Abstract
Disease resistance and sexual reproductive development are generally considered as separate biological processes, regulated by different sets of genes. Here we show that xa13, a recessive allele conferring disease resistance against bacterial blight, one of the most devastating rice diseases worldwide, plays a key role in both disease resistance and pollen development. The dominant allele, Xa13, is required for both bacterial growth and pollen development. Promoter mutations in Xa13 cause down-regulation of expression during host–pathogen interaction, resulting in the fully recessive xa13 that confers race-specific resistance. The recessive xa13 allele represents a new type of plant disease resistance.
Keywords: Bacterial blight, race-specific resistance, recessive gene, sexual development
Both disease resistance and sexual reproductive development are essential for plants to complete their life cycles. They are generally considered as separate and independent biological processes, regulated by different sets of genes (Martin et al. 2003; Ma 2005). Although >40 disease resistance (R) genes have been characterized (Martin et al. 2003), none of these genes was identified as being involved in reproductive development. It is generally accepted that the R proteins encoded by dominant R genes recognize specific pathogen effectors and trigger signal transduction cascades that lead to rapid disease resistance in the host plant (Dangl and Jones 2001; Martin et al. 2003; Belkhadir et al. 2004). However, the function of R proteins inherited recessively is not generally understood. R genes often show a constitutive expression pattern in either uninfected or infected plants, in agreement with their common role in the pathogen recognition step. In a few cases, specific additional induction of R gene expression is observed upon infection (Yoshimura et al. 1998; Piffanelli et al. 2002; Levy et al. 2004; Gu et al. 2005). These results suggest that, in most cases, the basal level of R proteins pre-existing in cells is sufficient for effective initiation of the resistance pathway while, in a few cases, more R protein is produced upon infection to help amplify the resistance response. Even with the R gene functioning as a mutation of a negative effector in disease resistance, induced expression upon infection has been observed (Buschges et al. 1997; Piffanelli et al. 2002).
Thirty R genes conferring resistance against various strains of Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial blight, have been reported in rice, of which nine were identified as recessively inherited. So far, five of the 30 R genes have been characterized as encoding four types of proteins, suggesting multiple mechanisms of R-gene-mediated Xoo resistance (Song et al. 1995; Yoshimura et al. 1998; Iyer and McCouch 2004; Sun et al. 2004; Gu et al. 2005). R gene xa13 is fully recessive, conferring resistance only in the homozygous status to Philippine Xoo race 6 (strain PXO99), which is virulent to most R genes (Khush and Angeles 1999). Here, by cloning and functional analysis of the xa13, we report that the product of the same gene plays key roles in both disease resistance and pollen development. The resistant (recessive) and susceptible (dominant) alleles of xa13 can encode identical proteins, but have crucial sequence differences in their promoter regions. Suppressing expression of either the dominant or recessive allele of xa13 enhanced the resistance, but caused male sterility of the plants, indicating that the product of this gene acted both as a bacterial growth-dependent modulator and as an essential constituent of pollen development.
Results and Discussion
We have previously mapped xa13 to a 14.8-kb fragment using three F2 populations from a cross between near-isogenic lines IRBB13 (resistant, carrying xa13) and IR24 (susceptible, carrying the dominant allele Xa13) (Chu et al. 2006). Using the same mapping populations and a newly developed molecular marker, ST12, we further mapped the xa13 locus to a DNA fragment of 9.2 kb, flanked by markers RP7 and ST12 (Fig. 1A). Analysis of the 9.2-kb sequence of BAC clone 14L03 from rice line IR64, carrying the dominant allele Xa13, revealed only one predicted gene, therefore regarded as the candidate of dominant Xa13 (GenBank accession no. DQ421395). The homologous fragment of the Xa13 candidate from IR24 (GenBank accession no. DQ421396) had an identical sequence with that of IR64. Transformation of IRBB13 with the DNA fragment encompassing the Xa13 candidate, including the promoter region isolated from IR64, produced 29 independent transformants. Sixteen of the 29 T0 transgenic plants were susceptible upon infection by PXO99, with average lesion lengths of 16.7 ± 2.95 cm as compared with 1.4 ± 0.23 cm measured for the untransformed control IRBB13 and 20.7 ± 2.74 cm measured for IR24. PCR analysis demonstrated that all of the susceptible transgenic plants had the band derived from the Xa13 candidate fragment, while none of the resistant plants amplified this band (Fig. 1B). To confirm that the susceptibility of the transgenic plants was caused by the transgene, T1 families derived from four of the susceptible T0 plants were investigated individually for resistance and the marker genotype. The susceptibility cosegregated perfectly with the transgene in all four families, indicating that the gene in this fragment was indeed Xa13 (Supplementary Table S1).
