Abstract
The mammalian cytoplasmic protein SirT2 is a member of the Sir2 family of NAD+-dependent protein deacetylases involved in caloric restriction-dependent life span extension. We found that SirT2 and its yeast counterpart Hst2 have a strong preference for histone H4K16Ac in their deacetylation activity in vitro and in vivo. We have pinpointed the decrease in global levels of H4K16Ac during the mammalian cell cycle to the G2/M transition that coincides with SirT2 localization on chromatin. Mouse embryonic fibroblasts (MEFs) deficient for SirT2 show higher levels of H4K16Ac in mitosis, in contrast to the normal levels exhibited by SirT1-deficient MEFs. The enzymatic conversion of H4K16Ac to its deacetylated form may be pivotal to the formation of condensed chromatin. Thus, SirT2 is a major contributor to this enzymatic conversion at the time in the cell’s life cycle when condensed chromatin must be generated anew.
Keywords: Histone deacetylation, chromatin, mitosis, SirT2, HsT2
Yeast Hst2 belongs to the Sir2 family of NAD+-dependent protein deacetylases. This family is defined by the presence of an ∼200-amino-acid sequence responsible for the NAD+-dependent deacetylase activity, also found in a wide variety of proteins from yeast to humans. Sir2 is required for the maintenance of silenced chromatin at the mating type loci, telomeres, and rDNA in yeast (Imai et al. 2000; Landry et al. 2000), and is associated with life span extension in both yeast and worms (Kennedy et al. 1997; Tissenbaum and Guarente 2001). The yeast Sir2 family includes four homologs termed Hst1–4, whose functions are still unclear, with the exception of Hst1, which regulates the expression of middle sporulation genes (Xie et al. 1999). Hst2p localizes to the cytoplasm and can affect nuclear silencing by an unknown mechanism (Perrod et al. 2001). Additionally, Hst2p is involved in caloric restriction (CR) dependent life span extension by a Sir2-independent mechanism (Lamming et al. 2005).
Among the seven Sir2 mammalian homologs (SirT1–7), SirT2 is the Hst2p counterpart and also localizes to the cytoplasm (Perrod et al. 2001). SirT2 seems to be overexpressed during mitosis, affecting mitotic exit (Dryden et al. 2003), and has been shown to deacetylate α-tubulin (North et al. 2003). However, since tubulin does not seem to be acetylated in yeast (Polevoda and Sherman 2002), the functional conservation of Hst2 (from yeast to human) is intriguing and strongly suggests another, more general function. Here we report that the general cytoplasmic localization of SirT2 is singularly exceptional during the G2/M phase when SirT2 is now present in the nucleus on chromatin. This correlates with a global decrease in the levels of acetylated H4K16 in the G2/M phase of the cell cycle. Cells deficient in SirT2 reflect this association, exhibiting increased levels as well as mislocalization of H4K16Ac and aberrancies in the integrity of their S phase.
Results and Discussion
SirT2 and Hst2p are NAD+-dependent histone deacetylases with preference for H4K16Ac in vitro and in vivo
The purification of human SirT2 rendered a homomultimer of ∼150 kDa (Supplementary Fig. 1A), which corresponds to a homotrimer as in the case for SirT1 (Vaquero et al. 2004). No stable SirT2 partner was detectable (see legend for Supplementary Fig. 1B; for specificity of the SirT2 antibodies, see Supplementary Fig. 2B,C). To determine a function of SirT2 that may explain its conservation through evolution, we took the approach of searching for common substrates. We first asked if SirT2, like Sir2, could utilize NAD+ in the presence of an acetylated protein by employing a nicotinamide exchange reaction (NER) (Supplementary Fig. 3, top panel) (Landry et al. 2000). The results showed that SirT2 was active in this reaction when acetylated bovine serum albumin (BSA) was used as a substrate but not with unacetylated BSA (Supplementary Fig. 3, left panel). Surprisingly, SirT2 was >10-fold more active than SirT1 in this NER assay (Supplementary Fig. 3, right panel), suggesting a difference in the enzymatic activities among the Sirtuins. This was not due to post-translational modifications, as SirT2 purified either from mammalian cells or after expression in Escherichia coli exhibited similar activity (data not shown).
