Abstract
Nucleolar dominance describes the silencing of one parental set of ribosomal RNA (rRNA) genes in a genetic hybrid, an epigenetic phenomenon that occurs on a scale second only to X-chromosome inactivation in mammals. An RNA interference (RNAi) knockdown screen revealed that the predicted Arabidopsis histone deacetylase, HDA6, is required for rRNA gene silencing in nucleolar dominance. In vivo, derepression of silenced rRNA genes upon knockdown of HDA6 is accompanied by nucleolus organizer region (NOR) decondensation, loss of promoter cytosine methylation, and replacement of histone H3 Lys 9 (H3K9) dimethylation with H3K4 trimethylation, H3K9 acetylation, H3K14 acetylation, and histone H4 tetra-acetylation. Consistent with these in vivo results, purified HDA6 deacetylates lysines modified by histone acetyltransferases whose substrates include H3K14, H4K5, and H4K12. HDA6 localizes, in part, to the nucleolus, supporting a model whereby HDA6 erases histone acetylation as a key step in an epigenetic switch mechanism that silences rRNA genes through concerted histone and DNA modifications.
Keywords: Chromatin, epigenetic, gene silencing
Nucleolus organizer regions (NORs) are the genetic loci at which eukaryotic ribosomal RNA (rRNA) genes transcribed by RNA polymerase I are clustered in long tandem arrays, typically spanning several megabases. Nucleoli form at NORs during interphase as a direct consequence of rRNA gene transcription (Grummt 2003; Grummt and Pikaard 2003; Shaw and Doonan 2005). In genetic hybrids of plants or animals, the NORs inherited from one parent are often inactivated, an epigenetic phenomenon known as nucleolar dominance (Reeder 1985; Pikaard 2000; Viegas et al. 2002). The molecular basis for nucleolar dominance is the selective silencing of one parental set of rRNA genes via an epigenetic switch mechanism that involves both DNA methylation and histone post-translational modifications (Chen and Pikaard 1997; Lawrence et al. 2004). If cytosine methylation is inhibited using 5-aza-2′-deoxycytosine (aza-dC) or if histone deacetylation is inhibited using Trichostatin A (TSA), the silenced rRNA genes are derepressed. Importantly, treatment with either aza-dC or TSA elicits the same outcome: Coincident with switching from off to on, rRNA gene promoter cytosines become hypomethylated, promoter-associated histone H3 Lys 9 dimethylation (H3K9me2) is lost, and the euchromatic mark, H3K4me3, is acquired (Lawrence et al. 2004). The fact that inhibiting cytosine methylation induces histone modifications and that inhibiting histone deacetylation causes the loss of cytosine methylation suggests a model whereby DNA methylation and histone deacetylation are each upstream of one another in a self-reinforcing pathway (Lawrence et al. 2004).
In Arabidopsis suecica, the allotetraploid hybrid of A. thaliana and A. arenosa (Fig. 1A), the A. thaliana-derived rRNA genes are selectively silenced (Chen et al. 1998). In A. suecica we have exploited RNA interference (RNAi) to knock down predicted chromatin modifying activities and then tested for loss of nucleolar dominance. Previously, we knocked down a small Arabidopsis gene family (the HDT gene family) of nucleolar proteins related to maize HD2 and identified HDT1 as a gene important for nucleolar dominance (Lawrence et al. 2004). HDT family proteins are putative plant-specific histone deacetylases (HDACs) (Lusser et al. 1997; Dangl et al. 2001) that are unusual in that they lack sequence similarity with all other classes of HDACs, which are conserved from plants to humans (Pandey et al. 2002). However, HDT proteins display similarity to FKBP proteins (Aravind and Koonin 1998), and there are examples of FKBPs that localize to the nucleolus, are required for silencing reporter genes integrated into NORs, that act as histone chaperones for chromatin assembly, or that interact with HDACs (Arevalo-Rodriguez et al. 2000; Yang et al. 2001; Kuzuhara and Horikoshi 2004). These findings raise the possibility that HDT1 may play a role in chromatin assembly and may interact with one or more HDACs, but may not be an HDAC itself. As yet, the TSA-sensitive HDAC(s) required for nucleolar dominance remain unidentified.
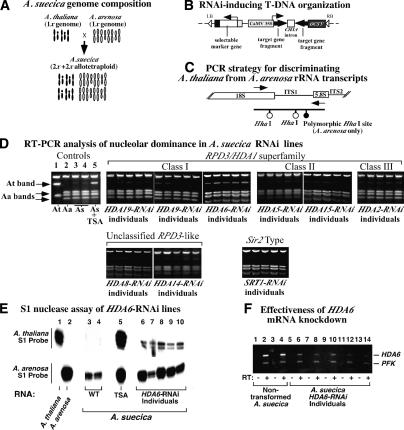
Figure 1.
HDA6 is required for rRNA gene silencing in nucleolar dominance. (A) Diagrammatic representations of A. thaliana, A. arenosa, and A. suecica chromosome compositions. (B) Organization of transferred DNAs (T-DNAs) containing RNAi-inducing transgenes. The T-DNA, delimited by left and right border sequences (LB and RB), contains a selectable herbicide-resistance gene and an inverted repeat of target gene cDNA fragments (500–700 bp), with a chalcone synthase (CHSA) intron spacer, expressed by the Cauliflower Mosaic Virus (CaMV) 35S promoter and terminated by octopine synthase (OCS) 3′ sequences. (C) rRNA gene organization surrounding internal transcribed spacer 1 (ITS1) with PCR primer (arrows) and HhaI sites indicated. (D) Screening A. suecica RNAi lines targeting predicted Rpd3-like and Sir2-like HDACs. (Lanes 1–5) Controls show HhaI-digested RT–PCR products of A. thaliana (At), A. arenosa (Aa), nontransformed A. suecica (As), or TSA-treated A. suecica. In the remaining panels, RNA from five independent RNAi lines targeting each HDAC was tested. (E) S1 nuclease protection analysis. Lanes 1 and 2 show A. thaliana and A. arenosa RNA controls, demonstrating probe specificity. In the remaining lanes, RNA of wild-type (lanes 3,4), TSA-treated (lane 5), or HDA6-RNAi lines of A. suecica were probed for A. thaliana-like or A. arenosa-like rRNA transcripts. (F) HDA6 mRNA levels are knocked down in RNAi lines. RNA from nontransformed or HDA6-RNAi lines was incubated ± RT. Resulting cDNA was amplified by PCR using primers for HDA6 and a PFK internal control.
Here, we report on a systematic knockdown screen of the non-HDT classes of predicted HDACs in Arabidopsis, including eight members of the protein superfamily related to yeast Rpd3p. We show that the Rpd3-like protein HDA6 is required for rRNA gene silencing in nucleolar dominance. Previous studies demonstrated roles for the HDA6 gene in transgene and transposable element silencing (Murfett et al. 2001; Aufsatz et al. 2002; Lippman et al. 2003; Probst et al. 2004) and in NOR condensation (Probst et al. 2004). However, no role for HDA6 in rRNA gene silencing was identified in prior studies nor has the biochemical specificity of HDA6 been defined. We show that HDA6 is a TSA-sensitive HDAC capable of removing acetyl groups from multiple lysines of multiple histones. Among the substrates of HDA6 are lysines of histones H3 and H4 whose acetylation is associated with gene activation. HDA6 is also required to maintain DNA hypermethylation at the promoters of silenced A. thaliana-derived rRNA genes. Collectively, these data identify HDA6 as a key component of the epigenetic switch mechanism that silences rRNA genes through concerted changes in histone modification and cytosine methylation.
