Abstract
The Na+/H+ exchanger regulatory factor 2 (NHERF-2) is a scaffold protein that regulates cellular signaling by forming protein complexes. Several proteins known to interact with NHERF-2 are abundantly expressed in the central nervous system, but little is known about NHERF-2 localization in the brain. By using immunohistochemistry combined with light and electron microscopy, we found that many populations of astrocytes, as well as some populations of neurons, were immunopositive for NHERF-2 throughout the mouse brain. Quantitative analysis of the subcellular distribution of NHERF-2 immunostaining in four brain structures, cerebral cortex, hippocampus, striatum, and cerebellar cortex, showed that NHERF-2 was expressed mainly in astrocytic processes but was also sometimes observed in both pre- and postsynaptic neuronal elements. NHERF-2 immunostaining was associated mainly with the plasma membrane of neurons and astrocytes. However, NHERF-2 immunoreactivity was also observed in association with synaptic vesicles in putative glutamatergic axon terminals. The subcellular localization of NHERF-2 in brain is consistent with a role for NHERF-2 in forming complexes between cell surface and cytosolic proteins, and the preferential expression of NHERF-2 in astrocytes suggests that this scaffold protein may play an important role in astrocytic physiology.
Indexing terms: PDZ, E3KARP, NHERF2, scaffolding, receptor, signaling
Scaffold proteins play an important role in regulating cellular signaling by physically tethering together key molecular components of signaling pathways. Many scaffold proteins consist of a series of modular domains that bind to specific motifs on target proteins. For example, NHERF-2 (also known as E3KARP, SIP-1, and TKA-1) is a scaffold protein that contains two PDZ domains and one ERM-binding domain (Yun et al., 1997). PDZ domains are named for the first three proteins in which they were described (postsynaptic density-95, discs large, zona occludens-1) and mediate protein–protein interactions by binding to the last few carboxyl-terminal residues of their target proteins (Hung and Sheng, 2002). ERM (ezrin, ra-dixin, moesin)-binding domains link NHERF-2 and the related scaffold NHERF-1 to the actin cytoskeleton by interacting with the ERM family of actin-binding proteins (Nguyen et al, 2001). Thus, via these multiple domains, NHERF-2 can link its interacting partners to each other and to the actin cytoskeleton.
The PDZ domains of NHERF-2 are known to interact with a handful of specific G protein-coupled receptors (GPCRs), growth factor receptors, ion channels, transporters, and cellular signaling effectors (Shenolikar et al., 2004). By recruiting protein targets and forming protein complexes, NHERF-2 can regulate the targeting and/or trafficking of its binding partners as well as their physiological functions. For example, the simultaneous interaction of NHERF-2 with phospholipase Cβ (PLCβ) and either the purinergic P2Y1 receptor or lysophosphatidic acid LPA2 receptor results in enhanced receptor signaling (Fam et al., 2005; Oh et al., 2004; Yun et al., 2005).
The distribution of NHERF-2 has been well described in the kidney (Wade et al., 2003), but nothing is currently known about NHERF-2 distribution in the brain. Many NHERF-2 binding partners are expressed in the central nervous system. However, it is not known whether NHERF-2 protein is expressed in the same subcellular compartments or even in the same cell types as the various known NHERF-2 binding partners. Thus, it is difficult to assess the potential physiological relevance of these NHERF-2 interactions in native brain tissue. In this study, we investigated the cellular and subcellular distribution of NHERF-2 in mouse brain in order to shed light on the potential functional roles that this scaffold protein may play in the central nervous system.
MATERIALS AND METHODS
Animals and tissue preparation
All procedures were approved by the animal care and use committees of Emory University and conformed to the U.S. National Institutes of Health guidelines. For Western blotting, brain tissue from three C57BL/6 mice was used. The rodents were rapidly decapitated, and the brains were removed from the skull and dissected on ice. For immunocytochemistry, eight C57BL/6 mice were used. They were deeply anesthetized with an overdose of pentobarbital (100 mg/kg i.p.) and transcardially perfused with ice-cold oxygenated Ringer’s solution, followed by a fixative solution containing 4.0% paraformaldehyde and 0.2% glutaraldehyde in phosphate buffer (PB; 0.1 M, pH 7.4). The brains were then removed from the skull, blocked, and postfixed in the same fixative for 2 hours at 4°C before being washed in phosphate-buffered saline (PBS; 0.01 M, pH 7.4) and cut into 50 – 60-μm-thick coronal sections with a vibrating microtome.
Generation and characterization of polyclonal antibody against NHERF-2
The sequence of human NHERF-2 corresponding to the carboxyl-terminal 106 amino acid residues (C-106) was generated by PCR, cloned into pET16b (Novagen, Madison, WI), expressed as a hexahistidine (His6)-tagged fusion protein in Escherichia coli, and affinity purified with Ni2+-nitrilotriacetic acid resin as suggested by the manufacturer (Qiagen, Valencia, CA). The anti-NHERF-2 antiserum against the C-106 fusion protein was produced by immunizing a rabbit (New Zealand white; Covance Research Products, Denver, PA). This polyclonal antiserum is referred to as Ab2570. Ab2570 was then affinity purified. Briefly, 1 ml Affi-Gel 10 (Bio-Rad, Hercules, CA), consisting of activated N-hydroxysuccinimide esters cross-linked to agarose gel bead support, was incubated with 2 mg of the fusion protein in 2 ml of PBS overnight at 4°C on a rocking shaker. Two hundred milliliters of 1 M ethanolamine were added to the suspension and incubated for 1 hour at 4°C to block any remaining activated esters. After four washes with distilled water, the beads with the bound fusion proteins were incubated with 1 ml of antiserum diluted 1:10 in 10 mM Tris-HCl, pH 7.5, for 2 hours at 4°C. The resulting resin was transferred into a column and subsequently washed with 20 ml of 10 mM Tris, pH 7.5, and 20 ml of 500 mM NaCl, 10 mM Tris, pH 7.5. Antiserum was eluted with 10 ml of 100 mM glycine, pH 2.5, and immediately neutralized with 1 ml of 1 M Tris-HCl, pH 8.0. The remaining bound antibodies were eluted with 10 ml of triethylamine, pH 11.5, followed by neutralization with 1 ml of 1 M Tris, pH 8.0. Acid- and base-eluted antibodies were combined together, dialyzed against PBS overnight at 4°C, and concentrated to a final volume of 4 ml. The specificity of this antiserum in cell lines transfected with NHERF-1 or NHERF-2 was previously tested (Yun et al., 1998). Additional control experiments for the antibody specificity, which are described below, were performed in this study.
Western blotting
Human embryonic kidney (HEK-293) cells were maintained in complete medium (D-minimum essential medium with 10% fetal bovine serum and 1% penicillin/ streptomycin) in 10-cm culture dishes at 37°C with 5% CO2. The cells at 70 – 80% confluence were transiently transfected with 2 μg of either rabbit Flag-tagged NHERF-1 or human Flag-tagged NHERF-2 in pBK plasmid mixed with Lipofectamine reagent (Invitrogen, Carlsbad, CA) in incomplete medium. Complete medium was added after 3– 4 hours of incubation. The cells were lysed 48 hours later in 1× SDS-PAGE sample buffer.