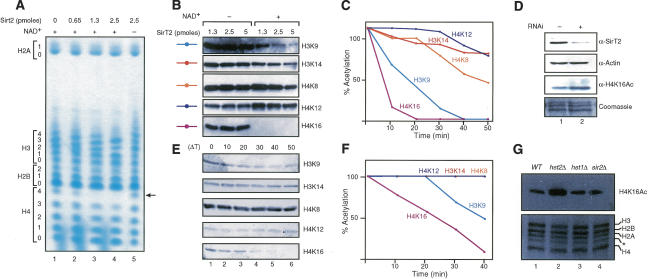
Since histones seem to be the natural substrate of Sir2 (Imai et al. 2000; Landry et al. 2000; Liou et al. 2005), we tested if SirT2 could deacetylate histones using hyperacetylated human core histones followed by their separation on TAU (triton, acetic acid, urea) gels. The results showed that SirT2 is an NAD+-dependent histone deacetylase in vitro, and deacetylates primarily one residue on histone H4 and to a lesser extent a single residue on histone H3 (Fig. 1A). Interestingly, H4 acetylation decreased by a single acetylated residue in the presence of the lowest amount of SirT2 tested (Fig. 1A, lane 2 vs. lane 1, see arrow). This was intriguing given that SirT2 is located in the cytoplasm (Perrod et al. 2001). To determine the specific residues affected by SirT2, we performed histone deacetylase complex (HDAC) assays followed by Western blots using specific antibodies against acetylated lysines. This analysis uncovered that SirT2 completely deacetylates H4K16Ac at the lowest concentration of enzyme used (Fig. 1B). The other residues affected at higher concentrations of enzyme were H3K9Ac and H4K8Ac. To assure that we were in the linear range of SirT2 activity, we performed a time course in the presence of limiting amounts of SirT2 followed by Western blots and obtained results similar to those of the previous experiment (Fig. 1C). Altogether, these experiments demonstrated that SirT2 primarily deacetylates H4K16Ac followed by H3K9Ac in vitro. Once H4K16Ac was identified as the residue preferentially deacetylated by SirT2 in vitro, we pursued the in vivo significance of this finding. RNA interference (RNAi) directed to SirT2 in 293 cells gave rise to higher levels of H4K16Ac (Fig. 1D). In contrast, H3K9Ac and H4K8Ac were not increased by the loss of SirT2, indicating that they are not valid substrates in vivo (data not shown).
Figure 1.
SirT2 and Hst2p are NAD+-dependent histone deacetylases with a preference for H4K16Ac in vitro and in vivo. (A) TAU gel analysis of hyperacetylated core histones treated with the indicated amounts of SirT2 in the presence and absence of NAD+. Levels of acetylation for each core histone are indicated. (B) Hyperacetylated core histones were incubated with the indicated amounts of SirT2 in the presence and absence of NAD+ followed by Western blot using antibodies against specific acetylated residues in H3 and H4. (C) Time-course experiment of core histone deacetylation by SirT2 in the presence of NAD+ analyzed as in B. Quantifications were determined as indicated previously. (D) RNAi experiments against SirT2 in 293 cells. Whole-cell extracts were probed for the presence of SirT2 and actin by Western blot. Histones were extracted with hydrochloric acid and tested for H4K16Ac levels by Western blot. Levels of total histones were visualized with Coomassie blue staining. (E) Western blot of a time-course experiment for deacetylation performed as in C but with purified Hst2p. (F) Graph of the quantifications of E determined as in C. (G, top) Levels of H4K16Ac and total histones purified from isogenic S. cerevisiae strains (wild type, W303-1b; hst2Δ, YRH45; sir2Δ, YRH15; hst1Δ LNYY315) analyzed by Western blot. The bottom panel shows the histones from the various mutants, stained with Coomassie. The asterisk indicates an H3 breakdown product commonly found in histones purified from yeast (Edmondson et al. 1996).
To study whether this function of SirT2 is a conserved one, we examined the histone specificity of yeast Hst2p. Similar to SirT2, purified Hst2p mediated deacetylation of core histones and exhibited a preference for H4K16Ac (Fig. 1E,F). Additionally, when we analyzed the levels of H4K16Ac in vivo we observed an increase in H4K16Ac in a yeast strain containing a deletion of hst2, relative to an isogenic wild-type strain. On the other hand, sir2Δ or hst1Δ mutants had levels of H4K16Ac comparable to wild type (Fig. 1G). This same large increase in H4K16Ac was also seen in two other hst2Δ strains (data not shown). Based on our results demonstrating increased levels of total H4K16Ac in yeast strains lacking Hst2p, we hypothesize that Hst2p functions globally while Sir2p function is more restricted.
