Abstract
Translation regulation is a critical means by which cells control growth, division, and apoptosis. To gain further insight into translation and related processes, we performed multifaceted mass spectrometry-based proteomic screens of yeast ribosomal complexes and discovered an association of 77 uncharacterized yeast proteins with ribosomes. Immunoblotting revealed an EDTA-dependent cosedimentation with ribosomes in sucrose gradients for 11 candidate translation-machinery-associated (TMA) proteins. Tandem affinity purification linked one candidate, LSM12, to the RNA processing proteins PBP1 and PBP4. A second candidate, TMA46, interacted with RBG1, a GTPase that interacts with ribosomes. By adapting translation assays to high-throughput screening methods, we showed that null yeast strains harboring deletions for several of the TMA genes had alterations in protein synthesis rates (TMA7 and TMA19), susceptibility to drugs that inhibit translation (TMA7), translation fidelity (TMA20), and polyribosome profiles (TMA7, TMA19, and TMA20). TMA20 has significant sequence homology with the oncogene MCT-1. Expression of human MCT-1 in the Δtma20 yeast mutant complemented translation-related defects, strongly implying that MCT-1 functions in translation-related processes. Together these findings implicate the TMA proteins and, potentially, their human homologs, in translation related processes.
Keywords: Mass spectrometry, proteomics, ribosome, Saccharomyces cerevisiae, translation
Protein translation is an essential cellular activity. In eukaryotic cells, it is an important mechanism for controlling gene expression. Regulation of gene expression at the level of translation allows for a rapid response to environmental stimuli, without the need for new transcription and nuclear export of mRNAs. The importance of translational control in eukaryotic gene expression is becoming more apparent, with the discovery that translational control plays a role in oncogenesis, viral infection, synaptic plasticity, and fragile X syndrome (for reviews, see Pe’ery and Mathews 2000; Dua et al. 2001; Calkoven et al. 2002; Schneider and Mohr 2003; Holland et al. 2004; Klann and Dever 2004; Rajasekhar and Holland 2004; Vanderklish and Edelman 2005).
The translation of mRNA into polypeptides is catalyzed by the ribosome, a large riboprotein complex comprised of two major subunits: a smaller 40S subunit and a larger 60S subunit. In eukaryotes, a large number of initiation factors are required in a multistep process to form the 80S initiation complex containing both ribosomal subunits plus the methionyl initiator tRNA base-paired with the AUG start codon of the mRNA (for reviews, see Kozak 1999; Hinnebusch 2000). After initiation, the 80S complex matches successive codons with their respective aminoacylated-tRNAs as the polypeptide chain elongates. Each new amino acid forms a peptide bond with the previously recruited amino acid. Elongation continues until a stop codon signals release of the polypeptide chain and dissociation of the ribosome subunits, thereby terminating translation.
The network of proteins regulating translation is complex, and many details remain to be discovered. For example, ∼10% of characterized yeast genes have been found to play roles in protein synthesis (Costanzo et al. 2000). Because two-thirds of the yeast ORFs have not been functionally characterized (Costanzo et al. 2000), a significant subset of these novel ORFs will probably have roles in mRNA translation. Historically, the majority of translation factors were purified using biochemical methods including sucrose gradients and ribosome salt washes and identified using Edman sequencing (Grossman and Moldave 1979). While largely successful, these methods would not be expected to identify proteins with subtle roles in translation or proteins of low abundance.
Additional factors were later identified in Saccharomyces cerevisiae using genetic suppressor screens, which are not dependent on a protein’s cellular abundance (Donahue 2000). Suppressor screens were particularly effective in dissecting components of the amino acid starvation response pathway and in elucidating mechanisms involved in translation start site selection (for review, see Donahue 2000; Hinnebusch 2000). Because proteins with subtle phenotypes may still be missed with genetic screens, it is impossible to rule out that all translation factors have been identified.
Numerous studies in the past several years have used mass spectrometry to discover new components of protein complexes (Pandey and Mann 2000). In one mass spectrometry approach termed Direct Analysis of Large Protein Complexes (DALPC), multidimensional microcapillary liquid chromatography and tandem mass spectrometry are coupled with genome-assisted data analysis to directly identify the composition of purified protein complexes (Link et al. 1999). DALPC allows the identification of proteins at the femtomole level and bypasses the detection, resolution, and extraction problems associated with conventional SDS-PAGE or 2D-electrophoresis protocols (Link et al. 1999). As such, the use of this sensitive technology allows the identification of translation factors that were not detected in gel-based studies. This was demonstrated with the discovery of a novel 40S ribosomal subunit, ASC1 (Link et al. 1999; Gerbasi et al. 2004). Similar approaches have been very successful in identifying components of preribosomal complexes and the RNA-processing machinery (Granneman and Baserga 2003, 2004; Milkereit et al. 2003; Takahashi et al. 2003).
The identification and characterization of all translation factors are critical for obtaining a better understanding of the biochemical mechanisms regulating protein synthesis. To address this issue, we have purified translation complexes using a variety of conventional approaches and applied state-of-the-art mass spectrometry to identify novel ribosome-associated factors. We then adapted established biochemical assays to high-throughput analysis to test the novel proteins identified in our proteomic screens for translation defects. Lastly, we identified a human homolog to one novel factor that can complement translation defects in yeast.
Results
Purification of ribosomal complexes from S. cerevisiae
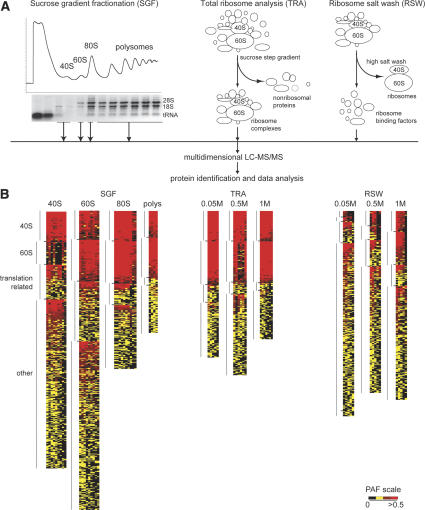
We carried out a multifaceted approach to identify components associated with the translation machinery using large-scale proteomic screens. Our initial goal was to define a comprehensive list of putative translation-machinery-associated (TMA) proteins, ordered by their relative abundance in the ribosome purifications. We combined several purification strategies to minimize the bias of any one approach and to increase the ability to detect true ribosome-interacting proteins (Fig. 1A). First, 40S, 60S, 80S, and polyribosomal complexes were fractioned using sucrose gradients (Link et al. 2005). Second, ribosomes were purified under increasing salt concentrations using discontinuous sucrose gradients. Third, ribosome salt washes (RSW) with three different salt concentrations were used to dissociate potential regulatory factors from core ribosomes (Link et al. 2005).
Figure 1.