All brain samples were prepared at 4°C. After dissections, total brain tissues were completely homogenized with a sonicator in an ice-cold buffer solution containing 10 mM HEPES, 0.1 mM EDTA, 50 mM NaCl, 1 mM benzamidine, 1.0% Triton X-100, and protease inhibitor cocktail (1 tablet per 50 ml; Roche Diagnostic GmbH, Manheim, Germany). After 1 hour of solubilization end over end, the homogenates were centrifuged for 5 minutes at 15,000 rpm to remove tissue debris and unsolubilized membranes. Total protein concentration of the lysates (supernatants) was measured by using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA). The lysates were then eluted with 6× SDS-PAGE sample buffer. The samples from transfected HEK-293 cells and 50 μg of brain lysates were resolved by SDS-PAGE (12% or 4 –20% Trisglycine gels; Invitrogen, Carlsbad, CA) and subjected to Western blot analysis with appropriate antibodies. Equal protein loading was assessed with a mouse anti-β-tubulin antibody (1:10,000; Sigma, St. Louis, MO). Immunoreactive bands were detected with the enhanced chemiluminescence detection system (Pierce, Rockford, IL) with horse-radish peroxidase-conjugated goat anti-rabbit or mouse secondary antibodies (1:4,000; Amersham Biosciences, Little Chalfont, United Kingdom).
Immunoperoxidase localization of NHERF-2 proteins for light microscopy
Tissue sections were treated with 1.0% NaBH4 in PBS for 20 minutes at room temperature (RT), followed by PBS rinses. The sections were then incubated for 1 hour at RT in PBS containing 0.3% Triton X-100 and 5% nonfat dry milk, followed by the primary antibody solution (rabbit anti-NHERF-2; 1:7,000) containing 0.3% Triton X-100 and 1% milk in PBS overnight at RT. After PBS rinses, the sections were incubated for 90 minutes in biotinylated goat anti-rabbit IgG (1:200; Vector Laboratories, Burlingame, CA), rinsed, and then incubated with avidin-biotin-peroxidase complex (1:200; Vector Laboratories). The sections were washed in PBS and Tris buffer (50 mM, pH 7.6) and transferred to a solution containing 0.025% 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma), 10 mM imidazole, and 0.005% hydrogen peroxide in Tris buffer for 10 minutes. After PBS washes, the sections were mounted on gelatin-coated slides, air dried, and dehydrated, and a coverslip was applied with Permount. The sections were examined with a Leica DMRB microscope (Leica Microsystems, Inc., Bannockburn, IL), and images were acquired with a CCD camera (Leica DC500) controlled by Leica IM50 software (version 1.20).
Immunoperoxidase localization of NHERF-2 proteins for electron microscopy
After NaBH4 treatment, all sections prepared for electron microscopy were placed in a cryoprotectant solution (0.05 M PB, pH 7.4, containing 25% sucrose and 10% glycerol), frozen at – 80°C, thawed, and returned to a graded series of cryoprotectant and PBS. Sections were incubated the same way as for light microscopy, except that no Triton X-100 was used in the antibody diluent, and the incubation in the primary antibodies lasted for 48 hours at 4°C. After immunostaining, the sections were transferred to 0.1 M PB, pH 7.4, for 10 minutes and treated with 1.0% OsO4 in PB for 20 minutes. They were then rinsed with PB and dehydrated in an ascending gradient of ethanol. Uranyl acetate (1.0%) was added to the 70% alcohol to enhance contrast. The sections were then treated with propylene oxide before being embedded in epoxy resin (Durcupan ACM; Fluka, Buchs, Switzerland) for 12 hours, mounted on microscope slides, and placed in the oven at 60°C for 48 hours. Samples of cerebral cortex (corresponding to the agranular cortex), rostral hippocampus, caudolateral striatum, and cerebellar cortex were cut out from the slides, glued on the top of resin blocks with cyanoacrylate glue, and cut into 60-nm-thick ultrathin sections with an ultramicrotome (Leica Ultracut T2). The ultrathin sections were serially collected on single-slot Pioloform-coated copper grids, stained with lead citrate for 5 minutes, and examined with a Zeiss EM-10C electron microscope (Carl Zeiss Microimaging Inc., Thornwood, NY). The micrographs were acquired with a CCD camera (DualView 300W; Gatan, Inc., Pleasonton, CA) controlled by DigitalMicrograph software (version 3.8.1; Gatan, Inc.). Micrographs were adjusted only for brightness and contrast, with the image resolution kept constant, in Photoshop 7.0 (Adobe Systems, Inc., San Jose, CA) to optimize the quality of the images for analysis.
Double-immunofluorescence labeling of NHERF proteins and astrocytic (glial fibrillary acidic protein) or pericyte (α-smooth muscle actin) cell markers
Sections were treated with 1.0% H2O2 in PBS for 10 minutes at RT. The sections were then incubated for 1 hour in PBS containing 5% nonfat dry milk, 0.3% Triton X-100, followed by a mix of the primary antibodies [rabbit anti-NHERF-2 with Alexa-488-conjugated mouse antiglial fibrillary acidic protein (GFAP; 1:1,000; Molecular Probes, Eugene, OR) or fluorescein isothiocyanate (FITC)-conjugated mouse anti--smooth muscle actin (1: 500)] in 1% nonfat dry milk/0.3% Triton X-100/PBS for 24 hours. After PBS rinses, the sections were incubated for 1 hour in donkey anti-rabbit IgG conjugated with Cy5 (1: 200; Jackson Immunoresearch Laboratories, West Grove, PA) in 1% nonfat dry milk/0.3% Triton X-100/PBS. The sections were then rinsed in PBS and incubated in a solution containing 10 mM CuSO4 and 50 mM NH4COOCH3 at pH 5.0 for 30 minutes, followed by PBS rinses. They were then mounted on glass slides, and coverslips were applied with Vectashield (Vector Laboratories). The mounted sections were stored in the dark at 4°C until examination with a Zeiss LSM410 confocal microscope with 515–540 nm and 600 –640 nm bandpass emission filters for GFP/Alexa-488 and Cy5, respectively. The micrographs were taken at random throughout the brain sections with a ×63 objective.
Control experiments: antibody specificity
Expression and purification of fusion proteins
Hexahistidine- and S-tagged truncations of NHERF-1 and NHERF-2 (PDZ2 + ERM), corresponding, respectively, to the last 206 and 188 amino acids of the two proteins, and GST-tagged full-length NHERF-1 and NHERF-2 were obtained by bacterial expression cDNA constructs in pET-30A (Novagen) and pGEX-4T1 (Amersham Pharmacia Biotech) vectors, respectively. His-tagged fusion proteins were then purified by using ProBond nickel resin (Invitrogen), and GST-tagged fusion proteins were purified with glutathione agarose beads (Sigma). His-tagged proteins were eluted from the resin, and the protein concentrations were determined by using the Bio-Rad Protein Assay. Protein amounts of GST-fusion proteins on agarose beads were determined by SDS-PAGE and Coomassie staining.