Results
Silenced rRNA genes subjected to nucleolar dominance are derepressed in HDA6-RNAi knockdown lines
To identify specific HDACs that are responsible for rRNA gene silencing, A. suecica plants were transformed with transgenes that express double-stranded RNAs matching targeted HDACs (Fig. 1B), thereby inducing their RNAi-mediated knockdown. Because A. thaliana and A. arenosa genes typically share >90% sequence identity, orthologous mRNAs of both progenitors are depleted (Lawrence and Pikaard 2003). There are 16 predicted Arabidopsis HDACs (Pandey et al. 2002). These include the four HDT family members targeted in our previous study (Lawrence et al. 2004), 10 members of the RPD3/HDA1 superfamily, and two genes related to the yeast NAD-dependent HDAC, Sir2p. The latter HDAC is known to suppress recombination among rRNA genes and is required for the silencing of protein-coding genes integrated into NORs (Gottlieb and Esposito 1989; Smith and Boeke 1997).
A systematic knockdown screen of the expressed members of the RPD3 and SIR2-related gene families showed that HDA6 is required for nucleolar dominance (Fig. 1). As a result of a single nucleotide polymorphism that generates a HhaI site (Fig. 1C), A. thaliana and A. arenosa transcripts that are reverse transcribed into DNA, amplified by PCR (RT–PCR), and then digested with HhaI yield diagnostic banding patterns (Fig. 1D, cf. lanes 1 and 2). In wild-type A. suecica (strain LC1), transcripts of the dominant A. arenosa-derived rRNA genes are readily apparent (Aa bands) whereas the underdominant A. thaliana rRNA gene transcripts are detected in only trace amounts (At band) (Fig. 1D, lanes 3,4). However, the silenced A. thaliana rRNA genes are readily derepressed upon treatment with TSA (Fig. 1D, lane 5). Using the RT–PCR assay, we next screened for HDACs whose knockdown could mimic the effects of TSA. As shown in Figure 1D, A. thaliana rRNA transcripts were readily detected in multiple RNAi lines targeting HDA6, but were not detected above wild-type levels in RNAi lines targeting other members of the RPD3/HDA1 superfamily (HDA2, HDA5, HDA8, HDA9, HDA14, HDA15, HDA19), or in lines targeting the two SIR2-like HDACs, SRT1 and SRT2 (the latter is not shown). Northern blot and massively parallel signature sequencing (MPSS) data indicate that, with the exception of SRT2, all of the HDAC genes that were targeted are expressed at some level (see http://www.chromdb.org and http://mpss.udel.edu for more information). It is noteworthy that the derepression of A. thaliana rRNA genes in HDA6-RNAi lines was not accompanied by any discernible visible phenotypes.
Derepression of A. thaliana-derived rRNA genes in HDA6-RNAi lines was confirmed using S1 nuclease protection (Berk and Sharp 1977) with species-specific probes (Fig. 1E). In all of the HDA6-RNAi lines, derepression of underdominant A. thaliana rRNA genes is correlated with decreased HDA6 mRNA levels, as determined by RT–PCR and comparison to an internal control, phosphofructokinase (PFK) (Fig. 1F). Note that A. thaliana rRNA genes are derepressed to a greater extent by TSA than by HDA6 knockdown in the RNAi lines (see Fig. 1E). A likely explanation is that RNAi does not result in an HDA6 null phenotype, but only knocks down HDA6 mRNA levels several-fold (see Fig. 1F). However, we cannot exclude the possibility that other HDACs are partially redundant with HDA6.
HDA6 localizes, in part, to the nucleolus
To determine where HDA6 is localized in the cell, we engineered a genomic HDA6 clone, under the control of its own promoter, to express HDA6 fused to the Flag epitope. HDA6-Flag protein was then detected in transgenic plants using anti-Flag monoclonal antibody (Fig. 2A). HDA6 is not uniformly distributed throughout the nucleus; instead, there are distinct nuclear regions in which HDA6 is concentrated. The most prominent of these HDA6-positive domains is the nucleolus, which is easily visualized as the region least stained by the fluorescent DNA-binding dye, DAPI (Fig. 2A). Native HDA6 was also localized to the nucleolus using an affinity-purified rabbit polyclonal antiserum raised against an HDA6 polypeptide expressed in Escherichia coli (data not shown). Because the nucleolus is the site of rRNA gene transcription, nucleolar localization of HDA6 is consistent with HDA6 playing a role in nucleolar dominance via direct modification of rRNA genes. Likewise, HDA6 concentration at other foci in the nucleus is consistent with the role of HDA6 in silencing other sequences (Murfett et al. 2001; Aufsatz et al. 2002; Lippman et al. 2003).
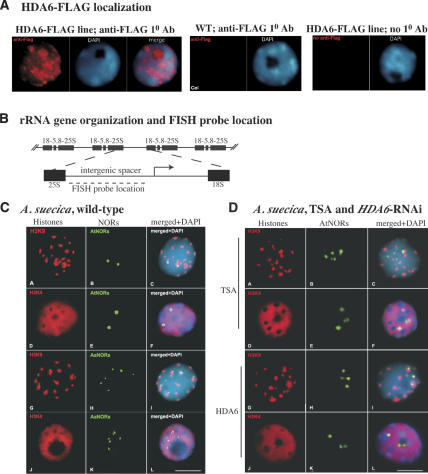
Figure 2.
HDA6 localizes to the nucleolus and is required for facultative heterochromatin formation and interphase condensation at underdominant NORs. (A) Immunolocalization of HDA6-Flag using an anti-Flag monoclonal primary antibody (red signal). The tagged protein was engineered by modifying a genomic HDA6 clone under control of its own promoter region. Nuclei were counterstained with DAPI (blue). Controls demonstrate the lack of Flag signal in a wild-type (nontransgenic) nucleus or a HDA6-Flag transgenic nucleus if the primary anti-Flag antibody is omitted and only the secondary antibody is used. (B) Organization of rRNA genes at NORs. Genes encoding 18S, 5.8S, and 25S structural rRNA precursors are separated by intergenic spacers. FISH probe and gene promoter (arrow) locations are indicated. (C, panels A–C) Immunolocalization of H3K9me2 (red signals), FISH localization of A. thaliana rRNA genes (AtNORs; green signals), and the merged image of panels A and B plus a DAPI-stained image (panel C) in meristematic root-tip cell interphase nuclei. (Panels D–F) H3K4me3 (red) and silent AtNORs. (Panels G–I) H3K9me2 (red) and dominant A. arenosa NOR (AaNOR) localization. (Panels J–L) H3K4me3 (red) and AaNORs. (D, panels A–C) H3K9me2 (red) and AtNORs (green) in interphase nuclei of meristematic root tip cells of TSA-treated plants. (Panels D–F) H3K4me3 (red) and AtNORs (green) in TSA-treated plants. (Panels G–I) H3K9me2 (red) and AtNORs (green) in an HDA6-RNAi plant. (Panels J–L) H3K4me3 (red) and AtNORs (green) in an HDA6-RNAi plant.
