylation, by testing the effect of drd1–6 on maintenance of DNA methylation at different endogenous loci. At the endogenous direct repeats present at FWA and MEA-ISR, drd1–6 lacked all non-CG methylation but did not affect CG methylation (Figure 4A). This phenotype is identical to mutants in the RNA polymerase IV/RDR2/DCL3/AGO4 RNAi pathway and drm1 drm2 at these loci [15,17]. Importantly, at the SINE element AtSN1, the drd1–6 mutant also lacked all non-CG methylation (Figure 4B). This is interesting because the drm1 drm2 or cmt3 mutants have only moderate effects on non-CG methylation at this locus, yet the drm1 drm2 cmt3 triple mutant shows a loss of all non-CG AtSN1 methylation [17]. This suggests that DRD1 can act through both DRM2 and CMT3 at endogenous genes such as AtSN1. Our finding is consistent with the fact that multiple mutant alleles of drd1 were isolated from a screen for plants that could not maintain non-CG DNA methylation and transcriptional gene silencing targeted by an inverted repeat of the soybean α′ promoter [21], yet neither DRM2 nor CMT3 were identified by this screen. This suggests that like AtSN1, the target of α′ siRNAs has non-CG DNA methylation that is controlled by DRD1, which acts through the redundant action of the DRM2 and CMT3 methyltransferases.
Figure 4. Role of DRD1 in Maintaining Non-CG DNA Methylation at Endogenous Loci.
(A) The drd1–6 mutant cannot maintain non-CG DNA methylation at the endogenous direct repeats FWA and MEA-ISR.
(B) drd1–6 loses all non-CG methylation at the SINE transposon AtSN1, and thus phenocopies drm1 drm2 cmt3. DNA methylation was measured by bisulfite genomic sequencing.
(C) Southern blot analysis of DNA methylation at the pericentromeric retrotransposon Ta3. HpaII digestion at CCGG is blocked by CG or CNG DNA methylation, whereas MspI digestion at CCGG is blocked by CNG methylation.
WT, wild type.
The DRD1-dependent non-CG DNA methylation at AtSN1, FWA, and MEA-ISR is associated with the presence of endogenous siRNAs corresponding to these loci. In contrast, the pericentromeric retrotransposon Ta3 lacks siRNAs, as shown by their absence from a very large small-RNA dataset compiled using the massively parallel signature sequencing technology [24]. This is also consistent with the fact that the ago4–1 mutation had no effect on DNA methylation at Ta3 [25]. Instead, at Ta3, CNG DNA methylation depends solely on the CMT3 DNA methyltransferase [17]. Importantly, drd1–6 mutants showed no defect in CNG DNA methylation at Ta3, as assayed by a Southern blot with the CNG methylation–sensitive restriction enzyme MspI (Figure 4C). This can be contrasted with the cmt3–11 mutant, in which digestion with MspI yields a far greater proportion of low-molecular weight bands consistent with restriction enzyme cleavage of DNA that lacks CNG methylation. Thus, Ta3 is a locus where CMT3 maintains CNG DNA methylation independent of siRNAs and of DRD1.
Developmentally Important Non-CG DNA Methylation Is Targeted by RNAi and DRD1
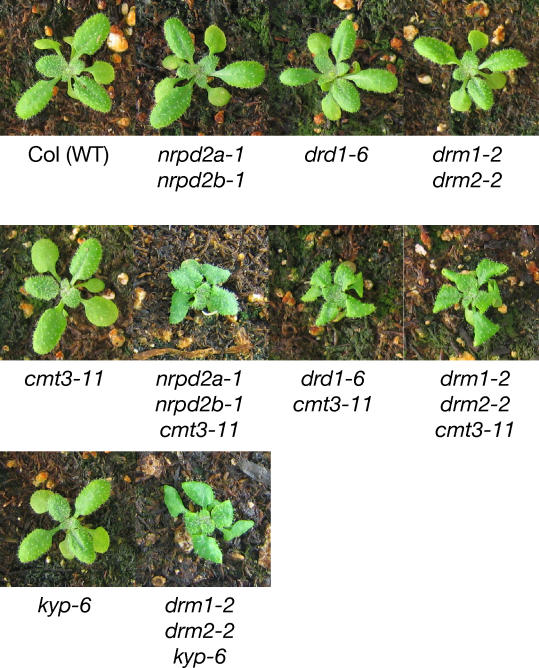
The fact that normal development is largely restored when DRM2 or CMT3 are reintroduced suggests that these enzymes are actively targeted by signals that persist in the drm1 drm2 cmt3 mutant. To test this model further, cmt3 was combined with null mutants in the RNA polymerase IV subunit–encoding gene NRPD2a and in DRD1, both of which are required for RNAi-directed DNA methylation [21,26–28]. NRPD2a encodes the second largest subunit of RNA polymerase IV, which acts together with and is necessary for the activity of NRPD1a and NRPD1b [26–29]. NPRD2b encodes a closely related gene copy which is likely nonfunctional [27,28]. We generated a nrpd2a-1 nrpd2b-1 cmt3–11 triple mutant utilizing T-DNA mutations all isolated in the Columbia (Col) wild-type background. We also constructed a drd1–6 cmt3–11 double mutant in the Col ecotype. As a control, we isolated new T-DNA mutations in DRM1, DRM2, and CMT3, and constructed the drm1–2 drm2–2 cmt3–11 triple mutant in the Col background. The Col drm1–2 drm2–2 cmt3–11 mutant has a phenotype that is similar to that of the drm1–1 drm2–1 cmt3–7 triple mutant in the Ler background, with minor differences. The short stature and sterility defects were more pronounced in Ler. However, the Col drm1–2 drm2–2 cmt3–11 mutant has a particularly strong leaf shape phenotype, in which the apical end of the rosette leaf is folded under the blade. The Col leaf shape phenotype was 100% penetrant in a population of more than 500 homozygous drm1–2 drm2–2 cmt3–11 plants. We found that both the nrpd2a-1 nrpd2b-1 cmt3–11 triple mutant and the drd1–6 cmt3–11 double mutant showed a developmental phenotype identical to that of drm1–2 drm2–2 cmt3–11 (Figure 5). These results show that the mutations in NRPD2 and DRD1 show the same effect as mutation of DRM2 when combined with mutation of CMT3. To confirm this we also constructed both the nrpd2a-1 nrpd2b-1 drm1–2 drm2–2 quadruple mutant and the drd1–6 drm1–2 drm2–2 triple mutant and found that these plants had a wild-type morphological phenotype (unpublished data). These results suggest that the role of DRM2 in developmental gene regulation requires RNAi and the RNA-directed DNA methylation factor DRD1. DRD1 does not control all developmental regulation by DRM2 and CMT3, however, because the single drd1–6 mutant has a wild-type morphological phenotype. This contrasts with AtSN1 non-CG methylation, where drd1–6 phenocopies drm1 drm2 cmt3. Since DRM2 requires DRD1 for establishment and maintenance of DNA methylation at all loci tested, we assume that CMT3 has a DRD1-independent targeting pathway, as exemplified by CNG methylation at the Ta3 retrotransposon (Figure 4C).
Figure 5. Developmentally Important Gene Regulation by DRM2 and CMT3 Requires RNAi, DRD1, and KRYPTONITE.
Rosette leaf shape defects in a variety of multiple mutant combinations are shown.
Col (WT), wild-type Columbia ecotype.
Developmentally Important Non-CG DNA Methylation Is Targeted by Histone H3K9 Methylation
The observation that the drm1–2 drm2–2 nrpd2a-1 nrpd2b-1 plants did not show the developmental phenotypes of drm1–2 drm2–2 cmt3–11 suggests that the control of normal gene expression by CMT3 is not solely directed by RNAi. We therefore tested whether histone H3K9 dimethylation (H3K9me2) is required to target CMT3 at developmental genes, by creating drm1–2 drm2–2 kyp-6 plants that lack the SET domain histone methyltransferase KRYPTONITE (KYP)/SUVH4. The kyp-6 allele is a newly isolated T-DNA allele such that the drm1–2 drm2–2 kyp-6 plants are in a pure Col background. We found that the drm1–2 drm2–2 kyp-6 plants displayed a very similar phenotype to drm1–2 drm2–2 cmt3–11 (Figure 5). This suggests that the loss of KYP-mediated H3K9me2 phenocopies the loss of CMT3, when combined with mutations in DRM genes. Thus, both RNAi pathways and the silencing-associated histone modification H3K9me2 can target non-CG DNA methylation to developmentally important genes.
A Model for the Targeting of Locus Specific Non-CG DNA Methylation
Our work shows that regulation of plant development by non-CG DNA methylation differs fundamentally from MET1-dependent CG DNA methylation that controls normal gene expression. Non-CG DNA methylation is directed in part by RNAi factors, in part by DRD1, and in part by histone H3 lysine 9 methylation through KYP. It is also efficiently restored after it is lost, implying that the targeting signals responsible for its propagation are persistent in plants that lack the DNA methyltransferase enzymes DRM2 and CMT3.