Preabsorption and competition assays
To control for nonspecific immunoreactivity, immunoblots and brain tissue sections were incubated with 1) primary NHERF-2 antibody preabsorbed with His-fusion proteins corresponding to 0, 2, 5, 10, and 100 times the amount of primary antibody by weight and 2) supernatant of the NHERF-2 antibody solution after pull-down of specific antibodies with a twofold excess quantity of GST-full length NHERF proteins.
Control experiments: immunohistochemistry
Immunoperoxidase labeling
To control for nonspecific staining by the secondary antibody and avidin-biotin complex, tissue sections were incubated in the absence of the primary and/or secondary antibodies in parallel with normal incubations. These control incubations resulted in the total absence of immunoreactivity at both light and electron microscopic levels.
Immunofluorescence labeling
To make sure that the secondary antibodies did not recognize endogenous nonspecific antigens in mouse brain sections, primary antibodies raised in mouse (anti-GFAP and anti-α-smooth muscle actin) were conjugated to a fluorophore. To control for cross-reactivity, the primary antibody against NHERF-2 was omitted from control incubations. Single incubations for NHERF-2, GFAP, and α-smooth muscle actin antibodies were also performed to test for possible competition between antibodies. Similar patterns of immunolabeling for each antibody were observed in single-and double-immunofluorescence-labeling experiments.
Analysis of material
Series of sections including cerebral cortex, basal ganglia, diencephalon, midbrain, brainstem, and cerebellum were examined at the light microscope level to analyze the overall pattern of NHERF-2 immunoreactivity. For the electron microscopic examination, tissue was randomly sampled from four different brain structures (cerebral cortex, hippocampus, striatum, and cerebellar cortex) from three animals. Electron micrographs were taken at ×12,500, ×16,000, and ×20,000 from ultrathin sections close to the surface, where the antibody penetration was optimal. For the quantitative analysis of the distribution of NHERF-2 immunoreactivity, observations were made from areas corresponding to 1,000 μm2 without small capillaries for each brain structure examined in the three animals used. All labeled elements in an area of observation were counted and categorized as dendrites, spines, axons, terminals, or astrocytes based on their ultrastructural features, as described elsewhere (Peters et al., 1991). Any elements that could not be identified because of lack of well-defined ultrastructural features were categorized as “unknown.” These “unknown” elements most likely consisted of small-caliber dendrites, unmyelinated axons, and glial processes. The “unknown” elements were included in the statistical analysis but were excluded from the data presented in Figure 7. Statistical differences in the sub-cellular distribution of NHERF-2 were analyzed with a two-way ANOVA, followed by post hoc Bonferroni tests. To estimate the relative proportion of immunoreactive and nonimmunoreactive elements in the cerebral cortex, a total surface area of 1,860 μm2 of cortical tissue was sampled from a total of three blocks of tissue from three different animals. Every labeled and unlabeled element was counted, and the mean percentages of immunoreactive structures were estimated for the three animals. In total, 544 dendrites, 426 spines, 875 axon terminals, and 1,009 glial processes were examined for this analysis.
Fig. 7.
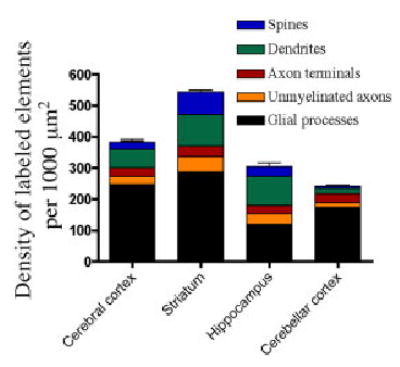
Quantification of the subcellular distribution of NHERF-2 labeling in four different mouse brain structures. Statistical analysis by a two-way ANOVA shows a significant difference (P < 0.05, F = 2.290, n = 3 animals) in the subcellular distribution of NHERF-2 labeling between brain structures, revealing that NHERF-2 distribution varies in different regions of the brain. However, in all regions examined, the majority of NHERF-2 labeled elements were glial processes.
RESULTS
Specificity of the rabbit anti-NHERF-2 antibody
The rabbit anti-NHERF-2 antibody recognized one band at the expected size (~40 kDa) in HEK-293 cells transfected with Flag-tagged NHERF-2. In Flag-tagged NHERF-1-transfected cells, no band corresponding to NHERF-1 at 50 kDa was detected, demonstrating the specificity of the antibody for NHERF-2. In brain lysates, a single major band was observed at approximately the same size as the transfected NHERF-2 (Fig. 1A). Preabsorption and competition assays, described in Material and Methods, resulted in a complete loss of NHERF-2 labeling in both Western blots and brain sections. Indeed, immunoreactivity was lost in the presence of His-NHERF-2 PDZ2-ERM and after pull-down with GST-NHERF-2, but not in the presence of His-NHERF-1 PDZ2 + ERM or after pull-down with GST-NHERF-1 (Fig. 1B,C). Thus, the anti-NHERF-2 antibody exhibited specific recognition of NHERF-2 in mouse brain tissue.
Fig. 1.
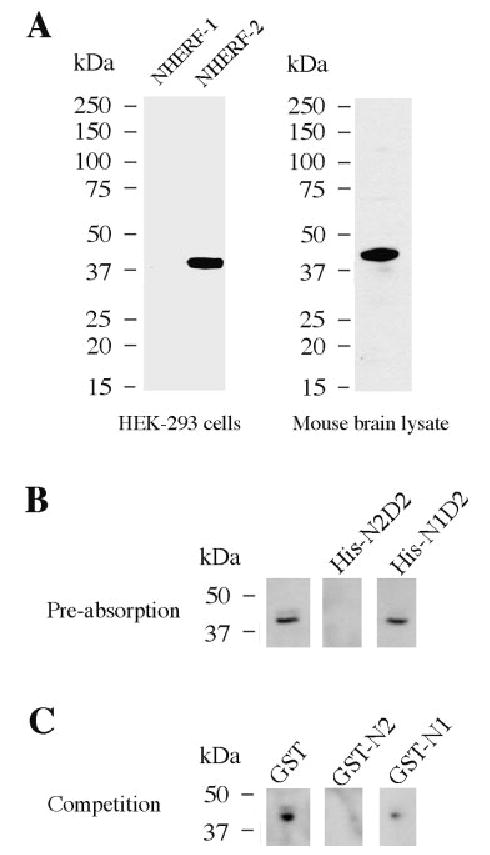
Rabbit anti-NHERF-2 antibody specifically recognizes transfected and endogenous brain NHERF-2 protein. A: Lysates from HEK-293 cells transfected with the two members of the NHERF family, NHERF-1 and NHERF-2, which share 62% similarity, and mouse brain lysate were immunoblotted with rabbit anti-NHERF-2 antibody at a 1:7,000 concentration. B: Mouse brain lysate was immunoblotted with the rabbit anti-NHERF-2 antibody after no preabsorption or preabsorption with the fusion protein His-NHERF-2 PDZ2 + ERM (His-N2D2) or His-NHERF-1 PDZ2 + ERM (His-N1D2). C: Mouse brain lysate was immunoblotted with supernatant of rabbit anti-NHERF-2 antibody pulled-down with GST, GST-NHERF-2 (GST-N2), or GST-NHERF-1 (GST-N1).