RNAi-mediated knockdown of HDA6 induces a heterochromatin-to-euchromatin transition at underdominant NORs
The two silenced A. thaliana-derived NORs and the six dominant A. arenosa NORs can be discriminated in A. suecica nuclei using fluorescence in situ hybridization (FISH) with species-specific probes (see Fig. 2B for probe location; Pontes et al. 2003). Sequential immunolocalization of H3K9me2, an epigenetic mark of heterochromatin (Richards and Elgin 2002), followed by FISH shows that the two silenced A. thaliana NORs colocalize with two prominent H3K9me2 foci in A. suecica interphase nuclei (Fig. 2C, panels A–C). Other major H3K9me2 foci correspond primarily to pericentromeric heterochromatin. Immunolocalization of H3K4me3, a mark of transcriptionally active euchromatin (Richards and Elgin 2002), yields a distinctly different pattern, namely a diffuse nuclear distribution that is punctuated by dark holes of condensed heterochromatin (Fig. 2C, panels D,J). Strikingly, two of these dark holes correspond to the two silenced A. thaliana NORs (Fig. 2C, panels D–F). Taken together, these results show that A. thaliana-derived NORs in A. suecica are highly condensed and associated with histones displaying the heterochromatin marker H3K9me2 but not the euchromatic marker H3K4me3. In contrast, the dominant A. arenosa-derived NORs in A. suecica colocalize with both H3K9me2 (Fig. 2C, panels G–I) and H3K4me3 (Fig. 2C, panels J–L), consistent with the interpretation that the dominant A. arenosa-derived NORs include subdomains consisting of active (H3K4me3-associated) or silent (H3K9me2-associated) genes (Lawrence et al. 2004).
Whereas the two A. thaliana NORs are highly condensed in wild-type A. suecica nuclei, yielding two large FISH signals (see Fig. 2C, panels B,E), three to five diffuse, lobed A. thaliana NOR FISH signals are observed in both TSA-treated plants and HDA6-RNAi lines (Fig. 2D, panels B,E,H,K). To interpret these observations, it is important to note that the NOR FISH signals we detect correspond to the portions of the NORs that are composed of condensed, inactive rRNA genes assembled in facultative heterochromatin. Whereas a silenced NOR is uniformly condensed to yield a single, large FISH signal, an active NOR includes both heterochromatic regions and decondensed, active regions (Pontes et al. 2003). The chromatin fibers consisting of active genes are too thin and dispersed to be detected using our methods. However, their existence can be inferred. In Figure 2, the transition from two inactive A. thaliana NORs, observed as two large FISH signals, in wild-type plants to three to five FISH signals, upon TSA treatment or HDA6 knockdown, indicates that internal decondensation has occurred within the NORs. Decondensed euchromatic regions flanked by blocks of condensed heterochromatin accounts for the increased number of FISH signals. Note also that the FISH signals themselves are qualitatively different in nuclei of TSA-treated and HDA6-RNAi plants, appearing less compact and more diffuse, presumably due to numerous local decondensations within the NORs.
In addition to decondensing, A. thaliana rRNA genes in nuclei of TSA-treated plants or HDA6-RNAi lines colocalize primarily with H3K4me3-enriched euchromatin (Fig. 2D, panels F,L) rather than with H3K9me2-enriched chromocenters (Fig. 2D, panels C,I) as is the case in untreated, nontransgenic plants (cf. Fig. 2C, panel C). Collectively, the data of Figures 1 and 2 suggest that HDA6 localizes within the nucleolus and is required to maintain the repression of the underdominant A. thaliana-derived rRNA genes in A. suecica. The chromosomal manifestation of this repression is the organization of the NORs into highly condensed facultative heterochromatin.
HDA6 is a broad-specificity HDAC
To explore the biochemical activity of HDA6, we affinity-purified Flag-tagged HDA6 from transgenic A. thaliana (Fig. 3A). The prominent polypeptide migrating on a SDS-PAGE gel (Fig. 3A, arrow) at the predicted size of ∼55 kD, and absent in nontransgenic (wild-type) plants, was confirmed to be HDA6 by tryptic fingerprinting and MALDI-TOF mass spectrometry, yielding eight matching peptides representing ∼40% of the HDA6 sequence (data not shown). We then used recombinant histone acetyltransferases (HATs) to acetylate purified Brassica oleracea (broccoli) core histones (which are identical between Arabidopsis and Brassica for histones H3 and H4), or histone N-terminal peptides (N-peptides), and asked if Flag-HDA6 could remove the acetyl groups. Three full-length His-tagged A. thaliana HATs, HAG1, HAG2, and HAG5 (Pandey et al. 2002), were expressed in E. coli and affinity-purified (Fig. 3B). Upon incubation with Brassica histones and 3H-acetyl CoA, we found that recombinant HAG2 and HAG5 specifically label histone H4 whereas recombinant HAG1 preferentially acetylates histone H3 and histone H2A and/or H2B (Fig. 3C). Purified Flag-HDA6 deacetylates the histones labeled by the three HATs whereas proteins of wild-type plants eluted from the anti-Flag resin (see Fig. 3A, left lane) lack HDAC activity (Fig. 3D). The three Arabidopsis HATs and HDA6 also displayed activity with histone N-peptides (Figs. 3E, 4; data not shown). For instance, HAG5 acetylates and HDA6 subsequently deacetylates a histone H4 N-peptide (Fig. 3E).
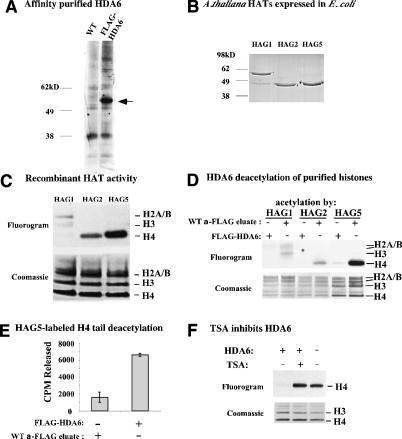
Figure 3.
Purified HDA6 has HDAC activity. (A) Affinity purification of Flag-HDA6 expressed in transgenic plants. Extracts of wild-type or Flag-HDA6-overexpressing A. thaliana was incubated with anti-Flag resin. A Coomassie blue-stained SDS-PAGE gel of proteins eluted using excess Flag peptide is shown. (B) Coomassie-stained SDS-PAGE gel of His-tagged recombinant Arabidopsis HATs HAG1, HAG2, and HAG5 after purification on nickel-agarose. (C) HAT activity of HAG1, HAG2, and HAG5. Broccoli histones were labeled using 3H-acetyl CoA and the resulting SDS-PAGE gel was Coomassie-stained and subsequently subjected to fluorography. Histone H3 and H4 bands were definitively identified using mass spectrometry. H2A and H2B variants with overlapping migration patterns precluded definitive assignment of H2A and H2B bands. (D) HDA6 deacetylates full-length histones acetylated by HAG1, HAG2, and HAG5. Histones labeled by the HATs were incubated with equal aliquots of Flag-HDA6 or wild-type protein eluted from anti-Flag resin. The fluorogram and Coomassie-stained histone bands are shown. (E) HDA6 deacetylates histone N-peptides. Anti-Flag resin incubated with extracts of wild-type or Flag-HDA6-expressing plants was washed extensively then incubated with HAG5-labeled H4 peptide immobilized on agarose beads. 3H (from 3H-acetyl CoA) released into the reaction buffer was measured by scintillation counting. (F) HDA6 is a TSA-sensitive HDAC. HAG5-labeled broccoli histones were incubated with HDA6 ± TSA and then subjected to SDS-PAGE and fluorography.