Recently, deacetylation of H4K16Ac and the resultant production of O-acetyl-ADP-ribose were reported to be necessary for the formation of a trimeric complex between Sir3 and Sir2/Sir4 in yeast (Liou et al. 2005). Thus the presence of H4K16Ac may actually stimulate the assembly of silent chromatin by supplying the necessary substrate for the production of O-acetyl-ADP-ribose. These results may explain the puzzling observation that overexpression of Hst2p weakened telomeric silencing significantly (Perrod et al. 2001). We suggest that such overexpression reduced the amount of H4K16Ac and hence led to less O-acetyl-ADP-ribose during assembly of silent chromatin. Interestingly, recent studies revealed that Hst2p functions in life span extension (Lamming et al. 2005). This role of Hst2p may be associated with its H4K16 deacetylation activity. This hypothesis is supported by the fact that Sir2p affects life span by silencing rDNA repeats in the nucleolus (Cockell and Gasser 1999), and one of the most notable features of Sir2-silenced regions is the presence of hypoacetylated H4K16 (Braunstein et al. 1993).
H4K16Ac levels peak at S phase and drop dramatically in the G2/M transition
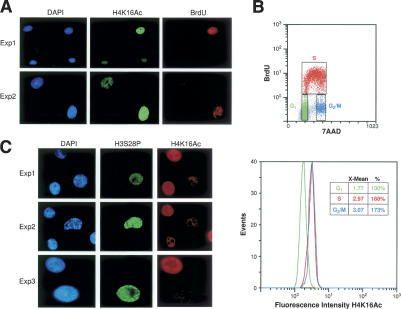
We next investigated how SirT2/Hst2 can affect H4K16Ac levels given that these are cytoplasmic proteins. To address this apparent paradox, we performed a series of experiments designed to study the dynamics of H4K16Ac levels through the mammalian cell cycle. Our previous results showed that the global levels of H4K16Ac peak around S phase and undergo a major decline between late S phase (G2) and mitosis (Rice et al. 2002). Interestingly, H4K16Ac in plants also peaks in S phase, but in this case the functional implications are clear. Acetylation of H4K5 and of H4K12 is related to histone deposition during S phase in mammals, and this process seems to include acetylation of H4K16 in plants (Belyaev et al. 1997). We first confirmed that the levels of H4K16Ac do change during cell cycle progression in vivo by immunofluorescence. Mammalian fibroblasts that exhibited positive staining with bromo-deoxyuridine (BrdU), and thus are in the replicative S phase, showed higher levels of H4K16Ac than the other cells (Fig. 2A).
Figure 2.
H4K16Ac levels peak at S phase and drop dramatically in the G2/M transition. (A) Mammalian fibroblasts were pulsed with BrdU, stained with H4K16Ac antibodies as indicated, and analyzed by immunofluorescence for colocalization of BrdU and H4K16Ac. Two different samples are shown (Exp1 and Exp2). (B) Same cells as in A were pulsed with BrdU, incubated with H4K16Ac antibody and 7AAD (DNA content marker), and analyzed by FACS for the levels of H4K16Ac at each stage of the cell cycle. (Upper panel) The distribution of the cells through the cell cycle is represented in relation to BrdU incorporation (Sasaki et al. 1986). Distribution of H4K16Ac levels in cells at each stage of the cell cycle is shown graphically and numerically, with 100% representing the levels of H4K16Ac in G1 phase. (C) Immunofluorescence as in A, but in this case with the mitotic marker H3S28P and H4K16Ac.
To accurately determine when these high levels of H4K16Ac drop, the BrdU-labeled cells were incubated with a marker for DNA content (7-amino-actinomycin D [7AAD]) and with antibodies specific for H4K16Ac. Cells were then analyzed by FACS. First, we used the 7AAD and BrdU signals to generate a horseshoe-like distribution to accurately distinguish cells in the different stages of the cell cycle (Sasaki et al. 1986): G1, S, and G2/M (Fig. 2B, upper panel). Then, we determined the levels of H4K16Ac in these three cell populations. The results show that G1 had considerably less H4K16Ac than S or G2/M (Fig. 2B, lower panel), strongly supporting the data presented above. Since the levels of H4K16Ac are the same for cells in S or G2/M, this result suggests that the global drop in H4K16Ac occurs either at the G2/M transition or in mitosis. Moreover, costaining with antibodies recognizing H3S28P and H4K16Ac showed that cells in early prophase already exhibit a drop in H4K16Ac levels (Fig. 2C). Overall, the data demonstrate that the decrease in H4K16Ac levels occurs during the G2/M transition.