Figure 1.
Identification of xa13. (A) Fine mapping of the xa13 locus on BAC clone 14L03. The figures between markers indicate the numbers of recombination events detected between the xa13 locus and corresponding markers. (B) Association between resistance and the transgene. All of the susceptible transgenic plants have the PCR fragment of Xa13 promoter as in IR24 carrying Xa13, while the resistant plants do not have this fragment. (C) Plasma membrane location of the GFP-XA13 protein revealed by transformation of calli from Zhonghua 11. Bars, 8.0 μm. (Left) GFP-XA13 expression on plasma membrane of callus cells. (Middle) Propidium iodide staining. (Right) Overlaying of left and middle images.
Xa13 consists of five exons and encodes a protein of 307 amino acids (Supplementary Fig. S1) that is located to the plasma membrane (Fig. 1C). Analysis of the xa13 sequence from IRBB13 (GenBank accession no. DQ421394) detected three nucleotide substitutions in the coding region that caused one amino acid change, as compared with the predicted product of Xa13. To determine whether this amino acid difference is the cause of the resistance, the DNA fragments corresponding to the Xa13 genomic region from 10 additional xa13-carrying varieties, Chinsurah Boro2 (11484), Tepa 1, Chinsurah Boro2 (11760), Aus 274, AC 19-1-1, Long Grain (35023), Kalimekri 77-5, Chinsurah Boro2 (50930), Long Grain (64950), and BJ 1 (Khush and Angeles 1999), and five additional Xa13 carrying varieties, Minghui 63, Zhonghua 11, Mudanjiang 8, 93-11, and Nipponbare, were isolated and sequenced. Sequence analysis revealed that although there were various substitutions, deletions, and insertions in the Xa13 region of the latter five varieties compared with IR24 and IR64, the Xa13 alleles of all the seven varieties encode an identical protein (Supplementary Fig. S1). The additional 10 xa13 carrying varieties showed approximately the same level of resistance as IRBB13 upon PXO99 infection (Supplementary Table S2). The xa13 alleles from only eight of the varieties encode proteins with one to three amino acid differences from that of the susceptible allele Xa13, while the predicted protein sequences of the xa13 alleles from Aus 274 and Kalimekri 77-5 were identical to that of Xa13 (Supplementary Fig. S1). These results suggest that amino acid change is not the determinant of xa13-mediated resistance.
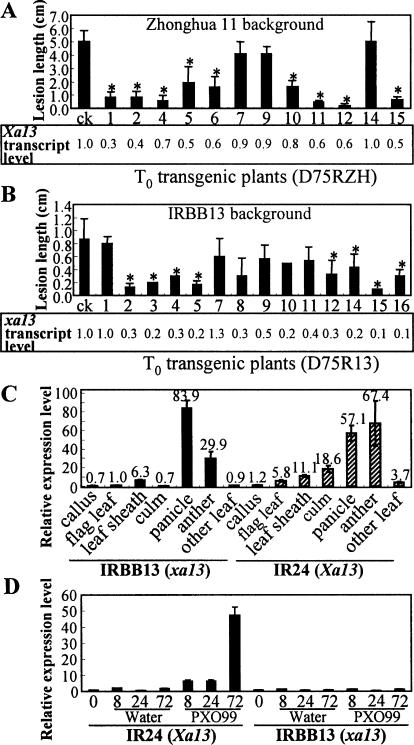
To characterize the function of xa13, we suppressed the expression of Xa13 or xa13 in different rice lines using an RNA interference (RNAi) strategy. Suppression of Xa13 expression in cultivar Zhonghua 11 significantly increased the resistance to PXO99 in nine of the 12 transgenic plants examined. The level of increased resistance was associated with the reduced accumulation of Xa13 transcripts (Fig. 2A). The correlation between lesion length and Xa13 expression level was 0.78, significant at P < 0.01 (n = 12). Similar results were also observed in Xa13-suppressed plants in the Minghui 63 genetic background (Supplementary Table S3). More interestingly, suppressing the expression of xa13 in IRBB13 further enhanced xa13-mediated resistance (Fig. 2B). The lesion length for eight of the 14 transgenic plants was significantly reduced upon PXO99 infection, and the correlation coefficient between the lesion length and the xa13 transcript level was 0.64 (P < 0.01, n = 14). These results suggest that resistance to PXO99 depends on the expression level of the gene, such that the lower the expression level of either xa13 or Xa13, the more resistant the plants become.