Cluster analysis of ribosome fractions. (A) Purification schemes used to purify ribosomes and ribosome-associated proteins. (B) Clustering of proteins identified in ribosome purifications. Heat map clustering proteins by their PAF within functional categories (40S, 60S, translation related). The most abundant proteins are bright red and cluster at the top, yellow is intermediate, and black indicates absence of a particular protein. Each column represents an independent purification, and each row represents an individual protein.
To identify purified proteins, each sample was digested with trypsin and analyzed by DALPC tandem mass spectrometry (Link et al. 1999, 2005). Relative protein abundances in each experiment were expressed as the total number of nonredundant tandem mass spectra that correlated significantly to each ORF normalized to the molecular weight of the cognate protein (×104) (Powell et al. 2004). We call this value a protein abundance factor (PAF) (see Supplemental Material). Proteins were clustered by their average PAF across the replicate experiments (Lewis et al. 2004; Powell et al. 2004). The results are displayed in heat maps using a range of colors to show patterns of enrichment. The most abundant proteins are shown as red, intermediate are yellow, and undetected proteins are black. Since controls for identifying nonspecific proteins interacting with the translation machinery were not feasible, this clustering method was used to identify proteins that showed a significant enrichment with ribosomes. Only proteins identified in at least 30% of the replicates are depicted in the heat maps.
As points of reference, known ribosomal proteins and ribosome-associated proteins were sorted into eight functional classes before clustering: 40S, 60S, translation initiation, elongation, and release factors, translation-related, ribosome biogenesis, and mitochondrial translation proteins. Proteins in these categories are known to or are predicted to copurify with ribosomes, and therefore can be used to validate our methods. Although placement of proteins into these categories was somewhat arbitrary, assignment was largely based upon information obtained from the Saccharomyces Genome Database (SGD) and Gene Ontology (GO) (Ashburner et al. 2000; Hong et al. 2005).
For the sucrose gradient fractionation (SGF) experiments, a minimum of nine independent purifications of 40S, 60S, and 80S and five for polysomes were analyzed (Fig. 1B). For the 40S, 60S, and 80S clusters, the data were further filtered to include only proteins that were identified in three or more experiments. For the polysome cluster, the cutoff was two or more experiments. The clustering clearly showed enrichment of ribosomal and translation-related proteins in the various purifications (Fig. 1B; Supplementary Tables S1–S4). All 33 components of the small ribosomal subunit were identified in the 40S purification. Similarly, we were able to identify 43 of 46 components of the large subunit in the 60S purification. Overall, the 60S purification was enriched in RPLs relative to the 40S and vice versa. There was some overlap because the sucrose gradient peaks partially overlapped and likely contained comigrating preribosomal complexes. As expected, the 80S and polysome purifications contained most of the 40S and 60S proteins. More translation factors and translation related proteins copurified with the 40S and 60S as compared with the 80S and polysomes, consistent with the large number of proteins required for translation initiation in contrast to elongation.
For the total ribosome analysis (TRA), proteins displayed in the cluster were identified in two or more purifications (at least five purifications for each salt concentration). Similar numbers of ribosomal components were identified here as compared with the SGF (Fig. 1B; Supplementary Tables S5–S7). At 0.05 M ammonium chloride, 78 of 79 ribosomal proteins were identified. RPL41 was the only exception. However, RPL41 is a 3.3-kDa protein that produces tryptic peptides two amino acids long, and therefore cannot generate fragmentation data that significantly match a peptide. Only two additional subunits (RPL29 and RPL37) were not identified when the salt concentration was increased to 1 M, demonstrating the stability of the ribosome. For each salt concentration, a subset of known translation-related proteins was also identified.
For the ribosome salt washes (RSW), ribosomes were pelleted by centrifugation, and the supernatants (RSWs) were saved for analysis. Proteins identified in two or more of at least five replicates were included in the cluster (Fig. 1B; Supplementary Tables S8–S10). Consistent with the ribosome stability observed in the TRA, we detected only a limited number of ribosomal proteins released by the 0.05 M salt wash. The number of released ribosomal proteins increased with higher salt concentrations, but the PAFs were lower as compared with those in the TRA, indicating that even the most stringent salt wash was not entirely effective at dissociating ribosomal proteins. Together, the RSWs contained a large subset of known ribosome-associated proteins, with PAFs similar to those seen in the 40S and 60S SGF experiments. Interestingly, the 0.05 M salt washes contained similar numbers of characterized translation-related proteins as compared with the 1 M salt wash, although they were relatively more abundant in the 1 M wash. This suggests that translation factors are not tightly associated or are transiently associated with the ribosomes.
Overall, ribosomal proteins were among the most highly abundant proteins in the purifications. Importantly, we also identified 23 of 28 canonical translation initiation factors. All of the elongation factors and one of two release factors were also identified. Interestingly, translation factors had a more variable relative abundance with lower PAFs in general than the ribosomal proteins, suggesting that only a subset of ribosomes is associated with translation factors. These combined analyses using PAFs clearly indicated that both ribosomes and known translation-related proteins were substantially represented in our purifications.
Identification of novel proteins that copurify with ribosomal complexes
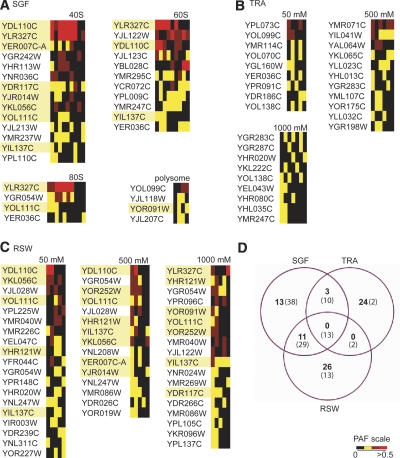
We next used PAFs to identify additional proteins enriched in our purifications. We chose to focus our efforts on uncharacterized ORFs. We applied the same filtering criteria to each data set and generated a list of expressed ORFs found in each purification approach (Fig. 2A–C). The majority of ORFs (76%) were identified in just a single purification scheme (Fig. 2D). Approximately 23% were identified in two types of purifications, and <1% were identified in all three purification strategies (Fig. 2D). The distribution of the ORFs was similar to that for the known translation proteins, but with more ORFs uniquely identified in the TRA and RSW and fewer identified in all three methods (Fig. 2D). Together the distribution data suggested that all three purification methods contributed to the identification of both known and novel translation factors.
Figure 2.
Identification of novel TMA proteins. Cluster analysis showing the relative abundances (PAFs) of uncharacterized ORFs identified in sucrose gradient fractions (A), total ribosome analysis (B), or ribosome salt washes (C). Proteins highlighted in yellow were selected for further analysis. (D) Venn diagram showing the numbers of ORFs (bold) or known translation factors (parentheses) identified in the three purification schemes.
To see whether the ORFs we identified were biased to a particular molecular weight or pI range, we plotted the values for each ORF and compared the distribution to that of the entire yeast proteome (Supplementary Fig. S1). The ORF distribution for both molecular weight and pI mirrored that of the entire proteome, confirming that our screens were not biased in this manner.