Localization of NHERF-2 immunolabeling in mouse brain
NHERF-2 immunolabeling was observed in all brain regions examined. In addition to immunopositive neuropil throughout the mouse brain, lightly immunoreactive neuronal cell bodies were observed in some brain structures, such as the cerebral cortex, striatum, and hippocampus (Fig. 2). In contrast, some neuronal populations, such as the granule and Purkinje cells in the cerebellar cortex (Fig. 3B) and the dopaminergic or GABAergic neurons in the substantia nigra (data not shown), were devoid of immunoperoxidase labeling. At the light microscopic level, populations of astrocytes, such as Bergmann glia in the cerebellum (Fig. 3B), were clearly immunoreactive for NHERF-2. The specificity of staining in these tissue sections was established in control experiments in which the anti-NHERF-2 antibody was preabsorbed with antigen (Fig. 3C). In addition, immunopositive astrocyte-like cells were observed in several brain regions, including layers 1 and 2 of the cerebral cortex (Fig. 3A) and the white matter (Fig. 3D). The astrocytic nature of these cells was confirmed by colocalization of NHERF-2 labeling with the astrocytic marker GFAP (arrows in Fig. 4B–D). However, the possibility of NHERF-2 labeling of microglia cannot be excluded.
Fig. 2.
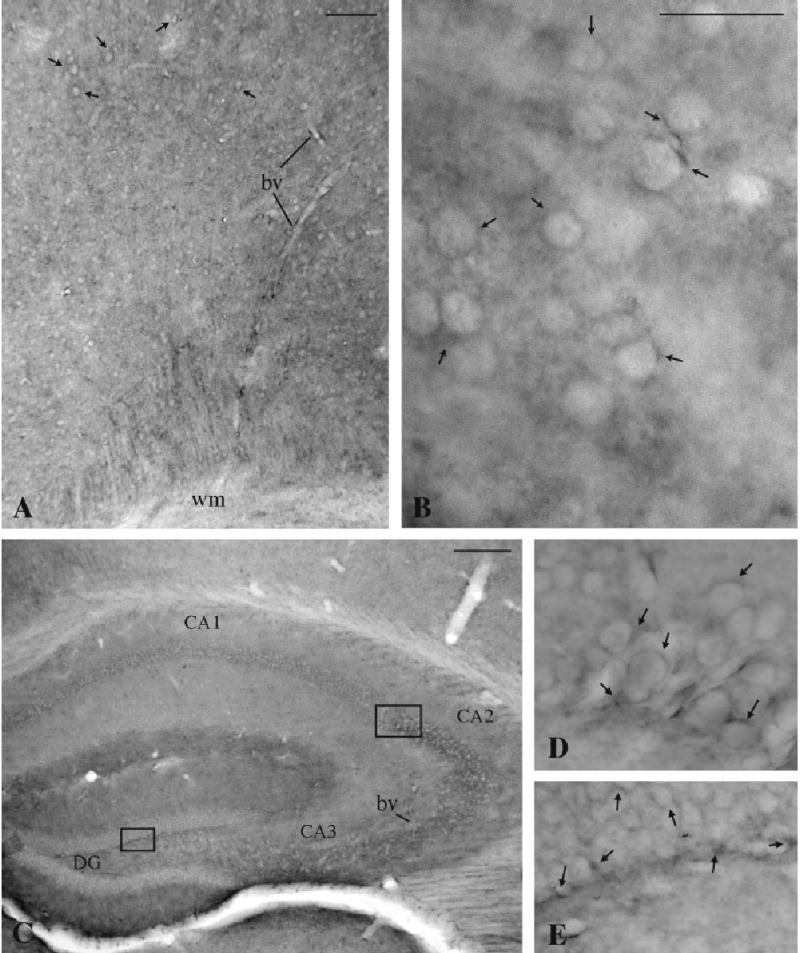
NHERF-2 immunoreactivity is associated with neuronal plasma membranes. A,B: In addition to NHERF-2 immunostained neuropil, lightly labeled neuronal cell bodies and proximal dendritic processes (arrows) were found throughout the cerebral cortex (A) and the striatum (B). Thin processes (arrows) in the cell body layer of hippocampal pyramidal neurons (C,D) and the dentate gyrus neurons (C,E) were also found to be immunopositive for NHERF-2. NHERF-2 labeling was often associated with blood vessels (bv), as shown in A and C. CA1–3, fields of the hippocampus; DG, dentate gyrus; wm, white matter. Scale bars = 500 μm in A,C; 50 = μm in B (applies to B,D,E).
Fig. 3.
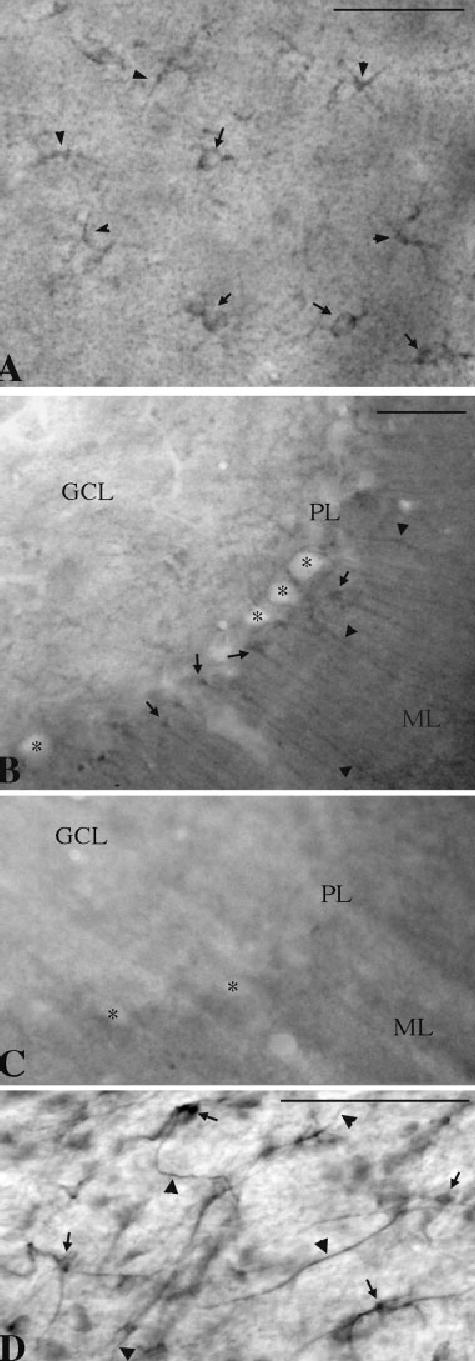
Astrocytes express NHERF-2 immunoreactivity throughout the brain. A: Labeled protoplasmic astrocytes (arrows) were observed in the upper layer of the cerebral cortex. B: In contrast to immunonegative granule and Purkinje cells (asterisks), cell bodies (arrows) and processes (arrowheads) of Bergmann glia were NHERF-2 immunopositive in the cerebellar cortex. C: Bergmann glia were devoid of NHERF-2 immunostaining after preabsorption of the antibody with a fusion protein corresponding to the antigen (His-NHERF-2 PDZ2 + ERM). D: Fibrous astrocyte-like labeled cells (arrows) and processes (arrowheads) were also occasionally found in the white matter. GCL, granule cell layer; ML, molecular layer; PL, Purkinje layer. Scale bars = 50 μm in A,D; 50 μm in B (applies to B,C).