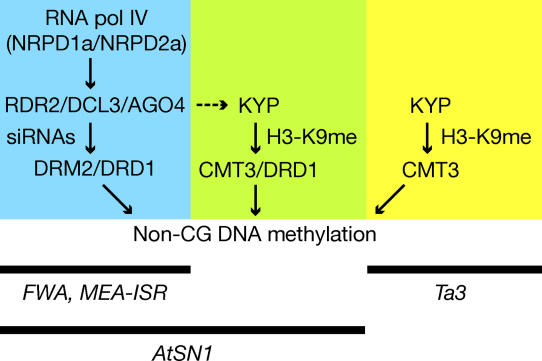
Figure 6 presents a model for the action of several targeting pathways that control the locus-specific propagation of non-CG DNA methylation patterns. In one branch of this pathway, the 24-nucleotide siRNA pathway acts together with DRD1 to target the DRM2 DNA methyltransferase. Certain loci like FWA and MEA-ISR appear to only use this pathway, since all non-CG methylation is lost at these loci in the RNAi mutants and in the drd1 and drm2 mutants. Other loci, such as AtSN1, appear to use a combination of the RNAi/DRD1/DRM2 pathway and a second pathway, in which CMT3 is guided by histone methylation through KYP. We propose that DRD1 acts in both of these pathways, which would explain why DRD1 can facilitate non-CG DNA methylation by both DRM2 and CMT3, even though these enzymes have locus-specific effects. DRD1 is a SNF2-related ATPase (from a plant-specific subfamily), suggesting that it is a chromatin remodeling protein, and such an activity may permit DRM2 and CMT3 to methylate nucleosomal DNA in vivo. In yet a third pathway, exemplified by the Ta3 locus, CMT3 propagates CNG DNA methylation without siRNAs or the need for DRD1.
Figure 6. A Model for the Inheritance of Non-CG DNA Methylation in Arabidopsis thaliana .
Background colors represent different pathways for targeting of non-CG DNA methylation. DRM2 is guided by an RNAi pathway initiated by DNA-dependent RNA polymerase IV. CMT3 is guided by histone H3 lysine 9 dimethylation (H3K9me2) that depends on KRYPTONITE/KYP. At some loci, histone H3 methylation is targeted by RNAi (represented by dotted arrow). DRD1 controls both DRM2 and CMT3, but there is also a DRD1-independent pathway that directs CMT3.
Ultimately, we hope to understand how evolution has co-opted non-CG DNA methylation to control developmentally important endogenous genes. DNA methylation may silence developmental regulators in a tissue-specific manner, or could be a general mechanism for repressing genes that have a deleterious effect on normal development when ectopically overexpressed. In particular, it will be interesting to investigate how genes controlled by RNAi, DRD1, DRM2, KYP and CMT3 differ from those whose normal regulation requires CG DNA methylation maintained by MET1. Furthermore, as exemplified by the FWA gene, loss of MET1-mediated CG DNA methylation can be associated with loss of non-CG DNA methylation. This indicates that there is feedback between CG and non-CG DNA methylation, and that some genes may be regulated by both mechanisms.
During evolution, the acquisition of regulation of endogenous genes by non-CG DNA methylation may involve local sequence repeats and/or the presence of homologous small RNAs. Furthermore, proximity to transposable elements was first suggested by Barbara McClintock as a mechanism for gene regulation during maize development [30]. It is possible that non-CG DNA methylation of transposons has contributed to the evolution of development in Arabidopsis and in other plants.
Materials and Methods
Plant materials.
Plants were grown under continuous light conditions. The drm1–1, drm2–1, cmt3–7, rdr2–1, drd1–6, rdr6–1, sgs3/sde2–1, sde3–1, dcl2–1, rdr1–1, nrpd2a-1, and nrpd2b-1 mutants have been previously described [18,20,23,27,31]. drm1–1 and drm2–1 are T-DNA alleles isolated in the Wassilewskija (WS) ecotype—both T-DNAs are predicted to disrupt essential catalytic domains. These mutations were backcrossed five times into Ler prior to this analysis. cmt3–7 is a point mutation isolated in the Ler ecotype that creates a stop codon, truncating the CMT3 protein after 27 amino acids. drm1–2, drm2–2, and cmt3–11 are T-DNA insertions in the predicted methyltransferase domains of DRM1, DRM2, and CMT3 that would be expected to create null mutations (T-DNAs SALK_031705, SALK_150863, and SALK_148381 respectively). kyp-6 is T-DNA SALK_041474. The phenotypes of drm1 drm2 cmt3 in the Ler ecotype were scored at two main stages. Twisted rosette leaf shape was scored at approximately 2 wk, prior to bolting. Short stature was scored at approximate 5–6 wk, once the plants had made the majority of their siliques.