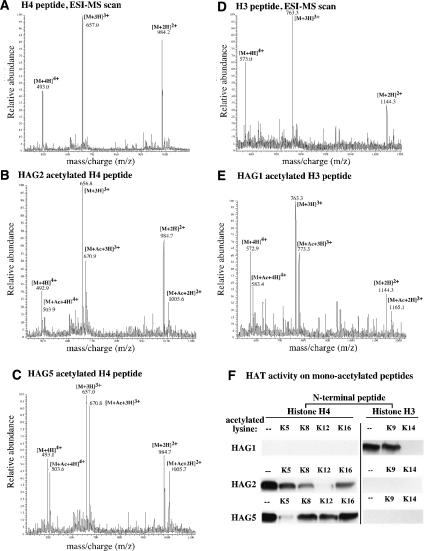
Figure 4.
HAG1, HAG2, and HAG5 monoacetylate histone H3 and H4 N-peptides. (A) Mass spectrum of unmodified H4 peptide (M) differentially protonated (H) to generate 2+, 3+, and 4+ charge states. (B,C) Mass spectra of H4 peptide acetylated by HAG2 or HAG5. Unmodified and monoacetylated peptides (M + Ac) were detected in the 2+, 3+, and 4+ charge states. (D) Mass spectrum of unmodified histone H3 peptide. (E) Mass spectrum of the HAG1-acetylated histone H3 peptide. (F) Determination of HAG1, HAG2, and HAG5 specificities. N-peptides bearing pre-existing, nonradioactive acetyl groups on individual lysines were incubated with HAG1, HAG2, or HAG5 and 3H-acetyl CoA, then subjected to SDS-PAGE and fluorography.
TSA blocks the ability of HDA6 to deacetylate full-length histone H4 (Fig. 3F) or H4 N-peptide (data not shown) acetylated by HAG5. The similar consequences of TSA treatment or HDA6 knockdown on NOR condensation and rRNA gene silencing (see Figs. 1, 2), together with these biochemical data, suggest that HDA6 can account for the effects of TSA on nucleolar dominance.
Determination of histone H3 and H4 acetyl-lysine substrates for HDA6
Determining the specificity of HATs whose acetylation can be removed by HDA6 provides a means for deducing the specificity of HDA6. We employed mass spectrometry as well as an independent biochemical assay to identify specific lysines of histone H3 or H4 N-peptides that are acetylated by HAG1, HAG2, or HAG5 in vitro (Fig. 4). In the first approach, acetylated N-peptides were subjected to electrospray ionization and quadrupole ion trap mass spectrometry. Ionized peptides appearing as [M + 2H]2+, [M + 3H]3+, and [M + 4H]4+ ions were detected in each of their ESI mass spectra (Fig. 4A–E). HAG2 and HAG5, which label full-length histone H4 (see Fig. 3C,D), each catalyze the monoacetylation of the H4 N-peptide (Fig. 4, cf. B,C and A). Likewise, HAG1, which can label full-length histone H3 (see Fig. 3), also catalyzes the monoacetylation of the H3 N-peptide (Fig. 4, cf. D and E).
To identify the amino acids monoacetylated by HAG1, HAG2, or HAG5, ionized acetylated species resolved in the initial ESI-MS scan (MS1) were selected and fragmented to produce daughter ions. Peaks in resulting product-ion spectra (MS/MS experiment) were further fragmented to yield higher-order product-ion spectra (MS3 experiments). This approach revealed the sites of acetylation and indicated that HAG2 acetylates histone H4 on Lys 12 (H4K12), HAG5 acetylates H4K5, and HAG1 acetylates H3K14 (data not shown). These specificities were also inferred from an independent assay in which we asked if pre-existing acetylation of a specific lysine would preclude HAT-catalyzed labeling of the peptide with 3H-acetyl CoA (Fig. 4F). HAG1 efficiently labeled a nonacetylated H3 peptide or an H3 peptide that had a pre-existing acetyl group on K9 (Fig. 4F, top row). However, prior acetylation of H3K14 prevented HAG1-catalyzed labeling with a radioactive acetyl group, consistent with H3K14 being the substrate for HAG1. Likewise, prior acetylation of H4K12 and H4K5 inhibited the labeling of the histone H4 peptide by HAG2 and HAG5, respectively, consistent with the HAG2 and HAG5 specificities that were determined by mass spectrometry. Note that HAG1 does not acetylate the H4 peptide, and HAG2 and HAG5 do not acetylate the H3 peptide, consistent with the specificities of the HATs on full-length histones. Based on the specificities of HAG1, HAG2, and HAG5 and the fact that HDA6 can remove the acetyl groups added to histones by these HATs, we deduce that the deacetylase activity of HDA6 includes Lys 14 of histone H3 and Lys 5 and Lys 12 of histone H4. Moreover, HDA6 deacetylates histone H2A and/or H2B at undefined amino acids that are acetylated by HAG1 (see Fig. 3D). Collectively, these data indicate that HDA6 is a broad-specificity deacetylase capable of erasing acetylation from multiple lysines of multiple histones.
In vivo effects of HDA6 knockdown on histone modifications and DNA methylation
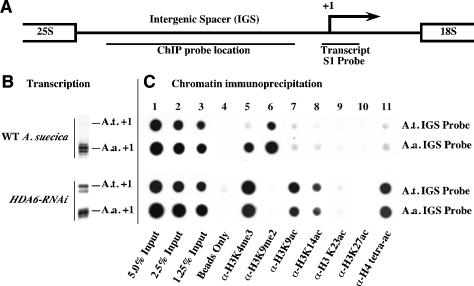
The in vivo consequences of HDA6 knockdown on histone modifications at rRNA gene promoters in A. suecica were tested using chromatin immunoprecipitation (ChIP) with antibodies specific for H3K4me3, H3K9me2, H3K9ac, H3K14ac, H3K23Ac, H3K27ac, or H4ac4 (H4 tetra-acetylated on Lys 5, Lys 8, Lys 12, and Lys 16). Following immunoprecipitation, purified DNA was dot-blotted, and resulting filters were hybridized to species-specific intergenic spacer (IGS) probes (see Fig. 5A).
Figure 5.
Transcriptional derepression of rRNA genes in A. suecica HDA6-RNAi lines is accompanied by changes in rRNA gene histone methylation and histone acetylation. (A) Diagram of an rRNA gene intergenic spacer highlighting the transcription initiation site (+1; arrow) and S1 and ChIP probe locations. (B) Pre-rRNA transcripts detected using S1 nuclease protection with A. thaliana- or A. arenosa-specific probes. (C) ChIP dot-blot analysis. Duplicate samples of wild-type or HDA6-RNAi plant chromatin, blotted in adjacent rows, were hybridized to A. thaliana- or A. arenosa-specific probes (see diagram). (Columns 1–3) Five percent, 2.5%, or 1.25% of the input chromatin in the ChIP reactions. (Column 4) Protein A beads in the absence of antibodies. (Columns 5–11) Chromatin immunoprecipitated with the indicated antibodies.