SirT2 is cytoplasmic during most of the cell cycle except in the G2/M transition and in mitosis, where it is localized in association with chromatin
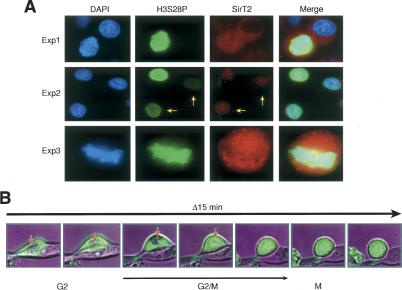
To study if the time frame of H4K16Ac decline correlates with SirT2-mediated deacetylation of H4K16Ac, we costained cells with antibodies directed against H3S28P and SirT2. The results clearly show that SirT2 is associated with chromatin exclusively during mitosis (Fig. 3A, Exp1–Exp3), and that SirT2 is chromatin associated by early prophase (Fig. 3A, Exp2, see arrows). Moreover, as previously published (Dryden et al. 2003), SirT2 does not seem to maintain its colocalization with chromatin by the time cells enter metaphase (Fig. 3A, Exp3).
Figure 3.
SirT2 is present in the cytoplasm during most of the cell cycle except in the G2/M transition and in mitosis, where it is found in the nucleus and in contact with chromatin. (A) Immunofluorescence performed as in Figure 2, showing levels of SirT2 and the mitotic marker H3S28P. Samples were derived from three independent experiments. Exp3 shows a cell in metaphase. (B) GFP-SirT2, a stable cell line derived from mammalian 293 cells, was followed during the cell cycle with a live cell microscope system for GFP localization. G2/M transition and M were identified as previously described and by timing with respect to cell division (A. Vaquero, M.B. Scher, and D. Reinberg, unpubl.; data not shown). The nucleus of the cell is indicated by an arrow during the end of G2 and beginning of G2/M transition, when the nucleus is still visually detectable.
To expand these results, we generated a GFP-SirT2 cell line to analyze GFP-SirT2 protein localization during a normal cell cycle using a live-cell microscopy system. SirT2 was found to be present exclusively in the cytoplasm during the cell cycle except around the G2/M transition, when, in <15 min, SirT2 is now also detected in the nucleus (Fig. 3B, see arrow). This result correlates with previous findings indicating that SirT2 is phosphorylated in the G2/M transition (Dryden et al. 2003), suggesting a possible mechanism by which SirT2 becomes associated with chromatin.
Loss of SirT2 produces higher levels of H4K16Ac in mitosis, a delay in S-phase entry and abnormal levels of H4K16Ac in heterochromatic foci in S phase
If our conclusions are valid and SirT2 is involved in the global drop of H4K16Ac levels during the G2/M transition, then loss of SirT2 should render mitotic cells with higher levels of H4K16Ac. This increase is expected to be specific during the mitotic phase of cells lacking SirT2. We previously showed that the loss of SirT1 gave rise to higher levels of H4K16Ac (Vaquero et al. 2004). However, our studies showed that the effect of SirT1 seemed to be, as in the case of yeast Sir2p and Hst1p, gene specific. Hence, the loss of SirT1 would not be expected to result in overall higher levels of H4K16Ac in mitotic cells.
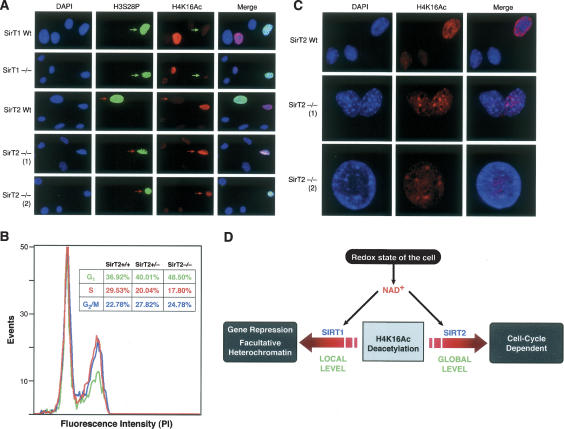
Indeed, when mouse embryonic fibroblasts (MEFs) obtained from wild-type and SirT1 or SirT2 knockout mice were stained with antibodies against H3S28P, higher levels of H4K16Ac were observed during mitosis in the case of cells deficient in SirT2 (indicated by red arrows), but not those deficient in SirT1 (indicated by green arrows), relative to wild type (Fig. 4A). The generation of the knockout SirT2 mice will be described elsewhere (H.-L. Cheng and F.W. Alt, unpubl.).