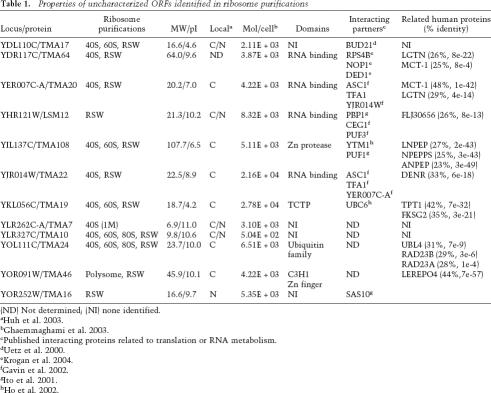
Of the novel ORFs identified in these screens, we chose 12 for further evaluation (Fig. 2, highlighted proteins; Table 1). The candidates represented different purification profiles, and therefore might represent proteins with different functions. Ten of the candidates were identified in the SGF. Among these 10 proteins, all but one were also identified in the RSWs. Because ∼10% of known translation-related proteins were identified solely by RSW purification (Fig. 2D), we also chose two ORFs from this category. Lastly, because we chose the candidates as the screens progressed, we did not always select the most enriched proteins in Figure 2. Screening of the remaining proteins is currently under way.
Table 1.
Properties of uncharacterized ORFs identified in ribosome purifications
(ND) Not determined; (NI) none identified.
cPublished interacting proteins related to translation or RNA metabolism.
Cosedimentation of expressed ORFs with ribosomal complexes is EDTA-sensitive
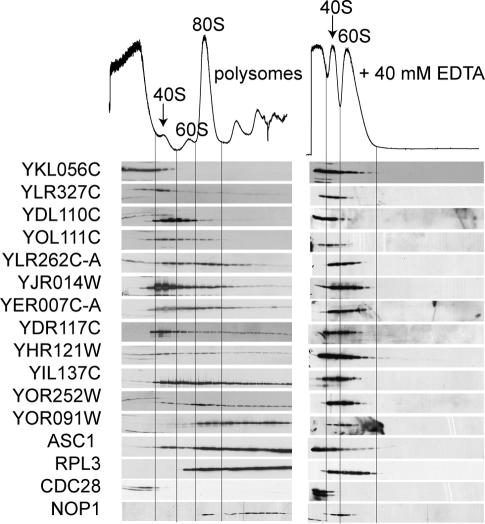
To confirm the mass spectrometry data identifying novel proteins comigrating with ribosomes in sucrose gradients, polysome profiles were generated for each strain expressing the corresponding ORF as a tandem affinity purification (TAP) fusion (Ghaemmaghami et al. 2003). Location of the ribosomes within the gradients was determined by measuring the OD254 and by Western blotting for RPL3 and ASC1 (Fig. 3). Anti-PSTAIR (CDC28) marked the nonribosomal fractions, and anti-NOP1 showed the migration of the small subunit processosome (Fig. 3). Sucrose gradient fractions from each TAP strain were immunoblotted using anti-TAP antibody. The sedimentation profiles essentially mirrored the purification results obtained in the initial mass spectrometry identification (Fig. 3). For example, nine of the 12 proteins we tested were identified in the 40S fractions by mass spectrometry. All but TAP-YKL056C were also detected in the 40S fractions by Western blotting. Although the distribution of the TAP fusion throughout the gradient was broader than detected for the native protein by mass spectrometry, the immunoblotting provided further evidence that the candidates cosedimented with ribosomes. In continuing to verify the mass spectrometry data, we have repeated the polysome immunoblots for an additional 39 TAP fusion proteins (Supplementary Fig. S2). For 28 of the 39 fusion proteins, we were able to detect a protein that migrated at the expected molecular weight. Of those, 86% colocalized with ribosomes, suggesting that mass spectrometry reliably identified proteins in these purifications.
Figure 3.
Novel proteins identified in the proteomic screens cosediment with ribosomes in an EDTA-dependent manner. Anti-TAP Western blots on sucrose gradient fractions in the absence (left) or presence (right) of EDTA. The locations of the ribosomal subunits in the gradients are indicated in the chromatograms and by Western blots of RPL3 and ASC1. CDC28 marks the nonribosomal fractions, and NOP1 shows the migration of a preribosomal complex
Treatment of cell lysates with EDTA dissociates the 40S and 60S ribosomal subunits. Therefore, in the presence of EDTA, ribosomal and ribosome-associated proteins are no longer found at the bottom of the gradient. As such, we tested the EDTA-sensitivity of each TAP-ORF sedimentation. As expected, in the presence of EDTA, RPL3 and ASC1 migrated only near the top of the gradient. NOP1 also shifted its sedimentation, indicating that preribosomal complexes are EDTA-sensitive. Following EDTA treatment, no TAP-ORF remained in high-molecular-weight fractions (Fig. 3). In concert with the mass spectrometry data, the immunoblotting provided compelling evidence that the selected ORFs interacted with the translation machinery. Therefore, we have named these expressed genes TMA, for translation machinery associated, followed by their approximate molecular weight (Table 1). While this work was in progress, the standard name LSM12 was issued for YHR121W, and we therefore have adopted this nomenclature (Albrecht and Lengauer 2004).
Identification of TMA-interacting proteins
To further characterize the TMA proteins, we next looked for interacting proteins. Each TAP-TMA fusion protein was purified three times, and the copurifying proteins were identified using DALPC mass spectrometry (Link et al. 2005). To identify proteins that associated nonspecifically with the IgG and calmodulin beads, five control purifications were carried out in parallel using lysates from cells that did not express a TAP fusion protein. For each protein, a relative abundance factor (RAF) was calculated by dividing the average PAF of the protein in the TAP purification by the average PAF of the protein in the control purification. The RAF was used to identify proteins that were enriched in the TAP fusion protein purification relative to the control purification (Lewis et al. 2004).
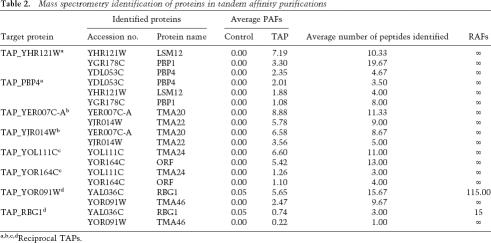
Several purifications contained proteins with PAFs at similar magnitudes as the TAP-TMA. These proteins were not found in the control purifications (RAF = ∞) or were enriched >100-fold as compared with the control. Importantly, the reciprocal TAP identified the TMA protein with similar PAF and RAF values (Table 2). LSM12 interacted with the PAB1-binding proteins PBP1 and PBP4. In addition to our confirmation of the LSM12–PBP4 interaction with the reverse TAP, a large-scale yeast two-hybrid study identified the LSM12–PBP1 interaction (Ito et al. 2001). We did not identify LSM12 in either of two TAP-PBP1 purifications; however, the known binding partners, PAB1 and PBP4, also were not identified, suggesting that the tag interferes with PBP1 interactions. Two of the novel ORFs we identified interacted with each other: TMA20 and TMA22. These interactions were also found in a large-scale TAP screen (Gavin et al. 2002). TMA46 interacted with RBG1, a GTPase (Li and Trueb 2000) that interacts with translating ribosomes (P. Wout and J. Maddock, unpubl.). Lastly, TMA24 interacted with YOR164C, an uncharacterized protein.