Fig. 4.
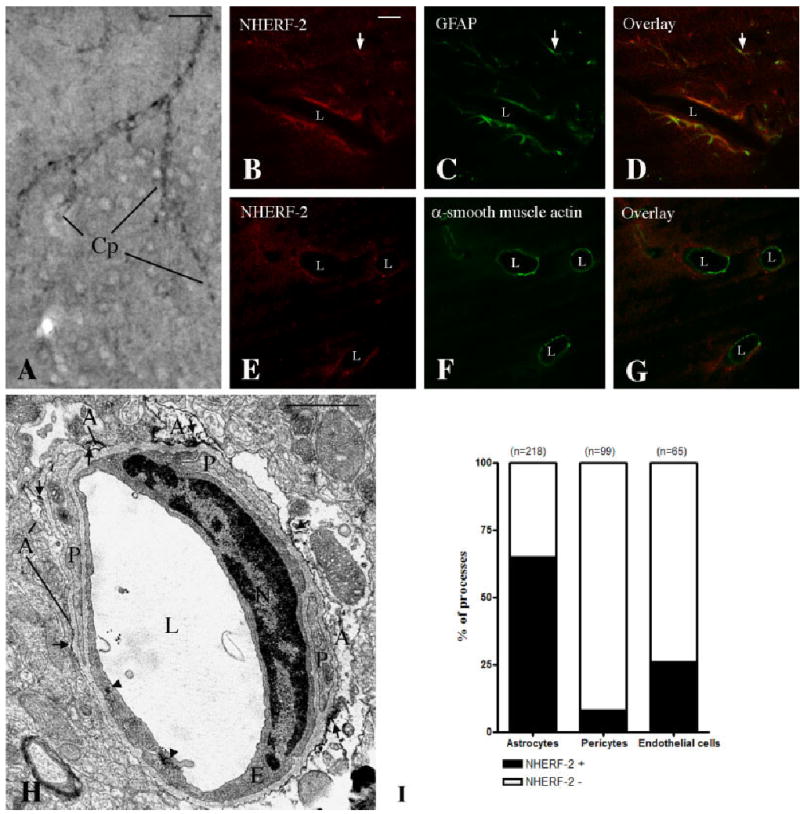
Capillary-associated NHERF-2 labeling is found mainly in astrocytic end-feet. A: Example of NHERF-2 immunoperoxidase labeling associated with capillaries (Cp) in the cerebral cortex. B–G: Double immunofluorescence of NHERF-2 with the astrocytic marker GFAP (B–D) and the pericyte marker α-smooth muscle actin (E–G) revealed that NHERF-2 is found in astrocytic processes but not in pericytes surrounding capillaries in the mouse brain. Note the fine astrocytic process labeled for both NHERF-2 and GFAP (arrows in B–D). H: Electron micrograph of a small capillary from the cerebral cortex. In this example, NHERF-2 immunostaining was found in astrocytic processes (arrows) and in the endothelial cell (arrowheads). I: Quantitative analysis of NHERF-2-positve (NHERF-2+) and NHERF-2-negative (NHERF-2−) astrocytes, pericytes, and endothelial cells surrounding small cortical capillaries. n = Number of astrocytic, pericytic, and endothelial processes examined. A, astrocytic process; E, endothelial cell; L, lumen of capillary; N, nucleus; P, pericyte process. Scale bars = 100 μm in A; 25 μm in B (applies to B–G); 1 μm in H.
Strong NHERF-2 immunoreactivity was consistently observed to be associated with blood vessels throughout the brain (Figs. 2A,C, 4A). The distribution of NHERF-2 labeling between the components of cerebral capillaries was studied at both the confocal and the electron microscopic levels, as shown in Figure 4. The astrocytic marker GFAP exhibited colocalization with NHERF-2 immunofluorescence around large capillaries (Fig. 4B–D). In contrast, NHERF-2 labeling typically appeared to surround the pericyte marker α-smooth muscle actin (Fig. 4E–G) but did not colocalize with it. Similar observations were made for small capillaries at the electron microscopic level (Fig. 4H,I), where 65% of the astrocytic processes surrounding the capillaries in the cerebral cortex were immunopositive for NHERF-2. In contrast, only 8% of the pericytes in the perivascular space and 26% of the endothelial cells were weakly labeled (Fig. 4I).
In addition to glial processes, both postsynaptic (Fig. 5) and presynaptic (Fig. 6) labeled neuronal elements were observed in the four brain structures investigated at the electron microscopic level. Figure 5A,C shows examples of NHERF-2-positive neuronal and astrocytic cell bodies, respectively, which are surrounded by labeled glial processes, dendrites, and dendritic spines. NHERF-2 immunoreactivity was generally associated with plasma membranes, endoplasmic reticulum, and microtubules (Figs. 5, 6). In axon terminals, NHERF-2 immunoreactivity was also associated with synaptic vesicles and the active zone of putative glutamatergic asymmetric synapses (Fig. 6B,C). As shown in Figure 7, the relative density of the different labeled elements varied significantly from one brain structure to another. The brain regions with labeled neuronal cell bodies, such as the cerebral cortex, striatum, and hippocampus, displayed a higher density of neuronal elements (spines, dendrites, unmyelinated axons, and axon terminals) than the cerebellar cortex, where the two main populations of neurons, the granule and Purkinje cells, were immunonegative. In all brain structures studied, glial processes, which are probably of astrocytic nature, were the most frequent NHERF-2-immunoreactive elements. In support of these observations, quantitative analyses of NHERF-2-labeled profiles in the cerebral cortex revealed that 39% ± 6% of the identified astrocytic processes were immunolabeled for NHERF-2, whereas only 12% ± 5% of the postsynaptic elements (spines and dendrites) and 5% ± 2% of the presynaptic elements (axon terminals) were NHERF-2 immunopositive.
Fig. 5.