Figure 4.
Loss of SirT2 in cells derived from knockout mice produces higher levels of H4K16Ac in mitosis, a delay in S-phase entry, and abnormal levels of H4K16Ac in heterochromatic foci in S phase. (A) Immunofluorescence of primary MEFs derived from SirT1 wild-type or knockout (−/−) mice (Cheng et al. 2003) and SirT2 wild-type or knockout (−/−) mice (H.-L. Cheng and F.W. Alt, unpubl.). Cells were stained with antibodies against H4K16Ac and H3S28P. Two different independent MEFs for SirT2−/− are shown [(1) and (2)]. (B) FACS analysis of the cell cycle distribution of MEFs wild type (+/+), heterozygote (+/−), and knockout (−/−) in SirT2 stained using propidium iodide. Average values of four different experiments are shown. Proportional levels of G1, S, and G2/M are indicated in the table. (C) Immunofluorescence of SirT2 wild-type and −/− cells tested in A using H4K16Ac antibodies. Two different cell lines [(1) and (2)] were used in the experiment. (D) Model showing two different levels of regulation of H4K16Ac by SirTs: SirT1 functions at a local level in the context of genes or loci whereas SirT2 functions at a global level dependent on the G2/M transition of the cell cycle.
Other reports have indicated that overexpression of SirT2 causes a lengthening of mitosis (Dryden et al. 2003) and a shortening of G1 (Bae et al. 2004). In order to understand the impact of SirT2 loss in cell cycle progression, we studied the distribution of wild-type, heterozygote, and double-knockout SirT2 MEFs by propidium iodide staining. The FACS analysis showed that loss of SirT2 results in a clear increase in the number of cells in G1 (cf. ∼37% of wild type and 48.5% of SirT2−/−), a decrease in the S-phase population (cf. 29% of wild type and ∼18% of SirT2−/−), and a very mild increase in the levels of cells in the G2/M phase (Fig. 4B).
That the loss of SirT2 affects S-phase levels of H4K16Ac may reflect a close relationship between H4K16Ac and S phase, as appears to be the case in plants (Belyaev et al. 1997). This connection between SirT2, H4K16Ac, and S phase is supported by additional evidence. The SirT2−/− MEFs not only exhibited higher levels of H4K16Ac during S phase, but a surprisingly high proportion of these S-phase cells also exhibited H4K16Ac mislocalized to heterochromatic regions that stain strongly with DAPI (Fig. 4C). This suggests that the changes generated in chromatin during mitosis in these cells may exact a toll on their proper entry and progress through S phase.
It is important to note that loss of Hst2p does not seem to affect viability of the yeast cell (Perrod et al. 2001), and that SirT2 knockout mice seem to be normal (H.-L.Cheng and F.W. Alt, unpubl.). These data suggest that a redundancy of functions may exist. Two different lines of evidence support the existence of a redundant activity that can substitute for SirT2 loss: First, treatment of cells with the class I and II HDAC inhibitor TSA also produced an increase in H4K16Ac in mitotic cells (A. Vaquero, M.B. Scher, and D. Reinberg, unpubl.; data not shown). Second, in experiments similar to those presented in Figure 2B, SirT2−/− cells showed lower levels of H4K16Ac in G1 compared with S and G2/M (A. Vaquero, M.B. Scher, and D. Reinberg, unpubl.; data not shown), which suggests that another activity can deacetylate H4K16Ac in late mitosis or G1. However, the abnormal localization of H4K16Ac in S phase (Fig. 4C), suggests that although redundant activities may establish global levels of H4K16Ac, they are not fully compensatory for a lack of SirT2. In the absence of SirT2, regions of constitutive heterochromatin appear not to be completely deacetylated in H4K16.