Table 2.
Mass spectrometry identification of proteins in tandem affinity purifications
a,b,c,dReciprocal TAPs.
Additional proteins with high PAFs and RAFs were identified, including ribosomal proteins (Supplementary Tables S11–S26); however, we have not yet validated these interactions by the reciprocal TAP. In addition to the strongly interacting proteins suggested by the PAFs, the TAPs identified a large number of lesser interactions. These proteins had PAFs that were 10-fold lower than the target protein and variable enrichment relative to the controls (Supplementary Tables S11–S26). Many of these proteins are known to be involved in translation or RNA processing. Again, additional experiments are required to determine whether they are true interacting proteins or nonspecific contaminants.
High-throughput translation screens identify defectsin yeast mutants lacking TMA proteins
Proteins that reproducibly and preferentially associate with whole ribosomes or ribosomal subunits are predicted to have roles in translation or related processes. Therefore, we wanted to extend our proteomic screens to identify TMAs required for normal protein synthesis. As none of the candidates chosen for validation were essential genes in yeast, we tested diploid yeast strains with homozygous null alleles for genes encoding the TMA proteins for translation phenotypes. In these experiments, we adapted established translation assays, including in vivo translation assays, drug susceptibility assays, and fidelity assays, to high-throughput screening methods. Cultures were typically grown and processed in 96-well plates, allowing simultaneous and rapid manipulation of several replicates or dilutions of the multiple strains.
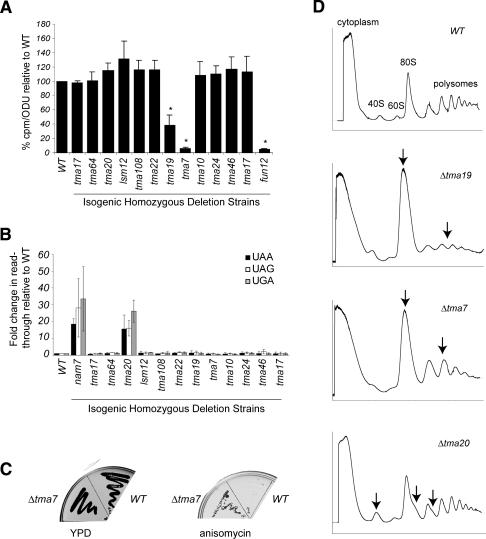
First, we screened for alterations in translation rates. Protein synthesis rates were measured as the amount of 35S-methionine incorporated into protein. The normalized rates of the deletion strains were plotted relative to wild type, designated as 100% (Fig. 4A). For comparison, 35S-methionine incorporation in the deletion strain for the translation initiation factor FUN12 was 5% of wild type (p = 1.8 × 10−5). FUN12 encodes the initiation factor eIF5B, a GTPase required for efficient protein synthesis (Choi et al. 1998; Lee et al. 2002). Two strains, Δtma19 and Δtma7, had significant decreases in 35S-methionine incorporation: 40% (p = 8.6 × 10−3) and 5% (p = 7.4 × 10−5) of wild type, respectively.
Figure 4.
Deletions of TMA proteins result in translation defects. (A) In vivo translation assay. Translation was measured as the amount of 35S-methionine incorporation in yeast strains deleted for the indicated TMA protein relative to the isogenic wild-type (WT) strain. The standard deviations between three independent samples are shown. (*) A p-value of <0.05. (B) Nonsense suppression assay. Translation of a β-gal reporter gene containing either UAA, UAG, or UGA mutations, in deletion strains relative to the wild-type strain (set as 1). The standard deviations from at least three independent experiments with three distinct colonies for each strain are shown. (C) Resistance to translation inhibitors. YPD or YPD + 50 μg/mL anisomycin plates streaked with wild-type or Δtma7 after incubation for 2 d at 30°C are shown. (D) Polysome profile analysis. Absorbance at 254 nm of sucrose gradient fractions from wild type, Δtma19, Δtma7, and Δtma20. Arrows indicate differences in the mutant profiles compared with wild type.
Next, the yeast strains were screened for alterations in translation fidelity. Nonsense suppression was measured as the amount of read-through translation of a reporter construct containing an in-frame nonsense mutation within lacZ relative to translation of the wild-type lacZ gene (Carr-Schmid et al. 1999). The Δtma20 strain had an average 19-fold higher read-through for each nonsense mutation (average p = 4.04 × 10−4) (Fig. 4B). By comparison, the deletion strain for NAM7, a RNA helicase implicated in translation termination (Wang et al. 2001; Keeling et al. 2004), had an average increase in stop codon read-through of 27-fold relative to wild type.
It has been shown that mutations in genes encoding translation factors cause cells to be more sensitive or resistant to translation inhibitors (Spahn and Prescott 1996; Dinman and Kinzy 1997). Therefore, we screened the deletion strains for altered growth in the presence of various translation inhibitors using a 96-well format dilution assay. Strains showing either sensitivity or resistance were retested by streaking onto the appropriate drug-containing medium. Resistance to anisomycin was detected for Δtma7 (Fig. 4C). The Δtma7 strain was not resistant to the other translational inhibitors we tested (data not shown), suggesting that Δtma7 is not deficient in drug transport or metabolism.
Lastly, the deletion strains were analyzed by polyribosome profiling. Consistent with decreased protein synthesis (Fig. 4A), both Δtma7 and Δtma19 strains had partial polysome runoff (Fig. 4D). While the profile was consistent for Δtma7, the magnitude of the runoff for Δtma19 was more variable. The variation might correlate with the magnitude of the decreased synthesis observed in the labeling experiment.
Additionally, Δtma20 consistently varied from the wild-type strain, with a larger 40S peak relative to the 60S peak, and half-mers on the 80S and polysome peaks (Fig. 4D). Because the phenotype was subtle, to determine whether the observed difference was significant, the 40S:60S ratio was calculated from seven independent profiles of wild-type and Δtma20 strains. The average 40S:60S was 1.5-fold higher in the Δtma20 strain, with values of 1.41 ± 0.20 and 2.08 ± 0.26 for wild type and Δtma20, respectively. A single-tailed nonpaired unequal-variance T-test indicated that the difference was significant, with a p-value of 8.19 × 10−5.