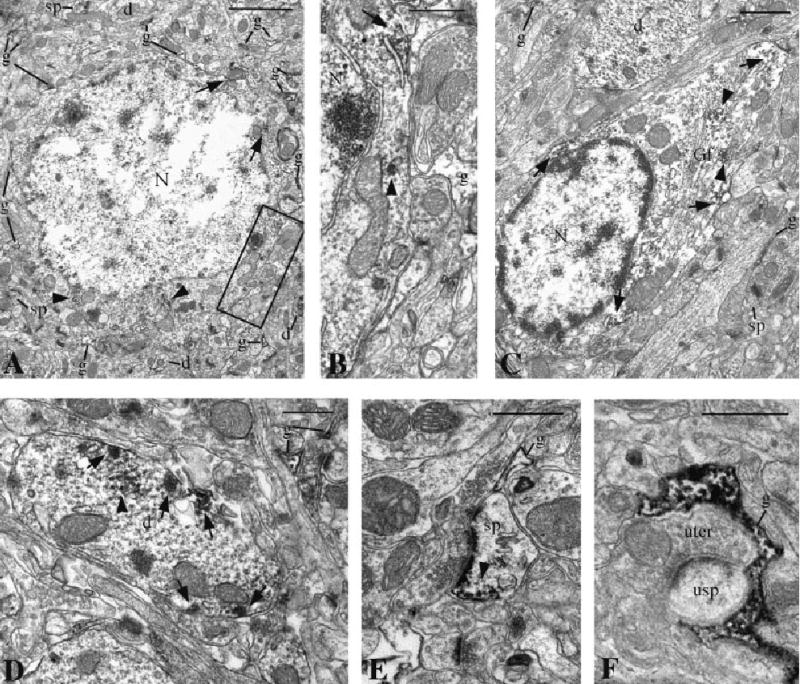
NHERF-2 labeling in postsynaptic neuronal elements and astrocytes. A,B: Example of an NHERF-2-labeled neuronal cell body in the cerebral cortex. B shows a higher magnification of the boxed area in A. Aggregations of immunoreactivity were associated with the plasma membrane (arrows) and the endoplasmic reticulum (arrowheads) of the perikaryon. NHERF-2-immunopositive glial processes (g), dendrites (d), and dendritic spines (sp) were also observed. C: Example of a NHERF-2 labeled astrocyte perikaryon, identified by its gliofilaments (Gf), in the hippocampus. Patches of immunoreactivity were associated with the plasma membrane (arrows) as well as the cytoskeleton (arrowheads). D: Hippocampal dendrite with patches of labeling associated with the plasma membrane (arrows) and microtubules (arrowheads). E: Example of a dendritic spine immunopositive for NHERF-2 (arrowhead) in the striatum. F: A NHERF-2 stained astrocytic process (g) wraps around an axospinous asymmetric synapse in the cerebral cortex. N, nucleus; usp, unlabeled dendritic spine; uter, unlabeled; t, axon terminal. Scale bars = 2 μm in A,C; 0.5 μm in B,D–F.
Fig. 6.
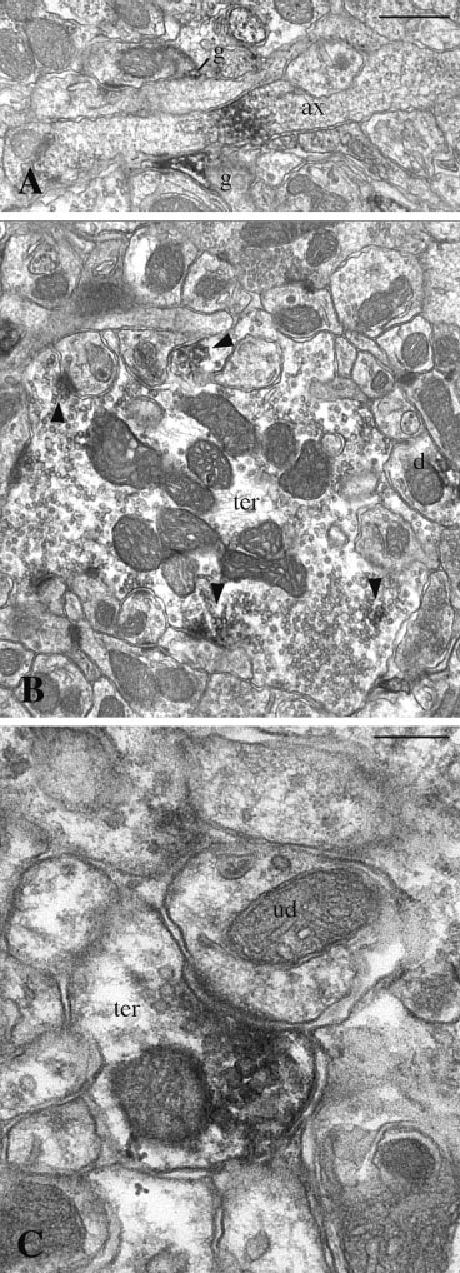
NHERF-2 expression in presynaptic neuronal elements. Examples from the cerebellar cortex are shown. A: An unmyelinated axon (ax) that displays a discrete patch of NHERF-2 immunoreactivity is shown. Glial processes are also identified (g). B: NHERF-2 immunolabeling was often associated with synaptic vesicles (arrowheads) in mossy fiber axon terminals (ter). C: Higher magnification of an axon terminal shows the association of NHERF-2 labeling with synaptic vesicles close to the active zone. d, Labeled dendrite; g, glial process; ud, unlabeled dendrite. Scale bars = 0.5 μ m in A (applies to A,B); 0.2 μm in C.
DISCUSSION
We investigated the localization of the scaffold protein NHERF-2 in the central nervous system. NHERF-2 was expressed in both pre- and postsynaptic neuronal elements but was predominantly found in astrocytes. It is well known that there is a high level of heterogeneity in both neuronal and glial populations in the brain (DeFelipe, 1993; Kimelberg, 1995; Porter and McCarthy, 1997). Accordingly, not all neurons or astrocytes in our studies displayed NHERF-2 immunoreactivity. For example, under our experimental conditions, no NHERF-2 labeling was detected in Purkinje and granule cells in the cerebellar cortex. Similarly, no labeling was observed in dopaminergic and GABAergic neurons of the substantia nigra, whereas striatal, cortical, and hippocampal neurons were labeled. Furthermore, not all glial processes present in the electron micrographs were labeled for NHERF-2. This observation is supported by the quantification of cortical astrocytic processes immunopositive for NHERF-2, which revealed that 39% of all astrocytic processes were NHERF-2 positive.
NHERF-2 and its interacting partners in astrocytes
Large numbers of NHERF-2 binding partners are known to be highly expressed in astrocytes. NHERF-2 interacts with G protein-coupled receptors such as the β2-adrenergic receptor (Hall et al., 1998), adenosine 2b receptor (Sitaraman et al., 2002), parathyroid hormone receptor (Mahon et al., 2002), LPA2 receptor (Oh et al., 2004; Yun et al., 2005), and P2Y1 receptor (Fam et al., 2005). All of these receptors exhibit robust astrocytic expression in the brain (Aoki, 1992; Fredholm and Altiok, 1994; Hashimoto et al., 1994; Struckhoff and Turzynski, 1995; Mantyh et al., 1995; Morán-Jiménez and Matute, 2000; Ye et al., 2002; Trincavelli et al., 2004). Moreover, other proteins known to associate with NHERF-2, including the cystic fibrosis transmembrane conductance regulator (CFTR; Hall et al., 1998; Sun et al., 2000), transient receptor potential channel 4 (TRPC4; Lee-Kwon et al., 2005), actin-binding protein ezrin (Yun et al., 1998), and enzyme phospholipase C-β (Hwang et al., 2000; Mahon et al., 2002) also exhibit significant astrocytic expression (Mizuguchi et al., 1991; Parkerson and Sontheimer, 2004; Song et al., 2005; Gronholm et al., 2005).