Transformation with DRM2 and CMT3.
The CMT3 genomic clone we used was a kind gift from Judith Bender [13]. The CMT3-encoding KpnI fragment was subcloned into pCAMBIA-1300 prior to transformation. The DRM2 gene and flanking intergenic regions were PCR amplified using Pfx (Stratagene, La Jolla, California, United States) from BAC clone T15N1 with primers JP2548 5′-GTAATGGAGATAGCTTCTCAGGATTATCATTAGC-3′ and JP2549 5′-AACCAGATTGGGGCAATATACATATAGAAGAGCC-3′. The PCR product was cloned into pCR4 (Invitrogen, Carlsbad, California, United States) and sequenced. The DRM2 gene was then cloned as an EcoRI fragment into the pCAMBIA-1300 binary vector.
FWA transformation and flowering time analysis.
Transformation of Arabidopsis with FWA and flowering time analysis were performed as described [18].
Bisulfite genomic sequencing.
Bisulfite sequencing was performed as described [15,17,25]. To create an FWA transgene that can be distinguished from endogenous FWA, we inserted an AT dinucleotide at position −780, changing a BglII site to an EcoRI site.
Southern blotting.
Southern blotting for Ta3 was performed as described [17].
Supporting Information
(57 KB XLS)
Accession Numbers
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) GeneID accession numbers for the genes and gene products discussed in the paper are AGO4 (817246), BAL/SNC1 (827397), CMT3 (843313), DCL2 (821300), DCL3 (823508), DDM1 (836808), DRD1 (816136), DRM1 (831390), DRM2 (831315), FWA (828658), MEA (MEA-ISR is downstream) (839422), MET1 (834975), NRPD1a (842605), NRPD2a (821960), NRPD2b (821334), RDR1 (838044), RDR2 (826714), RDR6/SDE1 (824112), SDE3 (837047), SGS3/SDE2 (832422), SUPERMAN (821888), and SUVH4/KRYPTONITE (831244).
Acknowledgments
We thank Marjori Matzke, Jim Carrington, Craig Pikaard, David Baulcombe, and Judith Bender for providing reagents, and members of the Jacobsen lab for helpful discussions.
Abbreviations
- Col
Columbia ecotype
- H3K9me2
histone H3K9 dimethylation
- Ler
Landsberg erecta ecotype
- RNAi
RNA interference
- siRNA
short interfering RNA
Footnotes
Author contributions. SW-LC and SEJ conceived and designed the experiments. SW-LC, IRH, XZ, GS, JS-CC, and SEJ performed the experiments. SW-LC and SEJ analyzed the data. SW-LC, IRH, XZ, and SEJ contributed reagents/materials/analysis tools. SW-LC and SEJ wrote the paper.
Competing interests. The authors have declared that no competing interests exist.
Funding. This research was funded by National Institutes of Health RO1 grant GM60398. SW-LC is a Department of Energy (DOE) Energy Biosciences fellow of the Life Sciences Research Foundation. IRH was supported by a European Molecular Biology Organization (EMBO) long-term fellowship. SEJ is an investigator of the Howard Hughes Medical Institute.
References
- Kakutani T. Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana . Plant J. 1997;12:1447–1451. doi: 10.1046/j.1365-313x.1997.12061447.x. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci U S A. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Scheid OM, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana . Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, et al. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Stokes TL, Kunkel BN, Richards EJ. Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 2002;16:171–182. doi: 10.1101/gad.952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol. 2001;11:747–757. doi: 10.1016/s0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, Van Der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation in Arabidopsis . Proc Natl Acad Sci U S A. 2002;99:16499–16506. doi: 10.1073/pnas.162371499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana . Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, et al. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci U S A. 2002;99:16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Cao X, Springer NM, Muszynski MG, Phillips RL, Kaeppler S, et al. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc Natl Acad Sci U S A. 2000;97:4979–4984. doi: 10.1073/pnas.97.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Kanno T, Aufsatz W, Jaligot E, Mette MF, Matzke M, et al. A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO Rep. 2005;6:649–655. doi: 10.1038/sj.embor.7400446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020104. e104. 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, et al. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis . Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfort NC. The tangled field: Barbara McClintock's search for the patterns of genetic control. Cambridge: Harvard University Press; 2001. 337. p. [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(57 KB XLS)