Figure 2.
Expression profiles of xa13 and Xa13 detected by quantitative RT–PCR and the relationship between resistance and levels of xa13- and Xa13-transcripts. Asterisks indicate significant (P < 0.05, t-test) reductions in lesion length between transgenic plants and wild-type plants (ck) measured at 14 d after PXO99 inoculation. Error bars are standard deviations. (A) Xa13-RNAi transformation of Zhonghua 11. Xa13 expression in leaves relative to the untransformed controls was detected at 3 d after inoculation. (B) xa13-RNAi transformation of IRBB13. The xa13 expression in leaves relative to the untransformed controls was detected before pathogen inoculation. (C) Expression levels of Xa13 and xa13 in different tissues, measured relative to xa13 expression in the flag leaves of IRBB13. (D) Expression levels of xa13 and Xa13 relative to the corresponding untreated (0) control in the leaves of near-isogenic lines IRBB13 and IR24, respectively, at 8, 24, and 72 h (8, 24, and 72) after PXO99 inoculation or mock (water) inoculation as the control.
To examine the expression profile of this gene, we analyzed the expression patterns of xa13 and Xa13 in the near-isogenic lines IRBB13 and IR24 by quantitative RT–PCR. The transcript levels of xa13 and Xa13 were very low in leaves but quite high in panicles and anthers (Fig. 2C). The expression of Xa13 was greatly elevated after PXO99 inoculation, but not mock inoculation (Fig. 2D). Xa13 expression in PXO99-challenged IR24 plants increased by sixfold at 8 h post-inoculation and 47-fold at 72 h post-inoculation compared with the nontreatment control. However, PXO99 inoculation did not induce the expression of xa13. We further compared the xa13- and Xa13-transcript levels in other rice lines. The Xa13-transcripts also accumulated upon PXO99 infection in rice lines Minghui 63 and IRBB4 in a similar pattern as in IR24, while the xa13-transcript level was not obviously increased by PXO99 in rice lines Tepa 1, Aus 274, AC19-1-1, Long Grain (35023), and Kalimekri 77-5 as in IRBB13 (Supplementary Fig. S2). These results further confirmed that a low expression level of this gene is the basis of xa13-mediated resistance.