TMA20 and MCT-1 are orthologous proteins
Because the translation machinery is highly conserved from yeast to humans, we next sought to identify homologs of TMA proteins. Protein or nucleotide BLAST searches identified highly related sequences in other fungi for TMA7, TMA10, TMA16, and TMA17 (Altschul et al. 1990). Proteins similar to TMA16 were identified in insects, worms, plants, rodents, and humans, although the expect values did not fall below 0.4 (Altschul et al. 1990). For the remaining eight yeast proteins, protein BLAST queries identified mammalian proteins with significant sequence conservation (Expect ≤ 7e-09) (Altschul et al. 1990), suggesting conserved functions in higher organisms (Table 1). At least 67% of our confirmed TMA proteins had mammalian homologs, supporting the idea that data from proteomic screens in yeast are applicable to studies of higher organisms. This number is higher than for yeast proteins in general (30%–40%) (Botstein et al. 1997), which might be expected given that translation is a highly conserved process.
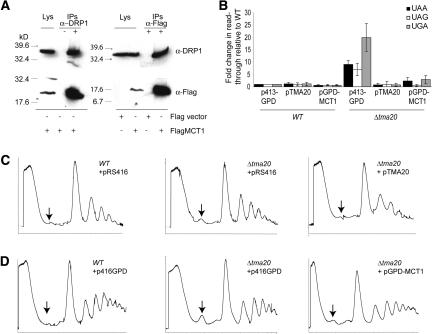
Because the Δtma20 strain had several translation defects and has a mammalian homolog, MCT-1, implicated in cancer, we focused on this protein in more detail. In yeast, TAP-TMA20 strongly interacted with TAP-TMA22 and vice versa (Table 2). TMA22 also has an apparent human homolog, DRP1. We first wanted to determine if the human homologs of TMA20 and TMA22 interacted. We tested whether human Flag-tagged MCT-1 interacted with endogenous DRP1 in human cells. DRP1 immunoprecipitates contained Flag-MCT1 (Fig. 5A). Similarly, Flag immunoprecipitates from cells transfected with Flag-MCT1 contained DRP1 (Fig. 5A), suggesting that the two proteins are in the same complex. Neither Flag-MCT1 nor DRP1 was identified in mock immunoprecipitates using beads only or in Flag immunoprecipitates from cells transfected with an empty Flag vector, suggesting that the interaction was specific. Together these interaction data provided evidence that human MCT1 and DRP1 interacted analogously to the yeast proteins TMA20 and TMA22.
Figure 5.
Human MCT-1 interacts with DRP1 and complements Δtma20 phenotypes. (A) α-DRP1 (top) or α-Flag (bottom) Western blots of cell lysates (Lys) and immunoprecipitates (IPs) from HEK293 cells transfected with Flag vector or Flag-MCT1. The lysate is 1/20 of the whole-cell extract used for the IP. (B) The amount of translation through nonsense mutations in a β-galactosidase reporter gene in wild-type (WT) and Δtma20 strains transfected with either empty expression vector (p413-GPD), yeast TMA20 (pTMA20), or the human MCT-1 gene expressed under control of the yeast GPD promoter (pGPD-MCT1). Error is reported as the standard deviation. (C) Polysome profiles from a wild-type yeast strain transfected with the empty pRS416 expression vector (left), the Δtma20 strain transfected with the empty pRS416 expression vector (middle), or Δtma20 transfected with an expression vector containing yeast TMA20 (pTMA20) (right). (D) Wild-type strain transfected with p416-GPD (left), Δtma20 strain transfected with p416-GPD (middle), or the Δtma20 strain transfected with pGPD-MCT1 (right). The arrow indicates the varying size of the 40S peak in the different profiles.
We next tested whether MCT-1 could complement the translation defects in the Δtma20 strain. As expected, reintroduction of yeast TMA20 under the control of its native promoter complemented the fidelity defect of Δtma20 (Fig. 5B) and resulted in a polysome profile that resembled the wild-type profile (Fig. 5C). In contrast, transformation of Δtma20 with the empty yeast expression vector did not rescue the phenotypes (Fig. 5B,C). Similarly, expression of a C-terminally V5 epitope-tagged MCT-1 expressed under the control of the relatively strong glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (pGPD-MCT1) rescued the nonsense read-through defects associated with deletion of TMA20 (Fig. 5B). The 40S peak for Δtma20 expressing pGPD-MCT1 was smaller than for Δtma20 + empty vector, similar to the wild type (Fig. 5D), indicating that expression of human MCT-1 rescued the defect. Together the bioinformatics, biochemical, and genetic analyses suggest that the oncogenic protein MCT-1 is the mammalian ortholog of TMA20.
Discussion
Here we describe comprehensive, large-scale, unbiased proteomic screens that identified novel proteins that copurify with S. cerevisiae ribosomes. By combining three purification strategies, two-dimensional liquid chromatography with tandem mass spectrometry, and clustering analysis, we have identified 77 uncharacterized ORFs as putative ribosome-interacting factors. Of these, we assayed 40 TAP-ORF fusion proteins and found that 90% copurified with ribosomes in sucrose gradients validating the initial mass spectrometry data. Each of the 11 TAP-TMAs tested further had an EDTA-sensitive distribution, consistent with ribosome-associated proteins. Through TAP purification, we linked two TMA proteins to translation-associated processes via their interacting partners. TAP-LSM12 interacted with the RNA-modifying proteins PBP1 and PBP4, and TAP-TMA46 pulled down RBG1, a ribosome-associated GTPase. Additionally, yeast strains containing deletions of tma7, tma19, or tma20 had phenotypes consistent with loss of proteins involved in translation or ribosome biogenesis, including decreased protein synthesis rates, resistance to translation inhibitors, decreased translation fidelity, and altered polysome profiles. Lastly, MCT-1, a human protein highly homologous to TMA20, complemented the translation defects associated with deletion of tma20 in yeast.
Our proteomic screens identified both ribosomal proteins and translation factors, validating the methodology. We identified 100% of the ribosomal proteins that can be detected by mass spectrometry and ∼87% of the canonical translation factors. Of the missing factors, components of the eIF2B complex were underrepresented in our purifications. However, eIF2B functions to exchange GDP for GTP on eIF2, a process that does not occur on ribosomes. Eliminating eIF2B components improved our identification to 96%. Only eIF1A (TIF11) was not detected in enough experiments to pass our stringent filtering criteria. For reasons that are not clear, we identified eIF1A in only two of nine 40S purifications and one of five RSWs.
A major concern with large-scale analyses is the purity of the samples being analyzed. We do not claim that the purification schemes used here yielded pure populations of ribosomes. Rather, the clustering data indicated an enrichment of ribosomal proteins and translation factors. Of the other proteins we identified, we found that approximately one-half to two-thirds of the copurifying proteins were highly abundant catalytic enzymes such as hydrolases and transferases. Although not currently linked to translation, we put forward the possibility that some catalytic enzymes may have dual functions in metabolism and protein synthesis. As an example, the S. cerevisiae PAS kinases regulate both sugar flux and translation, linking nutrient availability to protein synthesis (Rutter et al. 2002).