In interpreting the effects due to a loss in SirT2, it is notable that although cell cycle progression in yeast requires the H4 tail (Morgan et al. 1991), mutation of its individual lysines (including K16) had no effect; only simultaneous mutation of all four lysines produced a defect in G2/M (Megee et al. 1995). Considering all the data, we propose two different levels of regulation by H4K16Ac. The first is local (gene specific) wherein SirT1 (and possibly other SirTs) regulates the levels of H4K16Ac, thereby establishing facultative heterochromatin (Vaquero et al. 2004), and the second is global and dependent on the cell cycle, with high levels of H4K16Ac in S phase and G2 dropping before cells enter mitosis. This global deacetylation of H4K16 is mediated by SirT2 (Fig. 4D).
Overall, our results suggest a new function for the Sir2-family members and explain the conservation of cytoplasmic SirT2/Hst2p through evolution. SirT2 regulates global levels of H4K16Ac in the G2/M transition, a function that does not seem to be vital for mitotic progression but instead has repercussions in S phase.
Materials and methods
Plasmids and antibodies
All constructs were generated using a standard PCR-based cloning strategy, and the integrity of individual clones was verified through DNA sequencing using a ABI Prism DNA Analyzer.
Recombinant SirT1 and SirT2 constructs were described (Vaquero et al. 2004). GFP-SirT2 was constructed by inserting SirT2 cDNA into pcDNA3.1/NT-GFP (Invitrogen). Antibodies against SirT2 used for immunofluorescence were kindly provided by Dr. Michael A. Tainsky (Karmanos Cancer Institute, Detroit, MI) (Dryden et al. 2003). Additional polyclonal antibodies against the N terminus of SirT2 used for purification were prepared in rabbits as described (Harlow and Lane 1988). Antibodies against acetylated histones were purchased from Upstate, except H4K16Ac (Serotec), H3S28P (Sigma), and H4 (Cell signaling). Antibodies against Tubulin and Actin were obtained from Sigma.
Protein expression and purification
Recombinant proteins were generated as described (Vaquero et al. 2004). Hst2 was purified as described (Landry et al. 2000). Endogenous SirT2 was purified by fractionation of S100 fractions from HeLa cells (Loyola et al. 2001) over an S400 gel filtration column in buffer C (20 mM Tris-HCl at pH 7.9, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, 0.2 mM PMSF) containing 500 mM KCl. Fractions were analyzed as described.
Flag-SirT2 was purified from a 293F cell line stably expressing Flag-SirT2 under the CMV4 promoter. An S100 fraction was adjusted to buffer C (100 mM KCl), loaded on a DE52 cellulose column, and eluted with buffer C (1 M KCl). After dialysis to buffer C (100 mM KCl), the protein was loaded onto a phosphocellulose column. The flow-through was used for Flag affinity purification. Superose 6 gel filtration analysis was performed in buffer C containing 500 mM KCl.
Immunofluorescence assays, BrdU treatment, FACs, and live cell experiments
Immunofluorescence experiments were performed as described (Vaquero et al. 2004). For BrdU incorporation, cells were incubated with BrdU at a final concentration of 10 μM in cell culture complete media for 10 min. In immunofluorescence experiments, after washing in PBS, cells were fixed in 4% paraformaldehyde for 10 min at room temperature. Membrane permeabilization was achieved by incubation in buffer A (0.1% sodium azide PBS, 0.5% Triton-X, 0.5%–1% BSA) for 10 min at room temperature. Chromatin BrdU incorporation was detected by incubation for 45 min at 37°C in antibody/nuclease solution (Amersham Biosciences) that improves preservation of antigens. For FACs analysis, after BrdU treatment, cells were washed and harvested. A single suspension of the cells was fixed on ice-cold 70% ethanol. Cell permeabilization was performed with 2 N HCl/0.5% TX-100 at room temperature for 30 min, followed by washing in borate buffer and 1× PBS/1% BSA. Cells were incubated with α-H4K16Ac for 1 h at room temperature in 1× PBS/1% BSA/0.5% Tween. After washing, anti-rabbit PE secondary antibody was added (Molecular Probes) in combination with anti-BrdU fluorescein-conjugated mouse monoclonal antibody (BD Pharmigen) for 30 min. DNA was counterstained using 7AAD fluorescent dye (BD Biosciences). Data acquisition and analysis were performed using both flow cytometer (cell counter FC500) and software (XL-MCL) from Beckman Coulter. Cell cycle studies with Propidium Iodide (Sigma) were performed as described elsewhere (Dryden et al. 2003). Live cell experiments were performed on a Zeiss Axiovert 200M configured to observe live cells. Human 293 cells expressing GFP-SirT2 were followed for 26 h during an entire cell cycle. The experiment was repeated several times to accurately determine the transition of G2/M.