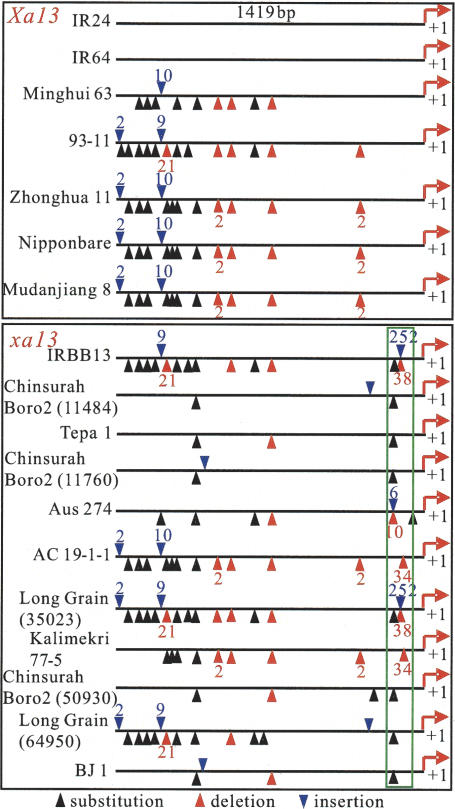
Since PXO99 infection can induce the expression of Xa13 but not xa13 in two rice lines with the same genetic background (Fig. 2D), it is highly likely that the differential expression of Xa13 and xa13 results from difference(s) in the promoter regions of the Xa13 and xa13 alleles of the gene. To examine this conjecture, we analyzed the promoter regions (∼1.4 kb) of Xa13 from seven rice lines and xa13 from 11 rice lines. The sequences of the Xa13 promoter region are identical between IR24 and IR64, but various nucleotide substitutions, deletions, and insertions were observed in the promoter regions of other rice lines, compared with IR24 and IR64 (Fig. 3). While the majority of the mutation sites are not allele specific, all of the 11 xa13-carrying lines had insertion, deletion, or substitution of 1–252 base pairs (bp) within a region of 18 bp corresponding to the −69 to −86 region of the Xa13 promoter in the seven rice lines (Fig. 3). Thus, mutations in this region abolished the induction activity of xa13 expression by pathogen infection, suggesting that this region is required for the Xa13 induction by the pathogen. This induction could be an indirect outcome of pathogen infection, or might be from direct interaction with a pathogen factor that up-regulates Xa13, as in the case of avirulence (avr) protein AvrXa27 from Xoo, which acts as a nuclear-localized regulator of rice R gene Xa27 expression (Gu et al. 2005). Related pathogen factors with features similar to transcription activators have also been identified both as elicitors and as virulence factors in host–pathogen interaction (White et al. 2000).
Figure 3.
Sequence comparison of the promoter regions of xa13 and Xa13 from different rice lines. Nucleotide substitution, deletion, or insertion are identified in different rice lines with reference to the sequence of IR24. The triangles with a figure indicate the numbers of nucleotides inserted or deleted and the triangles without a figure represent single-nucleotide insertions/deletions. The green-line-boxed regions represent the fragments relative to the −69 to −86 region of IR24.
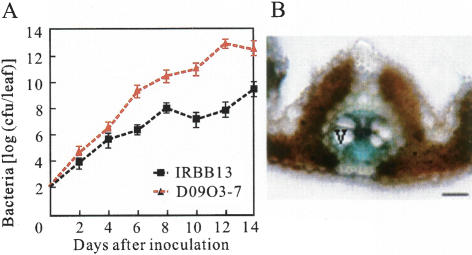
The xa13 protein has no sequence similarity with any known R proteins, but it shows 50% sequence identify and 68% sequence similarity (E value 10−52) to the product of a nodulin MtN3 gene (NCBI protein database accession no. CAA69976) that is induced by Rhizobium in legume during nodule development (Gamas et al. 1996). The differential expression of xa13 and Xa13 upon bacterial infection and the improved disease resistance achieved by suppressing xa13 or Xa13 suggest that this gene encodes a modulator required for bacterial invasion. Adequate expression of this modulator is critical for successful bacterial invasion. This inference is also supported by the result of bacterial growth analysis on wild-type IRBB13 carrying xa13 and transgenic line D09O3-7 carrying both xa13 and Xa13 (Fig. 4A). There were insignificant differences in growth rate of the bacterial strain PXO99 between the two lines in the first 4 d after inoculation, whereas the bacterial growth rate in D09O3-7 became 276-fold higher than in IRBB13 at 6 d after inoculation, and a 1847-fold difference was observed between the two lines at 14 d after inoculation (Fig. 4A). This is consistent with the accumulation rate of the Xa13 transcript as discussed above (Fig. 2D). Thus, loss of the ability to respond to PXO99 infection due to promoter mutations resulted in xa13, a gene that mediates a recessively inherited resistance. A recent study revealed that the dominant allele of barley recessive R gene mlo encodes a membrane protein functioning as an entry portal for successful pathogenesis of fungal pathogens (Bhat et al. 2005). But XA13 has no sequence similarity with MLO, thus it is unlikely to function in the same way. It is known that Xoo bacteria invade rice plants through hydathodes or wounds and live in the vascular systems (Huang and De Cleene 1989). Analysis of the promoter–GUS fusion revealed that Xa13 was preferentially expressed in the parenchyma cells surrounding the vascular vessels of leaves (Fig. 4B), suggesting that XA13 may be a bacterial growth-dependent modulator.