More likely, however, is that many of the additional proteins are components of larger complexes, and therefore may cosediment with free ribosomal subunits in sucrose gradients. Alternatively, the highly abundant proteins may simply be contaminants. For example, glyceraldehyde-3-phosphate dehydrogenase family members were consistently found in all preparations and are expressed at ∼105 molecules/cell (Ghaemmaghami et al. 2003). In contrast, the average number of molecules/cell for the 12 TMA proteins was nearly 50-fold lower, at 4.76 × 103 molecules/cell (1.25 × 102 to 3.75 × 104) (Table 1). Although we cannot exclude the possibility that some of the ORFs are not ribosome-associated, the abundance data suggest that we did not simply purify the most abundant proteins in the cell.
Importantly, using immunoblotting we were able to support the mass spectrometry data and show an EDTA-dependent colocalization with ribosomes in sucrose gradients for 11 of the 12 TMAs we tested. Overall, the immunoblotting revealed a broader sedimentation for each TAP-TMA than indicated by the mass spectrometry, especially for LSM12 and TMA16. These inconsistencies may reflect relative stoichiometries of the proteins in the ribosome complexes. Alternatively, because tandem mass spectrometry data are acquired for the three most intense ions in each full scan, the more stringent results seen by mass spectrometry may reflect the relative abundance of the protein of interest relative to the total numbers of proteins in the fraction. In contrast, the immunoblotting is less sensitive to the complexity of the sample. Despite these differences, the combined approaches indicated a strong connection for 11 TMAs with the translation machinery. Because the proteomic screens identified TMA19 in the 40S fraction, but TAP-TMA19 was absent from the 40S by immunoblotting, the 20-kDa TAP tag may have disrupted protein–protein interactions linking TMA19 to ribosomes. This interpretation, rather than TMA19 being a false-positive mass spectronomy hit, is supported by the fact that TMA19 was also identified in a second screen (RSW) and had translation phenotypes associated with its deletion.
The TAP data identified additional proteins linking the TMA proteins to complexes involved in translation, ribosome biogenesis, RNA processing, and RNA export (Table 2; Supplementary Tables S11–S26). Validation of each interaction is beyond the scope of this study. However, data from a large-scale TAP-tandem mass spectrometry project involving known translation factors, showed that the major components of any particular translation complex associated with the TAP-tagged target with PAFs >1.0 (C.M. Weaver, J.L. Jennings, D.T. Duncan, K.J. McAfee, V.R. Gerbasi, A.R. Farley, T.C. Fleischer, and A.J. Link, in prep.). Therefore, in our experience, proteins associating with TAP-ORFs with PAFs >1.0 are likely to be true interacting partners. This assumption holds true for TMA20-TMA22, LSM12-PBP1-PBP4, TMA24-YOR164C, and TMA46-RBG1, where the reciprocal TAP and/or published data (Tables 1, 2) corroborated the initial discovery.
The interaction of TMA46 with RBG1 provided another link for TMA46 to ribosomes, as RBG1 is a GTPase (Li and Trueb 2000) that interacts with translating ribosomes (P. Wout and J. Maddock, unpubl.). Likewise, the interaction of LSM12 with PBP1 and PBP4 connected LSM12 to RNA processing. LSM proteins, like SM proteins, are predicted to form heptameric rings and interact with RNA (for review, see Beggs 2005). One cytoplasmic LSM complex contains LSM proteins 1–7 and functions in mRNA degradation (Bouveret et al. 2000; Tharun et al. 2000). Interestingly, although LSM12 contains the SM domain, we did not find named LSM proteins in the TAP-LSM12 pull-down. However, we found a strong association with PBP1. Because PBP1 can self-interact (Mangus et al. 1998, 2004) and has an LSM domain, it is interesting to speculate that LSM12 forms an atypical ring comprised of multiple units of itself and PBP1. This ring may regulate various aspects of RNA processing via interactions with PBP4, PAB1, FIR1, UFD1, DIG1, and MKT1 (Mangus et al. 1998, 2004; Tadauchi et al. 2004).
Several published studies complement our findings and connect TMA proteins to additional translation-related proteins (Table 1; Uetz et al. 2000; Ito et al. 2001; Gavin et al. 2002; Ho et al. 2002; Krogan et al. 2004). These TMA-interacting proteins include ribosome subunits (RPS4, ASC1), components of the small ribosomal subunit processosome (BUD21, NOP1, SAS10), an RNA helicase required for translation initiation of all yeast mRNAs (DED1), an mRNA capping enzyme (CEG1), and proteins involved in deadenylation-dependent decay (PUF1, PUF3) and ribosome biogenesis (YTM1). Together, our data and published interaction data suggest roles for the TMA proteins in translation regulation or RNA metabolism, and support our proteomic data that the TMA proteins are translation machinery associated.
Four of the 12 TMA proteins have putative RNA-binding domains (Table 1), which could mediate interactions with a variety of different RNAs including mRNA or rRNA. TMA20 has the PUA RNA-binding motif, TMA22 has an eIF1/SUI1 RNA-binding domain, and TMA64 has both the eIF1/SUI1 and PUA domains. Given that Δtma20 has an altered 40S:60S ratio, we speculate that TMA20’s RNA-binding domain recognizes rRNA. Although we did not observe an altered polysome profile for the Δtma22 strain, rRNA binding is predicted for TMA22, due to its interaction with TMA20. The published interaction of TMA64 with the pre-rRNA processing proteins NOP1 and DED1 (Krogan et al. 2004) is again consistent with rRNA binding. LSM12 has an RNA-binding LSM (like SM) domain that we predict recognizes mRNA, given LSM12’s association with PBP1 and PBP4. However, SM and LSM domains are implicated in many aspects of RNA processing including pre-mRNA splicing and degradation, tRNA splicing, mRNA degradation, and rRNA processing (for review, see Albrecht and Lengauer 2004; Beggs 2005). Because these predicted domains are either not well characterized or have been shown to have variability in their type of RNA interactions, without further experimental data, it is difficult to extrapolate which of our additional uncharacterized proteins may be involved in rRNA or mRNA binding.