RNAi experiments
RNAi for SirT2 was generated and performed as described using a lentiviral system (Chuikov et al. 2004), in conjunction with the sequence 5′-GACTCCAAGAAGGCCTACA-3′. Cells were extracted for soluble and insoluble fractions. SirT2 and actin were present in the soluble fraction prepared as described previously (Vaquero et al. 2004) and histones were extracted from the insoluble fraction by hydrochloric acid treatment (von Holt et al. 1989). These fractions were analyzed by Western blot.
Yeast cells experiment
The yeast cells described (Lamming et al. 2005) were grown in YPD to exponential growth phase. Cells were then pelleted, and spheroplasts were obtained by zymolase digestion. Histones were extracted from spheroplasts by hydrochloric acid treatment (von Holt et al. 1989) and analyzed by Western blot.
In vitro NAD+-dependent enzymatic assays
Protocols for nicotinamide exchange reactions, deacetylation assays analyzed by TAU gel or by Western analysis were as described previously (Vaquero et al. 2004).
Mass spectrometry
The specific protocol was described previously (Vaquero et al. 2004).
Acknowledgments
We thank Dr. Michael A. Tainsky for α-SirT2 antibodies. We also thank Drs. Marc Gartenberg and Lynne Vales for comments on the manuscript and members of the Reinberg laboratory for helpful discussions. We thank Kettly Cabane for technical assistance. This work was supported by grants from NIH GM64844 (D.R.) and GM28220 (R.S.), the HHMI (D.R. and F.W.A), and the Ellison Foundation (F.W.A.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1412706
References
- Bae N.S., Swanson M.J., Vassilev A., Howard B.H. Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10. J. Biochem. (Tokyo) 2004;135:695–700. doi: 10.1093/jb/mvh084. [DOI] [PubMed] [Google Scholar]
- Belyaev N.D., Houben A., Baranczewski P., Schubert I. Histone H4 acetylation in plant heterochromatin is altered during the cell cycle. Chromosoma. 1997;106:193–197. doi: 10.1007/s004120050239. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose A.B., Holmes S.G., Allis C.D., Broach J.R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Cheng H.L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J., et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Cockell M.M., Gasser S.M. The nucleolus: Nucleolar space for RENT. Curr. Biol. 1999;9:R575–R576. doi: 10.1016/s0960-9822(99)80359-5. [DOI] [PubMed] [Google Scholar]
- Dryden S.C., Nahhas F.A., Nowak J.E., Goustin A.S., Tainsky M.A. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol. Cell. Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson D.G., Smith M.M., Roth S.Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. Antibodies: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K., Gotta M., Sinclair D.A., Mills K., McNabb D.S., Murthy M., Pak S.M., Laroche T., Gasser S.M., Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- Lamming D.W., Latorre-Esteves M., Medvedik O., Wong S.N., Tsang F.A., Wang C., Lin S.J., Sinclair D.A. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton A., Tafrov S.T., Heller R.C., Stebbins J., Pillus L., Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou G.G., Tanny J.C., Kruger R.G., Walz T., Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Loyola A., LeRoy G., Wang Y.H., Reinberg D. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes & Dev. 2001;15:2837–2851. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P.C., Morgan B.A., Smith M.M. Histone H4 and the maintenance of genome integrity. Genes & Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- Morgan B.A., Mittman B.A., Smith M.M. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B.J., Marshall B.L., Borra M.T., Denu J.M., Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Perrod S., Cockell M.M., Laroche T., Renauld H., Ducrest A.L., Bonnard C., Gasser S.M. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B., Sherman F. The diversity of acetylated proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-reviews0006. reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J.C., Nishioka K., Sarma K., Steward R., Reinberg D., Allis C.D. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes & Dev. 2002;16:2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Murakami T., Ogino T., Takahashi S., Kawasaki S. Flow cytometric estimation of cell cycle parameters using a monoclonal antibody to bromodeoxyuridine. Cytometry. 1986;7:391–395. doi: 10.1002/cyto.990070415. [DOI] [PubMed] [Google Scholar]
- Tissenbaum H.A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- von Holt C., Brandt W.F., Greyling H.J., Lindsey G.G., Retief J.D., Rodrigues J.D., Schwager S., Sewell B.T. Isolation and characterization of histones. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- Xie J., Pierce M., Gailus-Durner V., Wagner M., Winter E., Vershon A.K. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]