Figure 4.
Bacterial growth analysis and the expression pattern of Xa13 in leaves. (A) Growth of Xoo strain PXO99 in leaves of wild-type IRBB13 and T0 transgenic line D09O3-7 carrying Xa13. Bacterial population was determined from three leaves at each time point by counting colony-forming units (cfu) (Sun et al. 2004). (B) Xa13 promoter-β-glucuronidase (GUS) expression in transgenic rice plant. The blue color indicates that GUS was preferentially expressed in the parenchyma cells surrounding the vascular element (V). Bar, 30 μm.
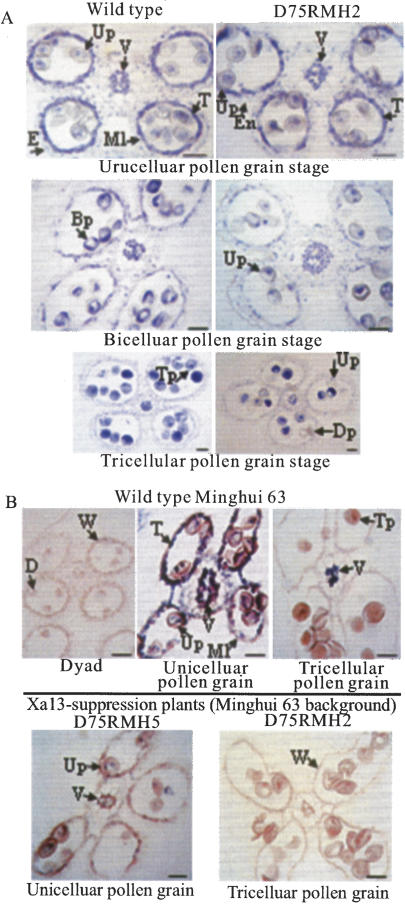
The high expression level of xa13 and Xa13 in panicles and anthers (Fig. 2C) suggests that this gene may play a role in the reproductive process. To assess the possible role of this gene in reproduction, we examined the fertility of xa13- and Xa13-suppressed transgenic plants. It was found that transgenic plants with significantly reduced expression of xa13 or Xa13 also had reduced spikelet fertility with an overall correlation of 0.59 (P < 0.01, n = 28) in the three crosses examined (Supplementary Table S3). The reduced fertility could be ascribed to male sterility as the xa13- and Xa13-suppressed plants had smaller anthers than the respective wild types and produced mostly abortive pollen (Supplementary Fig. S3). Histological analysis showed that while no abnormality was observed with the anther wall in Xa13-suppressed plants, the development of the microspores stopped at the unicellular pollen grain stage and gradually degenerated afterward (Fig. 5A). To assess whether the reduced fertility of these plants was associated with the reduced accumulation of Xa13 transcripts, we examined the expression of Xa13 in anthers by in situ hybridization. No Xa13 expression was detectable at early stages of anther development (e.g., microspore mother cell and dyad stage) (Fig. 5B). In wild-type plants, Xa13 transcripts accumulated to high levels in pollen grains, tapetal cells, and middle-layer cells of the anther wall. Moreover, it was also found at high levels in connective tissue cells and the connective vascular element at the unicellular-to-bicellular pollen grain stages and occurred only in the cells of connective vascular elements at the tricellular pollen grain stage (Fig. 5B). The hybridization signal in these tissues was very low or undetectable in the Xa13-suppressed transgenic plants (Fig. 5B). The coincidence in time between the accumulation of Xa13 transcripts in wild-type plants and the appearance of pollen abortion in Xa13-suppressed plants during pollen development suggests an indispensable role of Xa13 in pollen development.