Functional assays linked several TMA proteins to translation and translation-related processes. One interesting protein identified in our screens was TMA7, found in a 40S fraction from gradients cast in 1 M salt (data not shown). Loss of TMA7 was associated with multiple translation defects. One was resistance to the translation inhibitor anisomycin (Fig. 4C). A general defect in drug uptake or metabolism is unlikely, as we did not detect resistance to other translation inhibitors including cycloheximide, puromycin, rapamycin, or hygromycin (data not shown). Anisomycin competes with aminoacylated-tRNA binding to the A-site and thereby inhibits translation at the peptidyl transfer step (Barbacid and Vazquez 1974; Hansen et al. 2003). Elongation factor eEF1A mutant strains tef2-3 and tef2-13 are resistant to anisomycin (Dinman and Kinzy 1997). It is hypothesized that these mutations may increase the pause rate at the A-site (Dinman and Kinzy 1997). Similarly, the resistance to anisomycin in the absence of TMA7 (Fig. 4C) could result from translation rates that were slowed such that the aminoacylated-tRNAs could eventually bind the A-site, despite the anisomycin competition. This explanation is consistent with the dramatic decrease in 35S-methionine incorporation (Fig. 4A) and slow growth phenotype (data not shown) seen in Δtma7 strains. However, the polysome profile for Δtma7 showed a reduction in polysomes relative to monosomes, suggesting the translation defect is at the initiation stage rather than in elongation. In agreement, strains expressing mutant elongation factors eEF3 (F650S) or eEF1A (E286K) did not have altered polysome profiles when assayed in the presence of cycloheximide (Anand et al. 2003).
The tma19-null strain had a similar phenotype to Δtma7 (Fig. 4A,D), with decreased 35S-incorporation and reduced polysomes relative to monosomes, also suggesting a function for TMA19 in initiation. Although the reduced metabolic labeling was apparent in every experiment, the altered polysome profile was variable. The significance of this is unclear.
A Δtma20 strain had two translation defects: an altered polysome profile with a larger 40S peak and decreased translation fidelity (Fig. 4B,D). The altered 40S:60S ratio and polysome half-mers in Δtma20 suggest a role for TMA20 in ribosome biogenesis. As such, the 40S localization is more likely a preribosomal complex with similar sedimentation. In light of the defect in ribosome biogenesis, one interpretation of the increased read-through of nonsense mutations in Δtma20 strains is that the phenotype is indirect, resulting from aberrant ribosomes.
Defining normal TMA20 function in yeast may help shed light on the oncogenic aspects of its human homolog MCT-1. Overexpression of MCT-1 leads to increased cell proliferation, a shortened G1 phase, increased cyclin D1 expression, and an ability to grow in soft agar (Prosniak et al. 1998; Dierov et al. 1999). Recent data link MCT-1 with the DNA damage checkpoint and angiogenesis (Hsu et al. 2005; Levenson et al. 2005). However, the precise molecular function of this protein is still unclear. Our yeast complementation data would suggest that up-regulation of MCT-1 increases ribosome biogenesis, thereby supporting increased cell growth. Coordinately regulated responses to growth factors and mitogens might explain the data linking MCT-1 to other oncogenic functions. For example, PI-3K/AKT/mTOR-mediated signaling increases both ribosome biogenesis and translation (for reviews, see Meric and Hunt 2002; Holland et al. 2004; Ruggero and Sonenberg 2005).
Cumulatively, these four assays identified translation defects associated with one-quarter of the TMA proteins we screened. It is important to note that none of these TMA proteins was identified previously in similar biochemical assays using conventional methods, nor were they found in genetic screens for translation defects. Thus, we cannot rule out that the remaining candidates function in translation, perhaps in the translation of specific mRNAs or classes of mRNAs. We are expanding our repertoire of assays and are testing the deletion strains for altered GTP hydrolysis, RNA processing, and translation of various templates in vivo. Assays to analyze RNA processing may be especially informative given that more than half the TMA proteins including TMA19, TMA64, LSM12, TMA46, TMA7, TMA24, and TMA10, copurified with known rRNA- or mRNA-modifying proteins (Supplementary Tables S11–S26), although not all of these interactions have been confirmed. Another important consideration is that deletion of these candidates may only have observable phenotypes under specific physiological conditions, such as amino acid or glucose starvation.
In this study, we presented 77 uncharacterized proteins as potential ribosome-interacting proteins. Of these, we have data supporting ribosome interactions for at least 11 of the 12 novel ORFs tested. We are presently screening the remaining candidates using sucrose gradients, immunoblotting, and TAP. As we delineate the translation network, we take a critical step toward unraveling the complexities of eukaryotic protein synthesis. By testing the corresponding yeast deletion strains in high-throughput adaptations of conventional translation assays, we have begun the often-overlooked task of imparting functional significance to our interaction data. Collectively, our proteomics screens and translation assays have provided the framework for more rigorous investigations into the roles of these uncharacterized proteins in translation and related processes. Moreover, we have applied our studies in yeast to help elucidate the function of human disease proteins.
Materials and methods
Strains
BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0) was used for all protein purification experiments (Winzeler et al. 1999). Deletion strains in the BY4743 background had the pertinent ORFs replaced by kanamycin cassettes as described (Winzeler et al. 1999). Yeast strains with TAP-tagged genes have been described (Ghaemmaghami et al. 2003). We have generated the following strains: AL190 (BY4743 with pRS416), AL194 (Δtma20 + pRS416), AL195 (Δtma20 + pTMA20), AL259 (BY4743 + p416-GPD), AL260 (BY4743 + pGPD-MCT1), AL261 (Δtma20 + p416-GPD), and AL262 (Δtma20 + pGPD-MCT1).
Plasmids
To construct pTMA20, the endogenous S. cerevisiae YER007C-A promoter and ORF were PCR-amplified from yeast genomic DNA using primers 007gen694F (GATCGGATCCC GAGATTGTGTTTTTGCTGG) and 007gen2244R (GATCC TCGAGTCAGACTTTAATGTTGTGAACFGG) and cloned into the BamHI/XhoI sites of pRS416 (Sikorski and Hieter 1989). To construct pGPD-MCT1, a BamHI/PmeI fragment from pCDNA3-MCT1-V5-His (Shi et al. 2003) was cloned into BamHI/SmaI sites of p416-GPD (Funk et al. 2002). To construct FlagMCT1, an EcoRI/SalI fragment of PCR-amplified MCT-1 from pCDNA3-MCT1-V5-His was cloned into pFLAG-CMV-6a (Sigma). The primers to amplify MCT-1 were MCT_ATG_Eco_f (GATCGAATTCATGTTCAAGAAATTTGATG) and MCT_ TGA_Sal_r (GATCGTCGACTCATTTATATGTCTTCATAT GC). All clones were verified by sequencing.
Total ribosomal analysis
Yeast cultures were grown overnight at 30°C in 1 L of YPD to an OD600 between 2 and 3. Cells were pelleted at 2500 × g for 10 m and washed with 50 mL of ddH20. The cell pellet was resuspended in 25% (w/v) standard buffer (10 mM Tris at pH 7.4, 5 mM 2-mercaptoethanol [βME], 50 mM ammonium chloride, 5 mM magnesium chloride, 100 μg/mL cycloheximide, 500 μg/mL heparin, and protease inhibitors [Roche Complete EDTA-free]). Cells were lysed by the addition of 1/2 volume 0.5-mm glass beads (Bio-Spec Products) and the use of a BeadBeater (BioSpec Products) for 4 min (four cycles of 30 sec on/30 sec off). The lysate was centrifuged at 20,000 × g for 30 min, transferred to fresh tubes, and centrifuged at 150,000 × g for 2 h. The ribosome pellet was resuspended in 24 volumes of wash buffer (10 mM Tris at pH 7.4, 5 mM βME, 10 mM magnesium acetate, and the indicated amount of ammonium chloride) supplemented with 100 μg/mL cycloheximide, 500 μg/mL heparin, and protease inhibitors (Roche Complete EDTA-free). A 6-mL aliquot was layered on a 6-mL 5%:20% discontinuous sucrose gradient and centrifuged at 40,000 × g for 18 h. The ribosome pellet was resuspended in 25 volumes of standard buffer and centrifuged at 7100 × g for 10 min.