In wild-type A. suecica, in which A. arenosa rRNA genes are expressed and A. thaliana rRNA genes are silenced (Fig. 5B), the A. thaliana genes are preferentially associated with H3K9me2-modified histones (Fig. 5C, column 6, top row), in agreement with cytological observations (see Fig. 2). In contrast, the dominant A. arenosa rRNA genes are associated with both H3K4me3 and H3K9me2 (Fig. 5C, columns 5 and 6, second row), also in agreement with the cytological evidence (see Fig. 2) and consistent with the interpretation that the dominant set of genes includes both active and inactive gene subsets. HDA6 knockdown alters the chromatin modification profiles and eliminates the differences between the A. arenosa and A. thaliana genes with respect to transcription (Figs. 1, 5B) and chromatin modification (Fig. 5C, lower two rows). In HDA6-RNAi lines, decreased association of both parental sets of rRNA genes with H3K9me2 is accompanied by a corresponding increase in H3K9 acetylation and H3K4 trimethylation (Fig. 5C, columns 5–7, lower two rows). Notable increases in H3K14 acetylation and H4 tetra-acetylation also occur in HDA6-RNAi plants (Fig. 5C, columns 8 and 11), consistent with our biochemical evidence that HDA6 deacetylates H3K14, H4K5, and H4K12. These latter H3 and H4 acetylation marks are detected at much lower levels in wild-type A. suecica, suggesting a dramatic change in steady-state histone acetylation/deacetylation activity in HDA6-RNAi lines.
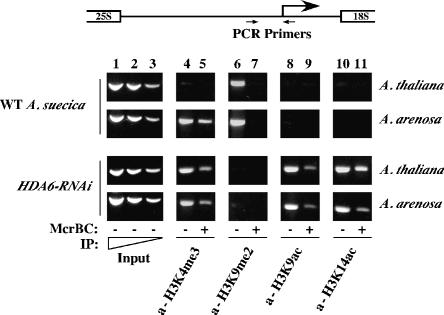
rRNA gene promoter cytosines become demethylated in HDA6-RNAi lines
To examine simultaneously the histone modification and cytosine methylation status of rRNA gene promoters, we employed ChIP–chop PCR (Fig. 6; Lawrence et al. 2004). Whereas hypomethylated DNA templates survive McrBC treatment, hypermethylated DNA is “chopped” by McrBC and cannot be amplified by PCR. In wild-type A. suecica, silenced A. thaliana rRNA gene promoters are immunoprecipitated by H3K9me2 antibodies (Fig. 6, column 6) and have hypermethylated promoters such that McrBC digestion precludes subsequent PCR amplification (Fig. 6, column 7). In contrast, the A. arenosa rRNA gene promoters associate with both H3K9me2 and H3K4me3 (Fig. 6, columns 4–7). Those A. arenosa rRNA gene promoters associated with H3K4me3 are hypomethylated and resistant to McrBC (Fig. 6, cf. columns 4 and 5) but A. arenosa rRNA gene promoters associated with H3K9me2 are hypermethylated and digested by McrBC (Fig. 6, cf. columns 6 and 7). In HDA6-RNAi lines, neither A. thaliana nor A. arenosa rRNA gene promoters associate with H3K9me2 (Fig. 6, lower two rows, columns 6 and 7). Instead, the promoters are found in association with H3K4me3, K3K9ac, and H3K14ac and are resistant to McrBC digestion (Fig. 6, columns 4–5 and 8–11), indicating that cytosine hypomethylation is linked to the occurrence of these specific, euchromatic histone modifications at active rRNA gene promoters.
Figure 6.
HDA6 is required for cytosine hypermethylation at silenced promoters. ChIP–chop PCR was used to evaluate cytosine methylation density within A. thaliana- and A. arenosa-derived promoters in wild-type and HDA6-RNAi lines. Ten percent of the immunoprecipitated chromatin dot-blotted in Figure 5 was incubated with (lanes 5,7,9,11) or without (lanes 4,6,8,10) McrBC, then PCR was used to amplify A. thaliana or A. arenosa rRNA gene promoter regions. Hypermethylated DNA digested by McrBC is not amplified. (Lanes 1–3) Input controls used 0.1%, 0.05%, or 0.025% of the chromatin subjected to ChIP and show that the assay is semiquantitative.
Discussion
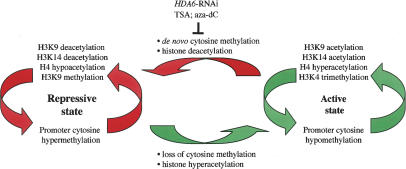
Based primarily on the observation that the DNA methylation inhibitor, aza-dC, and the HDAC inhibitor TSA both cause rRNA gene derepression, promoter demethylation, loss of promoter-associated H3K9me2, and acquisition of the euchromatic mark H3K4me3, we previously proposed a model whereby DNA methylation and histone deacetylation are each upstream of one another in a circular, self-reinforcing pathway (Lawrence et al. 2004). Our evidence that HDA6 is a TSA-sensitive HDAC whose knockdown phenocopies the effects of TSA to cause rRNA gene derepression, NOR decondensation, and loss of promoter cytosine methylation fits the predictions of this model. Moreover, our latest data indicate that acetylated H3K9, acetylated H3K14, and tetra-acetylated H4 are histone modifications associated with active rRNA gene promoters whose cytosines are hypomethylated, further extending the model (Fig. 7).
Figure 7.
A model for the epigenetic control of rRNA gene on/off states. ChIP–chop PCR data indicate that active and silenced rRNA genes are marked by distinctive DNA and histone modifications that are mutually reinforcing. The model predicts de novo cytosine methylation and histone deacetylation as key events in switching from the transcriptionally permissive to the repressive state. Likewise, loss of promoter cytosine methylation and histone hyperacetylation are likely key events in switching from the silent to the active state.
In vitro, purified HDA6 deacetylates full-length histones or histone tail peptides that are acetylated on H3K14, H4K5, and H4K12 by three Arabidopsis HATs whose specificities were previously undefined—namely, HAG1, HAG2, and HAG5. Collectively, the evidence suggests that HDA6 is a broad-specificity HDAC. The in vitro HDAC specificity of HDA6, the in vivo hyperacetylation of histones at corresponding lysines in HDA6-RNAi lines, and the nucleolar localization of HDA6 are all consistent with the hypothesis that HDA6 directly deacetylates rRNA gene promoter histones to bring about rRNA gene silencing.
It is noteworthy that HDA6 is abundant inside the nucleolus, where the decondensed, active subset of rRNA genes is transcribed, and is not appreciably colocalized with the highly condensed, heterochromatic portions of the NORs located outside the nucleolus. We interpret this as evidence that HDA6 plays a role in switching rRNA genes from an active state to the inactive state, which would explain the presence of HDA6 inside the nucleolus where the active genes are located. In contrast, if HDA6 only played a role in maintaining the silent state, we would expect HDA6 to colocalize with the condensed, inactive portions of the NORs peripheral to the nucleolus. HDA6 may play similar roles at transgenes and transposable elements that can switch between active and inactive states, which is a defining characteristic of facultative heterochromatin, thereby driving the steady-state balance toward gene silencing.