NHERF-2 was first characterized as a protein in the kidney capable of regulating the activity of the Na+/H+ exchanger NHE3 (Yun et al., 1997; Donowitz et al., 2005). Astrocytes are known to exert a strong influence on pH and ion homeostasis in the brain (Kettenmann and Ransom, 1995), so it is possible that astrocytic NHERF-2 may be involved in brain NHE regulation, analogous to the role that NHERF-2 plays in NHE3 regulation in the kidney. However, it is not clear at present whether NHERF-2 can regulate the NHE isoforms that are found in the brain, or whether instead NHERF-2 regulation of NHE is NHE3 specific (Donowitz et al., 2005). Of course, astrocytes play many other important roles beyond the control of pH and ion homeostasis, including the regulation of neuronal development and metabolism, neurotransmitter reuptake, release of neurotransmitters, and phagocytosis (Kettenmann and Ransom, 1995). These functions are all dependent on the activity of specific astrocyte-expressed receptors, transporters, and exchangers, many of which are known to be regulated by NHERF-2. Thus, NHERF-2 may play a central role in the regulation of diverse aspects of astrocytic physiology.
NHERF-2 and the blood– brain barrier
The blood– brain barrier is a diffusion barrier essential for normal function of the central nervous system. Endothelia, basal lamina, pericytes, and astrocyte end-feet compose the blood– brain barrier of the cerebral microvasculature. Intercellular signaling among endothelial cells, pericytes, and astrocytes is necessary to maintain the integrity of the blood– brain barrier (Ballabh et al., 2004). We observed that NHERF-2 immunostaining was present in all three of these cellular components, although more frequently in astrocytes than in the other two cell types. Na+/H+ exchanger activity has been reported to regulate cerebral arteriolar tone in rats (Saesue et al., 2004), which may suggest a role for NHERF-2 in the regulation of local cerebral blood flow.
Pericytes, which reside between basal laminae in the perivascular space (Peters et al., 1991), and endothelial cells have been previously reported to express NHERF-2 in kidney (Lee-Kwon et al., 2005). However, we observed, in contrast to these findings from renal tissue, more sporadic labeling of pericytes (8%) and endothelial cells (25%) in the cerebral cortex, whereas 65% of the astrocytic end-feet were NHERF-2 immunopositive. These observations suggest that the microvascular expression of NHERF-2 may vary from one tissue to another. However, we cannot rule out the possibility that the different polyclonal antibodies used in the kidney vs. brain studies might account for the observed differential patterns of microvascular staining.
Subcellular localization of NHERF-2 labeling
The regulatory functions of NHERF-1 and NHERF-2 on their target proteins are believed to be achieved mainly by physically tethering cytosolic proteins, such as phospholipases and protein kinases, to transmembrane proteins, such as receptors, transporters, and channels. However, there is also evidence that the NHERF proteins may play important roles in the trafficking of their transmembrane binding partners to and from the plasma membrane (Shenolikar et al., 2004). In our study, NHERF-2 immunoreactivity was associated mainly with the plasma membrane of both neurons and astrocytes, which is consistent with the fact that NHERF-2 avidly associates with a number of transmembrane proteins. In addition, NHERF-2 labeling was also often observed to be associated with synaptic vesicles in putative glutamatergic axon terminals. In renal proximal tubules, NHERF-2 is preferentially localized in vesicle-rich regions (Wade et al., 2003). These observations suggest that NHERF-2 might play a role in vesicular trafficking. It is possible that NHERF-2 interacts with specific vesicle-associated proteins, or, alternatively, this NHERF-2 association with vesicles might simply reflect the interactions of NHERF-2 with certain transmembrane proteins that undergo extensive vesicle-mediated trafficking.
In conclusion, the scaffold protein NHERF-2 was found to be present in populations of neurons and astrocytes throughout the mouse brain. NHERF-2 immunolabeling was most frequently found in astrocytic processes, where a large number of NHERF-2 binding partners are also known to be expressed. These findings suggest that NHERF-2 may play defined roles in certain neuronal populations and a central role in the regulation of astrocytic physiology.
Acknowledgments
The authors thank Susan Maxson, Jean-François Paré, Marc Verreault, and Kristopher Bough for their technical assistance and advice.
Footnotes
Grant sponsor: National Institutes of Health (to R.A.H., Y.S., and Yerkes National Primate Research Center); Grant sponsor: W.M. Keck Foundation (to R.A.H.).
References
- Aoki C. β-Adrenergic receptors: astrocytic localization in the adult visual cortex and their relation to catecholamine axon terminals as revealed by electron microscopic immunocytochemistry. J Neurosci. 1992;12:781–792. doi: 10.1523/JNEUROSCI.12-03-00781.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabh P, Braun A, Nedergaard M. The blood– brain barrier: an overview structure, regulation, and clinical implications. Neurobiol Dis. 2004;16:1–13. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- DeFelipe J. Neocortical neuronal diversity: chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell surface molecules. Cereb Cortex. 1993;3:273–289. doi: 10.1093/cercor/3.4.273. [DOI] [PubMed] [Google Scholar]
- Donowitz M, Cha B, Zachos NC, Brett CL, Sharma A, Tse C-M, Li X. 2005. NHERF family and NHE regulation. J Physiol [epub]. [DOI] [PMC free article] [PubMed]
- Fam S, Paquet M, Castleberry A, Oller H, Lee C, Traynelis SF, Smith Y, Yun CC, Hall RA. P2Y1 receptor signaling is controlled by interaction with the PDZ scaffold NHERF-2. Proc Natl Acad Sci U S A. 2005;102:8042– 8047. doi: 10.1073/pnas.0408818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredholm BB, Altiok N. Adenosine A2B receptor signalling is altered by stimulation of bradykinin or interleukin receptors in astroglioma cells. Neurochem Int. 1994;25:99 –102. doi: 10.1016/0197-0186(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Gronholm M, Teesalu T, Tyynela J, Piltti K, Bohling T, Wartiovaara K, Vaheri A, Carpen O. Characterization of the NF2 protein merlin and the ERM protein ezrin in human, rat and mouse central nervous system. Mol Cell Neurosci. 2005;28:683– 693. doi: 10.1016/j.mcn.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Hall RA, Ostedgaard LS, Premont RT, Blitzer JT, Rahman N, Welsh MJ, Lefkowitz RJ. A C-terminal motif found in the β2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc Natl Acad Sci U S A. 1998;95:8496 –8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Aino H, Ogawa N, Nagata S, Baba A. Identification and characterization of parathyroid hormone/parathyroid hormone-related peptide receptor in cultured astrocytes. Biochem Biophys Res Commun. 1994;200:1042–1048. doi: 10.1006/bbrc.1994.1555. [DOI] [PubMed] [Google Scholar]
- Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. J Biol Chem. 2002;277:5699 –5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- Hwang J-I, Heo K, Shin K-J, Kim E, Yun CC, Ryu SH, Shin H-S, Suh P-G. Regulation of phospholipase C-β3 activity by Na+/H+ exchanger regulatory factor 2. J Biol Chem. 2000;275:16632–16637. doi: 10.1074/jbc.M001410200. [DOI] [PubMed] [Google Scholar]
- Kettenmann H, Ransom BR. 1995. Neuroglia. NewYork: Oxford University Press.