Figure 5.
Association of Xa13 expression and pollen development. Bars, 30 μm. (Bp) Bicellular pollen grain; (D) dyad; (Dp) degenerated pollen grain; (E) epidermis; (En) endothecium; (Ml) middle layer; (Up) unicellular pollen grain; (T) tapetum; (Tp) tricellular pollen grain; (V) vascular element; (W) anther wall. (A) Pollen development at different stages in wild-type Minghui 63 and Xa13-suppressed T0 plant D75RMH2, showing that there is no apparent difference between the wild-type and transgenic plants at the unicellular pollen grain stage, whereas various abnormalities are observed at bi- and tricellular pollen grain stages. (B) Expression of Xa13 in anthers of wild-type Minghui 63 and T0 plants D75RMH2 and D75RMH5 examined by in situ hybridization. The dark blue color in the pollen grains, the cells of anther wall, and the connective vascular element indicates hybridization of Xa13 transcripts.
Thus, xa13, in addition to being a fully recessive R gene functioning by expressional loss-of-function from rice, is also an essential gene for pollen development. The dual function of this gene provides a unique opportunity for exploring the cross-talk between disease resistance and male gamete development pathways. Although there have been a few reports on the functional overlap between pathogen-induced defense signaling and plant development (Holt et al. 2002; Montoya et al. 2002; Godiard et al. 2003; Nakashita et al. 2003), none of these genes have major effects on race-specific disease resistance. The complete recessivity of the resistance conferred by xa13 indicates that Xoo resistance may only be a fortuitous by-product of this gene. Thus, mutation of the regulatory element(s) of an essential gene for pollen development results in a novel recessive gene for disease resistance in rice. Although it is not clear how these processes might be linked, the similarities in the mechanisms of pollen/pathogen recognition and development should be investigated in future studies.
Materials and methods
Characterization of resistant and susceptible alleles of xa13
A cleaved amplification polymorphism sequence marker, ST12, detected by PCR amplification using the primers ST12F (5′-AGTAGCAGTAGC CGCACACA-3′) and ST12R (5′-TGGATCGAAGGAGGAGAATG-3′) and digestion by Sau3AI, was used to screen the five recombinant F2 individuals that defined the location of xa13 locus (Chu et al. 2006) for fine mapping of this gene. A DNA fragment containing the dominant allele Xa13 and its upstream (5021 bp) and downstream (6134 bp) regions from rice line IR64 was inserted into pCAMBIA1301 vector and introduced to rice line IRBB13 with Agrobacterium-mediated transformation (Ge et al. 2006). For detecting the transgene in the transgenic plants, a pair of PCR primers, X13-seq2F and X13-seq2R (Supplementary Table S4), was designed to display the 289-bp difference between IR24 (or IR64) and IRBB13 in the promoter region of Xa13 and xa13. The coding sequence of Xa13 was determined by comparing the cDNA sequences from rice lines Minghui 63 and Nipponbare (GenBank accession nos. AK070510 and AK106127). For comparative sequencing, genomic DNA fragments containing the xa13 or Xa13 locus were isolated from different rice lines by PCR amplification using primers X13-FulF and X13-FulR (Supplementary Table S4). The subcellular localization of the XA13 protein was determined by inserting the GFP (green fluorescent protein)-Xa13 fusion gene driven by a maize ubiquitin promoter into pCAMBIA1300 vector to transform rice cultivar Zhonghua 11 by Agrobacterium-mediated transformation. To verify the plasma membrane location of the protein, onion epidermal cells were transformed with the same GFP-Xa13 fusion by particle bombardment. The bombarded cells were incubated in the dark at 22°C–24°C for 12–24 h to allow transient expression of the protein, and then treated with 0.5 M NaCl for 3–10 min to separate cell walls and protoplasts. The GFP location was viewed with a confocal microscope.