It is interesting that HDA6 knockdown derepresses silenced rRNA genes subjected to nucleolar dominance in A. suecica whereas hda6 mutations had no discernible effect on rRNA gene transcription in A. thaliana (Probst et al. 2004). One trivial explanation may be that with ∼1500 essentially identical rRNA genes in diploid A. thaliana (Copenhaver and Pikaard 1996), an estimated one-third to one-half of which are active (Lawrence et al. 2004), increased rRNA gene transcription is undetectable against the backdrop of pre-existing active genes. In contrast, sequence differences among the A. thaliana and A. arenosa-derived rRNA genes in A. suecica, combined with the nearly complete silencing of the former, makes it easy to detect the derepression of underdominant genes in the hybrid. Alternatively, the number of active rRNA genes may change in A. thaliana hda6 mutants without changing the overall level of transcription. In Saccharomyces cerevisae, the number of active rRNA genes can change more than twofold without changing steady-state rRNA transcript levels owing to compensatory changes in the average number of RNA polymerases engaged in transcription on each gene (French et al. 2003). The latter explanation is attractive given that NOR decondensation, increased H3K4 methylation, and subtle changes in DNA methylation at uncharacterized locations within the rDNA were observed in A. thaliana hda6 mutants (Probst et al. 2004), consistent with our findings using A. suecica HDA6-RNAi lines.
The lack of an effect on nucleolar dominance upon knocking down putative HDAC genes related to yeast SIR2 is noteworthy given that the role of Sir2p in rRNA gene regulation is controversial. Although Sir2p is needed to represss protein coding genes integrated into rRNA genes, Sir2p is tethered to the genes via Net1p, which stimulates RNA polymerase I transcription (Shou et al. 2001). Moreover, silencing of protein-coding genes, which are transcribed by RNA polymerase II, depends on transcription of adjacent rRNA genes by RNA polymerase I (Buck et al. 2002; Cioci et al. 2003). Therefore, it is not clear that Sir2p represses yeast rRNA gene transcription nor is there evidence that Arabidopsis SRT proteins play roles in rRNA gene silencing in nucleolar dominance.
Thus far, we have identified two genes that are required for nucleolar dominance in A. suecica: HDT1 and HDA6. The proteins encoded by both of these genes localize, at least in part, to the nucleolus, consistent with their roles in rRNA gene silencing. As mentioned previously, HDT1 is a putative plant-specific HDAC related to maize HD2 (Lusser et al. 1997). However, HDT1 that is expressed in bacteria or as an epitope-tagged recominant protein in transgenic plants is unable to deacetylate histones using methodologies similar to those employed for HDA6 (K. Earley and C.S. Pikaard, unpubl.). Perhaps we have not yet supplied HDT1 with the proper acetylated histone substrates in our assays. However, it is intriguing that HDT1 lacks similarity with other known classes of HDACs but shares limited sequence similarity with FKBP proteins (Aravind and Koonin 1998; Dangl et al. 2001). FKBP proteins have been shown to interact with HDACs, including yeast Rpd3p (Arevalo-Rodriguez et al. 2000; Yang et al. 2001), an HDAC that regulates the accessibility of rRNA genes to the DNA cross-linking agent psoralen (Sandmeier et al. 2002). HDA6 is a member of the Rpd3-like protein family in Arabidopsis. Furthermore, fission yeast FKBP localizes to the nucleolus, is required for silencing protein-coding genes integrated into the NOR, and acts as a histone chaperone during chromatin assembly in vitro (Kuzuhara and Horikoshi 2004). Thus far, we have been unable to detect a direct interaction between HDT1 and HDA6 in vitro. However, the similar HDT1 and HDA6 knockdown phenotypes with respect to rRNA gene derepression, promoter cytosine methylation, histone modifications, and NOR condensation (Lawrence et al. 2004; this study) implicate both proteins in the same rRNA gene repression pathway. We suspect that functionally conserved FKBP–HDAC partnerships may play important roles in chromatin-mediated regulation of rRNA genes in plants, yeast, and possibly other eukaryotes.
Materials and methods
Plant materials
A. suecica (strain LC1) plants for ChIP experiments were grown on sterile MS medium (Sigma-Aldrich) ± 4 μM TSA (Sigma-Aldrich) for 18 d, 22°C, 24-h photoperiod. Greenhouse-grown plants were used for cytology. Agrobacterium-mediated genetic transformation of A. suecica or A. thaliana was by the floral dip method (Clough and Bent 1998; Lawrence et al. 2004). RNAi vectors are described at http://www.chromdb.org. Genomic A. thaliana HDA6 was PCR-amplified using Platinum Pfx (Invitrogen) polymerase and primers 5′-CACCCTTTTACTACTT TACTCTCAAGTCAACC-3′ and 5′-AGACGATGGAGGATT CACG-3′ and cloned into pENTR-D TOPO (Invitrogen). The resulting HDA6 insert was recombined into pEarleyGate 302 (Earley et al. 2006) to add a C-terminal Flag tag (amino acids DYKDDDDK) for immunolocalization studies. Construction of a Flag-HDA6 cDNA overexpressed from the CaMV 35S promoter was described previously (Earley et al. 2006).
RNA isolation and transcription analysis
S1 nuclease protection using 5′-end-labeled probes was as described previously (Lawrence et al. 2004). Briefly, probes and RNA were hybridized overnight at 50°C and resulting probe–RNA hybrids were digested with 750 U/mL S1 nuclease (Invitrogen) at 50°C for 45 min. Resulting digestion products were resolved on a sequencing gel, dried onto filter paper, and exposed to X-ray film.
Immunological techniques
ChIP and ChIP–chop PCR analyses were as described previously (Lawrence et al. 2004). Briefly, nuclei of seedlings cross-linked in 1% formaldehyde were sonicated, and soluble chromatin was subjected to immunoprecipitation using anti-histone antibodies specific for H3K9me2, H3K4me3, H3K9ac, H3K14ac, or tetra-acetylated H4. Antibodies were purchased from Upstate Cell Signaling solutions, except anti-H3K4me3, purchased from Abcam (Abcam Ltd.). Chromatin–antibody complexes were captured on protein A-agarose beads, washed, then eluted with 1% SDS, 0.1 M NaHCO3. DNA–protein cross-links were reversed at 65°C overnight. Purified DNA was dot-blotted onto Genescreen Plus membranes (Perkin-Elmer) and hybridized to labeled probes. For ChIP–chop PCR, 10% of the immunoprecipitated DNA was digested with 10 U of McrBC (New England Biolabs) and 10% of the digestion reaction (equivalent to 1.0% of the total immunoprecipitated material) was PCR-amplified (28 cycles) using primers flanking an ∼400-base-pair (bp) region that includes the rRNA gene promoter. All ChIP and ChIP–chop PCR experiments were repeated in at least three independent experiments.