- Kimelberg HK. Receptors on astrocytes—what possible functions? Neurochem Int. 1995;26:27– 40. doi: 10.1016/0197-0186(94)00118-e. [DOI] [PubMed] [Google Scholar]
- Lee-Kwon W, Wade JB, Zhang Z, Pallone TL, Weinman EJ. Expression of TRPC4 channel protein that interacts with NHERF-2 in rat descending vasa recta. Am J Physiol Cell Physiol. 2005;288:C942–C949. doi: 10.1152/ajpcell.00417.2004. [DOI] [PubMed] [Google Scholar]
- Mahon MJ, Donowitz M, Yun CC, Segre GV. Na+/H+ exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signalling. Nature. 2002;417:858 – 861. doi: 10.1038/nature00816. [DOI] [PubMed] [Google Scholar]
- Mantyh PW, Rogers SD, Allen CJ, Catton MD, Ghilardi JR, Levin LA, Maggio JE, Vigna SR. Beta 2-adrenergic receptors are expressed by glia in vivo in the normal and injured central nervous system in the rat, rabbit, and human. J Neurosci. 1995;15:152–164. doi: 10.1523/JNEUROSCI.15-01-00152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi M, Yamada M, Kim SU, Rhee SG. Phospholipase C isozymes in neurons and glial cells in culture: an immunocytochemical and immunochemical study. Brain Res. 1991;548:35– 40. doi: 10.1016/0006-8993(91)91103-8. [DOI] [PubMed] [Google Scholar]
- Morán-Jiménez M-J, Matute C. Immunohistochemical localization of the P2Y1 purinergic receptor in neurons and glial cells of the central nervous system. Brain Res Mol Brain Res. 2000;78:50 –58. doi: 10.1016/s0169-328x(00)00067-x. [DOI] [PubMed] [Google Scholar]
- Nguyen R, Reczek D, Bretscher A. Hierarchy of merlin and ezrin N-and C-terminal domain interactions in homo- and heterotypic associations and their relationship to binding of scaffolding proteins EBP50 and E3KARP. J Biol Chem. 2001;276:7621–7629. doi: 10.1074/jbc.M006708200. [DOI] [PubMed] [Google Scholar]
- Oh YS, Jo NW, Choi JW, Kim HS, Seo SW, Kang KO, Hwang J-I, Heo K, Kim SH, Kim YH, Kim IH, Kim JH, Banno Y, Ryu SH, Suh P-G. NHERF2 specifically interacts with LPA2 receptor and defines the specificity and efficiency of receptor-mediated phospholipase C-β3 activation. Mol Cell Biol. 2004;24:5069 –5079. doi: 10.1128/MCB.24.11.5069-5079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkerson KA, Sontheimer H. Biophysical and pharmacological characterization of hypotonically activated chloride currents in cortical astrocytes. Glia. 2004;46:419 – 436. doi: 10.1002/glia.10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Palay SL, Webster H. 1991. The fine ultrastructure of the nervous system: neurons and their supporting cells. New York: Oxford University Press. 486 p.
- Porter JT, McCarthy KD. Astrocytic neurotransmitter receptors in situ and in vivo. Prog Neurobiol. 1997;51:439 – 455. doi: 10.1016/s0301-0082(96)00068-8. [DOI] [PubMed] [Google Scholar]
- Saesue P, Horiuchi T, Goto T, Tanaka Y, Hongo K. Functional role of the Na+/H+ exchanger in the regulation of cerebral arteriolar tone in rats. J Neurosurg. 2004;101:330 –335. doi: 10.3171/jns.2004.101.2.0330. [DOI] [PubMed] [Google Scholar]
- Shenolikar S, Voltz JW, Cunningham R, Weinman EJ. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology. 2004;19:362–369. doi: 10.1152/physiol.00020.2004. [DOI] [PubMed] [Google Scholar]
- Sitaraman SV, Wang L, Wong M, Bruewer M, Hobert M, Yun CH, Merlin D, Madara JL. The adenosine 2b receptor is recruited to the plasma membrane and associates with E3KARP and Ezrin upon agonist stimulation. J Biol Chem. 2002;277:33188 –33195. doi: 10.1074/jbc.M202522200. [DOI] [PubMed] [Google Scholar]
- Song X, Zhao Y, Narcisse L, Duffy H, Kress Y, Lee S, Brosnan CF. Canonical transient receptor potential channel 4 (TRPC4) colocalizes with the scaffolding protein ZO-1 in human fetal astrocytes in culture. Glia. 2005;49:418 – 429. doi: 10.1002/glia.20128. [DOI] [PubMed] [Google Scholar]
- Struckhoff G, Turzynski A. Demonstration of parathyroid hormone-related protein in meninges and its receptor in astrocytes: evidence for a paracrine meningo-astrocytic loop. Brain Res. 1995;676:1–9. doi: 10.1016/0006-8993(95)00088-8. [DOI] [PubMed] [Google Scholar]
- Sun F, Hug MJ, Lewarchik CM, Yun CC, Bradbury NA, Frizzell RA. E3KARP mediates the association of ezrin and protein kinase A with the cystic fibrosis transmembrane conductance regulator in airway cells. J Biol Chem. 2000;275:29539 –29546. doi: 10.1074/jbc.M004961200. [DOI] [PubMed] [Google Scholar]
- Trincavelli ML, Marroni M, Tuscano D, Ceruti S, Mazzola A, Mitro N, Abbracchio MP, Martini C. Regulation of A2B adenosine receptor functioning by tumor necrosis factor a in human astroglia cells. J Neurochem. 2004;91:1180 –1190. doi: 10.1111/j.1471-4159.2004.02793.x. [DOI] [PubMed] [Google Scholar]
- Wade JB, Liu J, Coleman RA, Cunningham R, Steplock DA, Lee-Kwon W, Pallone TL, Shenolikar S, Weinman EJ. Localization and interaction of NHERF isoforms in the renal proximal tubule of the mouse. Am J Physiol Cell Physiol. 2003;285:C1494 –C1503. doi: 10.1152/ajpcell.00092.2003. [DOI] [PubMed] [Google Scholar]
- Ye X, Fukushima N, Kingsbury MA, Chun J. Lysophosphatidic acid in neural signaling. Neuroreport. 2002;13:2169 –2175. doi: 10.1097/00001756-200212030-00002. [DOI] [PubMed] [Google Scholar]
- Yun CC, Oh S, Zizak M, Steplock D, Tsao S, Tse C-M, Weinman EJ, Donowitz M. cAMP-mediated inhibition of the epithelial brush border Na+/H+exchanger, NHE3, requires an associated regulatory protein. Proc Natl Acad Sci U S A. 1997;94:3010 –3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CC, Lamprecht G, Forster DV, Sidor A. NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J Biol Chem. 1998;273:25856 –25863. doi: 10.1074/jbc.273.40.25856. [DOI] [PubMed] [Google Scholar]
- Yun CC, Sun H, Wang D, Rusovici R, Castleberry A, Hall RA, Shim H. LPA2 receptor mediates mitogenic signals in human colon cancer cells. Am J Physiol Cell Physiol. 2005;289:C2–C11. doi: 10.1152/ajpcell.00610.2004. [DOI] [PubMed] [Google Scholar]