Pathogen inoculation
Three to five uppermost fully expanded leaves of each plant were inoculated with Philippine Xoo race 6 (PXO99) using the leaf-clipping method (Kauffman et al. 1973) at tillering to booting stages. Lesion length (centimeters) was measured at 14–21 d after inoculation, based on which the mean and standard deviation were calculated.
Analyses of xa13 function
To construct an RNAi vector for Xa13, a 650-bp cDNA fragment of Xa13 was amplified using primers ds1-2F and ds1-2R (Supplementary Table S4) from cDNA clone EI74B06 of rice line Minghui 63 and was inserted into the pDS1301 vector, which was constructed by inserting the cassette-producing double-strand RNA of the pMCG161 vector (McGinnis et al. 2005) into the pCAMBIA1301 vector. The expression levels of Xa13 and xa13 were determined by quantitative RT–PCR. RT–PCR was carried out in a two-step reaction (Zhou et al. 2002) using gene-specific primers (Supplementary Table S4). Quantitative PCR was performed using ABI 7500 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s protocol. The expression level of each gene in each sample was calculated relative to the expression level of the actin gene and was the average of three repetitions.
Xa13 promoter–GUS analysis
The promoter region of Xa13 from IR24 (∼1.4 kb) (Fig. 3) was fused with the GUS gene and cloned into pCAMBIA1381 vector. The vector was introduced to rice line IRBB13 with Agrobacterium-mediated transformation. GUS histochemical staining of the leaves from T0 transgenic plants was assayed as described previously (Wu et al. 2003). The stained leaves were then sectioned using a razor blade.
In situ hybridization and histological staining
Anthers were fixed in a solution containing 5% glacial acetic acid, 1.85% formaldehyde, 63% ethanol, and 30.15% water at room temperature for 24 h. After dehydration and infiltration, the samples were embedded in paraffin and cut into 10-μm-thick sections. The expression of Xa13 in anthers was assayed using in situ hybridization analysis (Drews 1998). For synthesizing the hybridization probe, a 552-bp DNA fragment was amplified from Minghui 63 cDNA clone 74B06 using Xa13-specific primers X13-QtF, and X13-Rt3R (Supplementary Table S4) and ligated into vector pGEM-T. A digoxigenin-labeled antisense RNA probe was synthesized using SP6 RNA polymerase and digoxigenin-labeled sense RNA (control) probe was synthesized using T7 RNA polymerase. Hybridization was as described previously (Drews 1998). The tissue samples were stained with Ehrlich’ haematoxylin solution before dehydration for structural analysis and with 1% eosin in 95% ethanol during dehydration for in situ hybridization.
Acknowledgments
We thank the Center for the Application of Molecular Biology to International Agriculture for transformation vectors. This work was supported by grants from the Ministry of Science and Technology of China and the National Natural Science Foundation of China.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1416306
References
- Belkhadir Y., Subramaniam R., Dangl J.L. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 2004;7:391–399. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bhat R.A., Miklis M., Schmelzer E., Schulze-Lefert P., Panstruga R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. 2005;102:3135–3140. doi: 10.1073/pnas.0500012102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschges R., Hollricher K., Panstruga R., Simons G., Wolter M., Frijters A., van Daelen R., van der Lee T., Diergaarde P., Groenendijk J., et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- Chu Z., Fu B., Yang H., Xu C., Li Z., Sanchez A., Park Y.J., Bennetzen J.L., Zhang Q., Wang S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2006;112:455–461. doi: 10.1007/s00122-005-0145-6. [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Drews G.N. In situ hybridization. Methods Mol. Biol. 1998;82:353–371. doi: 10.1385/0-89603-391-0:353. [DOI] [PubMed] [Google Scholar]