Immunolocalization was performed in paraformaldehyde-fixed root tip cells according to Houben et al. (Houben et al. 1996). For histone immunolocalization experiments, primary antibodies were diluted 1:2000 in PBS, 1% BSA and detected using Cy3-conjugated secondary antibodies, as described previously (Lawrence et al. 2004). DNA was counterstained with DAPI (4′,6′-diamidino-2-phenylindole hydrochloride) in CITIFLUOR antifade buffer (AF1; Agar Scientific). Epifluorescence microscopy images were recorded using a Zeiss AxioCam digital camera. FISH was performed using A. thaliana- or A. arenosa-specific intergenic spacer probes labeled with digoxygenin-dUTP, as described previously (Pontes et al. 2003). For detection of Flag-HDA6, overnight incubation with mouse monoclonal anti-Flag primary antibody (Sigma-Aldrich) diluted 1:400 in PBS and 0.5% blocking reagent (Roche) was followed by washing in PBS and incubation with anti-mouse Rhodamine (1:100; Sigma-Aldrich) secondary antibody. Nuclei were counterstained with 2 μg/mL DAPI (Sigma-Aldrich) in Vectashield (Vector Laboratories). Nuclei were examined using a Nikon Eclipse E800i epifluorescence microscope and images collected using a Photometrics Coolsnap ES Mono digital camera. Images were pseudocolored, merged, and processed using Adobe Photoshop (Adobe Systems).
B. oleracea histone isolation
One-hundred grams of broccoli inflorescence were homogenized in a Waring blender at high speed (three pulses of 15 sec) at 4°C in 200 mL of 20 mM HEPES-KOH (pH 7.4), 10 mM MgCl2, 0.44 M sucrose, 1.25% Ficoll (type 400), 2.5% Dextran T40, 0.5% Triton X-100, and 0.5 mM DTT, to which protease inhibitors were added just before use (1.0 mM PMSF, 2.5 μg/uL antipain, 0.5 μg/mL bestatin, 0.5 μg/mL leupeptin, 4.0 μg/mL pepstatin A). The homogenate was filtered through two layers of Miracloth (Calbiochem) and subjected to centrifugation at 5000 × g, 4°C, 15 min. Pellets were resuspended in 50 mL of nuclear lysis buffer (50 mM sodium phophate buffer at pH 6.8, 0.6 M NaCl, 1.0 mM DTT, 1.0 mM PMSF). Ten grams of dry Bio-Gel HTP resin (Bio-Rad) were added. The resulting slurry was rocked for 1 h at 4°C, then poured into a column and washed with 10 column volumes of nuclear lysis buffer. Bound histones were eluted in 50 mM sodium phophate buffer (pH 6.8), 2.0 M NaCl, 1 mM DTT, and 1 mM PMSF, and peak protein fractions were pooled, sealed in dialysis tubing (5 kD cutoff), and concentrated ∼20-fold to a final concentration of ∼1–2 mg/mL by immersion in solid sucrose granules. Concentrated samples were dialyzed against 1 L of 10 mM Tris-HCl (pH 8.0), 10% glycerol, 0.1 mM EDTA, 1.0 mM DTT, and 1.0 mM PMSF. Aliquots frozen in liquid nitrogen were stored at −80°C.
HAT expression, purification, and activity assays
GNAT/MYST superfamily HATs HAG1 (gene locus identifier At3g54610), HAG2 (At5g56740), and HAG5 (At5g09740) name designations were according to Pandey et al. (2002). HAG5 has recently been redesignated HAM2 at the chromatin database (http://www.chromdb.org) to reflect the fact that it is a member of the MYST family. ORFs of HAG1, HAG2, and HAG5/HAM2 full-length cDNAs were amplified by PCR using Platinum Pfx (Invitrogen) polymerase and primers HAG1 F (forward) (5′-ATG GACTCTCACTCTTCCC-3′) and HAG1 R (reverse) (5′-CTAT TGAGATTTAGCACCAGATTGG-3′), HAG2 F (5′-CACCAT GGTTCAGAAGCAGCAAGC-3′) and HAG2 R (5′-TCAGA CTTTTAGCGTTTGACC-3′), and HAG5 F (5′-CACCATGG GATCGTCAGCGAATAC-3′) and HAG5 R (5′-TTAACTCTG GTCCTTGTAAGGTG-3′). PCR products were cloned into pENTR-D TOPO (Invitrogen), and inserts were then recombined into pDEST17 (Invitrogen). For protein overexpression, pDEST17–HAT constructs in E. coli BL21-AI cells were inoculated into 100 mL LB medium at 37°C. When cultures reached an A600 of 0.5–1.0, protein expression was induced by addition of 0.2% (w/v) L-arabinose and incubation, with shaking, for 12–16 h at 16°C. Cells collected by centrifugation at 5000 × g, 15 min, 4°C, were resuspended in 10 mL of 10 mM Tris (pH 7.6), 500 mM NaCl, 5 mM imidazole, sonicated (four pulses, 30 sec at medium power on ice) to lyse the cells, and centrifuged at 12,000 × g, 15 min, 4°C. Soluble His-tagged HATs were purified by nickel-agarose chromatography resin (Novagen). Affinity-purified HATs, eluted in 10 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 250 mM imidazole, were frozen in liquid nitrogen and stored at −80°C. HAT assays were conducted in 200-μL reactions containing 200 ng of purified HAT enzyme, 20 μg of broccoli histones, and 1 μCi of 3H-acetyl CoA (200 mCi/mmol; Perkin-Elmer Life Science) in HAT reaction buffer (10 mM Tris-HCl at pH 8.0, 10% glycerol, 0.1 mM EDTA, 1.0 mM DTT, 1.0 mM PMSF). After 1 h at 30°C, reactions were subjected to SDS-PAGE on 4%–12% Tris-Glycine gels (Invitrogen). Gels were soaked in En3hance (Perkin-Elmer Life Science), vacuum dried onto filter paper, and exposed to Biomax MS film (Kodak). Large-scale, 10-mL histone-labeling reactions for HDAC assays included 200 μg of purified HAT, 2 mg of broccoli histones, and 10 μCi 3H-acetyl CoA in HAT reaction buffer. Labeled histones (10 mL) were then loaded onto a 500-μL Biorex 70 column, washed with 20 column volumes of 50 mM Tris-HCl (pH 7.5), 2 M NaCl, 10% glycerol, 0.1 mM EDTA, 1 mM DTT, and 1.0 mM PMSF, and eluted with high-salt buffer (50 mM Tris-HCl at pH 7.5, 2 M NaCl, 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 1.0 mM PMSF). Peak fractions were concentrated against solid sucrose to ∼1 mg/mL, dialyzed against 1 L of HDA6 reaction buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 10% glycerol), and stored at −80°C. Biotinylated histone peptides (Upstate) were labeled in 200-μL reactions containing 200 ng of purified HAT enzyme, 20 μg of peptide, and 1 μCi of 3H-acetyl CoA (200 mCi/mmol; Perkin-Elmer Life Science) for 1 h at 30°C. Peptides were subjected to SDS-PAGE on 4%–12% Tris-Glycine gels (Invitrogen) and exposed to film or captured on streptavidin beads (Upstate) for HDAC assays.
HDAC expression, purification, and assay
Aboveground tissues of 2- to 4-wk-old A. thaliana plants expressing Flag-HDA6 were harvested and ground to a powder in liquid nitrogen. The powder was resuspended in 2 vol (w/v) of Cell Lytic P (Sigma-Aldrich), amended with plant-specific protease inhibitor cocktail (Sigma-Aldrich) diluted 1:100 (v/v) and 1 mM PMSF. Homogenates were filtered through two layers of Miracloth (Calbiochem) and subjected to centrifugation at 6000 × g, 15 min, 4°C. The supernatant was incubated with anti-Flag-conjugated agarose (Sigma-Aldrich) for 1 h at 4°C. The resin was then washed three times with 10 vol of HDAC buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 10% glycerol) and eluted with HDAC buffer containing Flag peptide (200 μg/mL). Purified HDA6 was then incubated 3 h at 30°C with ∼1 μg of 3H-acetyl-labeled biotinylated peptide captured on streptavidin agarose in 200-μL reactions. Beads were collected by centrifugation, and CPM released into the soluble fraction was determined by scintillation counting. For HDAC assays using B. oleracea-labeled histones, Flag-HDA6 was incubated in 100-μL reactions including HDAC reaction buffer and 20 μg of labeled histones for 3 h at 30°C. Reactions were subjected to electrophoresis on 4%–12% gradient Tris-Glycine SDS–polyacrylamide gels (Invitrogen), and gels were incubated with En3hance (Perkin-Elmer Life Science) prior to exposure to Biomax MS film (Kodak).
Mass spectrometry
Histone H3 and H4 N-peptides (Upstate Cell Signaling Solutions) were acetylated in 100-μL reactions containing 4 μg of peptide, 800 ng of HAT enzyme (HAG1, HAG2, or HAG5), and 10 μM acetyl CoA in HAT reaction buffer, 30°C, 45 min. Trifluoroacetic acid (TFA) was then added to a final concentration of 0.1% (v/v), and peptides were purified by reverse-phase chromatography using ZipTipC18 (Millipore) columns. Peptides eluted in 10 μL of 50% (v/v) acetonitrile were diluted with 100 μL of 50% (v/v) acetonitrile, 0.1% (v/v) formic acid. Samples were then infused into a Thermo Finnigan LCQ classic, LC quadropole ion trap mass spectrometer at a flow rate of 5 μL/min and subjected to electrospray ionization. Resulting mass spectra and product-ion mass spectra taken in the MS/MS (MS2) and MS3 modes were acquired in sequence. Acquired spectra were summed from 20 independent scans. Only peaks displaying at least 3:1 signal:noise were considered. Peaks whose m/z agreed within 0.1 mass units of theoretical ions predicted by using the Protein Prospector database (http://prospector.ucsf.edu) were considered matches. Some peaks had several potential matches.
Acknowledgments
Data for Figures 1, 5, and 6 were generated by Rick Lawrence; cytology was by Olga Pontes and Manuela Silva; data for Figures 3 and 4F were generated by Keith Earley; Rachel Reuther did the mass spectrometry. Nuno Neves, Wanda Viegas, Michael Gross, and Craig Pikaard supervised various aspects of the work. We thank Douglas Chalker (Washington University) for use of his microscope. Olga Pontes was supported by a fellowship (SFRH/BPD/17508/2004) from the Fundação para a Ciência e Tecnologia (Portugal). Pikaard laboratory work was supported primarily by National Institutes of Health grant R01-GM60380. RNAi vector development and HAT specificity experiments were supported by the National Science Foundation under grants 9975930 and 0421619, respectively. Mass spectrometry was supported by NIH grant P41-RR00854. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or National Institutes of Health. Research in the Viegas laboratory was supported by the Fundação para a Ciência e Tecnologia (project POCTI/BCI/38557/2001 to N.N.) and Fundo Comunitário Europeu FEDER.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1417706
REFERENCES
- Aravind L., Koonin E.V. Second family of histone deacetylases. Science. 1998;280:1167a. [Google Scholar]
- Arevalo-Rodriguez M., Cardenas M.E., Wu X., Hanes S.D., Heitman J. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 2000;19:3739–3749. doi: 10.1093/emboj/19.14.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W., Mette M.F., Van Der Winden J., Matzke M., Matzke A.J. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A.J., Sharp P.A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Buck S.W., Sandmeier J.J., Smith J.S. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell. 2002;111:1003–1014. doi: 10.1016/s0092-8674(02)01193-5. [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Pikaard C.S. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes & Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Comai L., Pikaard C.S. Gene dosage and stochastic effects determine the severity and direction of uniparental rRNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. 1998;95:14891–14896. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci F., Vu L., Eliason K., Oakes M., Siddiqi I.N., Nomura M. Silencing in yeast rDNA chromatin: Reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell. 2003;12:135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Copenhaver G.P., Pikaard C.S. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J. 1996;9:273–282. doi: 10.1046/j.1365-313x.1996.09020273.x. [DOI] [PubMed] [Google Scholar]
- Dangl M., Brosch G., Haas H., Loidl P., Lusser A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta. 2001;213:280–285. doi: 10.1007/s004250000506. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- French S.L., Osheim Y.N., Cioci F., Nomura M., Beyer A.L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R.E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Grummt I. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes & Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- Grummt I., Pikaard C.S. Epigenetic mechanisms controlling RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Houben A., Belyaev N.D., Turner B.M., Schubert I. Differential immunostaining of plant chromosomes by antibodies recognizing acetylated histone H4 variants. Chromosome Res. 1996;4:191–194. doi: 10.1007/BF02254958. [DOI] [PubMed] [Google Scholar]
- Kuzuhara T., Horikoshi M. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 2004;11:275–283. doi: 10.1038/nsmb733. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Pikaard C.S. Transgene-induced RNA interference: A strategy for overcoming gene redundancy in polyploids to generate loss-of-function mutations. Plant J. 2003;36:114–121. doi: 10.1046/j.1365-313x.2003.01857.x. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A., Brosch G., Loidl A., Haas H., Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- Murfett J., Wang X.J., Hagen G., Guilfoyle T.J. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13:1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Muller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C.S. The epigenetics of nucleolar dominance. Trends Genet. 2000;16:495–500. doi: 10.1016/s0168-9525(00)02113-2. [DOI] [PubMed] [Google Scholar]
- Pontes O., Lawrence R.J., Neves N., Silva M., Lee J.H., Chen Z.J., Viegas W., Pikaard C.S. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. 2003;100:11418–11423. doi: 10.1073/pnas.1932522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R.H. Mechanisms of nucleolar dominance in animals and plants. J. Cell Biol. 1985;101:2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J., Elgin S.C. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Sandmeier J.J., French S., Osheim Y., Cheung W.L., Gallo C.M., Beyer A.L., Smith J.S. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 2002;21:4959–4968. doi: 10.1093/emboj/cdf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P., Doonan J. The nucleolus. Playing by different rules? Cell Cycle. 2005;4:102–105. doi: 10.4161/cc.4.1.1467. [DOI] [PubMed] [Google Scholar]
- Shou W., Sakamoto K.M., Keener J., Morimoto K.W., Traverso E.E., Azzam R., Hoppe G.J., Feldman R.M., DeModena J., Moazed D., et al. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell. 2001;8:45–55. doi: 10.1016/s1097-2765(01)00291-x. [DOI] [PubMed] [Google Scholar]
- Smith J.S., Boeke J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Viegas W., Neves N., Caperta A., Silva M., Morais-Cecílio L. Nucleolar dominance: A ‘David and Goliath’ chromatin imprinting process. Curr. Genomics. 2002;3:563–576. [Google Scholar]
- Yang W.M., Yao Y.L., Seto E. The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. EMBO J. 2001;20:4814–4825. doi: 10.1093/emboj/20.17.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]