- Gamas P., Niebel F.C., Lescure N., Cullimore J.V. Use of a subtractive approach to identify new Medicago truncatula genes induced during root nodula development. Mol. Plant Microbe Interact. 1996;9:233–242. doi: 10.1094/mpmi-9-0233. [DOI] [PubMed] [Google Scholar]
- Ge X., Chu Z., Lin Y., Wang S. A tissue culture system for different germplasms of indica rice. Plant Cell Rep. 2006 doi: 10.1007/s00299-005-0100-7. (in press) [DOI] [PubMed] [Google Scholar]
- Godiard L., Sauviac L., Torii K.U., Grenon O., Mangin B., Grimsley N.H., Marco Y. ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 2003;36:353–365. doi: 10.1046/j.1365-313x.2003.01877.x. [DOI] [PubMed] [Google Scholar]
- Gu K., Yang B., Tian D., Wu L., Wang D., Sreekala C., Yang F., Chu Z., Wang G.-L., White F.F., et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- Holt B.F., Boyes D.C., Ellerstrom M., Siefers N., Wiig A., Kauffman S., Grant M.R., Dangl J.L. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell. 2002;2:807–817. doi: 10.1016/s1534-5807(02)00174-0. [DOI] [PubMed] [Google Scholar]
- Huang J.S., De Cleene M. Bacterial blight of rice. International Rice Research Institute; Manila: 1989. How rice plants are infected by Xanthomonas campestris pv. oryzae? pp. 31–42. [Google Scholar]
- Iyer A.S., McCouch S.R. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 2004;17:1348–1354. doi: 10.1094/MPMI.2004.17.12.1348. [DOI] [PubMed] [Google Scholar]
- Kauffman H.E., Reddy A.P.K., Hsieh S.P.Y., Merca S.D. An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae pv. oryzae. Plant Dis. Rep. 1973;57:537–541. [Google Scholar]
- Khush G.S., Angeles E.R. A new gene for resistance to race 6 of bacterial blight in rice, Oryza sativa L. Rice Genet. Newslet. 1999;16:92–93. [Google Scholar]
- Levy M., Edelbaum O., Sela H. Tobacco mosaic virus regulates the expression of its own resistance gene N. Plant Physiol. 2004;135:2392–2397. doi: 10.1104/pp.104.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- Martin G.B., Bogdanove A.J., Sessa G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- McGinnis K., Chandler V., Cone K., Kaeppler H., Kaeppler S., Kerschen A., Pikaard C., Richards E., Sidorenko L., Smith T., et al. Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 2005;392:1–24. doi: 10.1016/S0076-6879(04)92001-0. [DOI] [PubMed] [Google Scholar]
- Montoya T., Nomura T., Farrar K., Kaneta T., Yokota T., Bishop G.J. Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRl1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell. 2002;123:3163–3176. doi: 10.1105/tpc.006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashita H., Yasuda M., Nitta T., Asami T., Fujioka S., Arai Y., Sekimata K., Takatsuto S., Yamaguchi I., Yoshida S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003;33:887–898. doi: 10.1046/j.1365-313x.2003.01675.x. [DOI] [PubMed] [Google Scholar]
- Piffanelli P., Zhou F., Casais C., Orme J., Jarosch B., Schaffrath U., Collins N.C., Panstruga R., Schulze-Lefert P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.-Y., Wang G.-L., Chen L.-L., Kim H.-S., Pi L.-Y., Holsten T., Gardner J., Wang B., Zhai W.-X., Zhu L.-H., et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sun X., Cao Y., Yang Z., Xu C., Li X., Wang S., Zhang Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- White F.F., Yang B., Johnson L.B. Prospects for understanding avirulence gene function. Curr. Opin. Plant Biol. 2000;3:291–298. doi: 10.1016/s1369-5266(00)00082-0. [DOI] [PubMed] [Google Scholar]
- Wu C., Li X., Yuan W., Chen G., Kilian A., Li J., Xu C., Li X., Zhou D.-X., Wang S., et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35:418–427. doi: 10.1046/j.1365-313x.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Yamanouchi U., Katayose Y., Toki S., Wang Z.X., Kono I., Kuruta N., Yano M., Iwata N., Sasaki T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. 1998;95:1663–1668. doi: 10.1073/pnas.95.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Peng K., Chu Z., Wang S., Zhang Q. The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci. China C Life Sci. 2002;45:449–467. doi: 10.1360/02yc9050. [DOI] [PubMed] [Google Scholar]