Ribosome salt wash
Ribosome salt washes were carried out as described (Link et al. 2005). Briefly, yeast whole-cell lysates were centrifuged to remove cellular debris. Ribosomes were then pelleted by ultracentrifugation and resuspended by sonicating in buffer supplemented to 0.05 M, 0.5 M, or 1 M potassium acetate. Ribosomal proteins were again pelleted by centrifugation, and the supernatant (ribosome salt wash) was saved for analysis.
Sucrose gradient fractionation
A high-resolution purification of ribosome complexes was employed using sucrose gradient fractionation (Link et al. 2005). The method was used to analyze the polysome profiles of the yeast deletion and complemented strains with the following modifications. Strains were grown to log phase in 100 mL of SC-URA medium with the final OD600 ∼ 0.8. Briefly, cycloheximide (50 μg/mL) was added directly to each culture, and then the culture was chilled on ice for 10 min. Cells were lysed in chilled Breaking Buffer (10 mM Tris-HCl at pH 7.5, 100 mM NaCl, 30 mM MgCl2, 50 μg/mL cycloheximide, 200 μg/mL heparin, and protease inhibitors [Roche Complete EDTA-free]), and 20 OD260 of the cleared extract was loaded onto 12-mL 7%–47% sucrose gradients. The gradients were centrifuged in an SW-41 rotor for 18 h at 15,000 rpm at 4°C.
Immunoprecipitation and immunoblotting
Aliquots (30 μL) of 0.5-mL sucrose gradient fractions were run on Tris-glycine gels, transferred to Immobilon-P (Millipore), and immunoblotted with indicated antibodies. For immunoprecipitations, HEK293 cells were seeded at 1.0 × 106 and 16 h later were transfected with 5 μg of either pFLAG-CMV-6a (Sigma) or Flag-MCT1 using calcium phosphate (Kingston et al. 1996). After 24 h, cells were lysed in 0.5 mL of L-buffer (PBS, 0.1% NP-40) containing EDTA-free complete mini-protease inhibitors (Roche), 50 μg/mL cycloheximide, 1 mM dithiotreitol, and 40 U/mL RNAsin (Promega), by freezing on dry ice and thawing at 37°C. Nuclei and debris were pelleted by centrifugation for 5 min at 13,000g. Cytoplasmic extracts were immunoprecipitated for 1 h at 4°C with 20 μL of Flag-M2 agarose (Sigma) or 20 μL of ImmunoPure Immobilized Protein A (Pierce) with or without 5 μL of α-DRP1 antibody. Lysates and immunoprecipitates were resolved on 12% Tris-glycine gels, transferred to Immobilon-P (Millipore), and immunoblotted with indicated antibodies. Interactions were detected using chemiluminescence (ECL Plus; Amersham Biosciences).
Antibodies
Anti-TAP (Open Biosystems), anti-Flag M2-peroxidase (HRP; Sigma), anti-DRP1 (BD Transduction Laboratories), anti-NOP1 (EnCor Biotechnology, Inc.), and anti-PSTAIR (Sigma) antibodies are commercially available. Anti-RPL3 (Vilardell and Warner 1997) and anti-ASC1 (Gerbasi et al. 2004) have been described. Anti-rabbit HRP and anti-mouse HRP were from Promega.
Tandem affinity purification
TAP fusion proteins were purified using IgG and calmodulin affinity resins essentially as described (Link et al. 2005).
DALPC
To identify the proteins found in purified ribosomes or TAP purifications, samples were trypsin-digested and analyzed by off-line SCX fractionation coupled to RP-LC-ESI-MS/MS as described (Link et al. 2003, 2005). Data processing of theSEQUEST output files into a list of proteins has been previously described (Link et al. 1999).
[35S]-methionine incorporation
Overnight cultures grown in YPD were diluted 1:10 into 1 mL of SC-MET (OD595 ∼ 0.5) in triplicate in a 96-well 2-mL culture dish and grown for 3 h at 30°C. The OD595 of one replicate was measured to determine relative cell numbers. The remaining two samples were labeled with 6 μCi/mL [35S]-methionine/cysteine (EXPRE35S35S Protein Labeling Mix; >1000 Ci/mmol; NEN Research Products) for 15 min at 30°C. Labeling was stopped by the addition of 1/10 volume of 100% TCA to each culture and heating the entire 96-well dish at 100°C for 30 min. TCA precipitates were collected on GFC filters (Whatman), washed sequentially with 2 mL of each 10% TCA and 95% ethanol, and counted in 5 mL of UniverSol (ICN). For each sample, the counts per minute/OD595 was normalized to wild type, set as 100%. Three independent experiments were graphed with the error bars representing the standard deviation. The P-values shown were determined using a single-tailed nonpaired unequal-variance T-test.
Drug dilution assay
Cells were grown overnight in 1 mL of YPD. The OD595 was measured, and an equal number of cells for each strain (up to 24 different strains/plate) was aliquoted into a 96-well plate, and then serially diluted, fivefold each, into YPD. Using a 96-pin multiblot replicator (V&P Scientific, Inc.), the master dilution plate was replica-plated in duplicate onto YPD plates containing no drug, 5 ng/mL rapamycin, 50 ng/mL rapamycin, 0.2 μg/mL cycloheximide, 2.0 μg/mL cycloheximide, 20 μg/mL anisomycin, 50 μg/mL anisomycin, 10 μg/mL hygromycin, or 100 μg/mL hygromycin. Plates were incubated at 30°C for up to 4 d. All strains were tested for sensitivity or resistance to each drug concentration in at least two independent experiments. Pertinent strain/drug combinations were verified in at least three experiments by streaking overnight cultures onto duplicate drug plates and growing at 30°C.
Nonsense suppression assay
Nonsense suppression was assayed as described (Carr-Schmid et al. 1999). Production of β-galactosidase (β-gal) was measured using the Tropix Gal-Screen chemiluminescent reporter gene system (Applied Biosystems) with 50 μL of cells and 50 μL of Reaction Buffer B. Luminescence was measured for 20 sec in a Turner Designs TD-20/20. Assays were repeated at least twice with three independent colonies for each strain.
Acknowledgments
We thank Amar Kar for assistance with the translation fidelity experiments, R.B. Gartenhaus for plasmid MCT-V5, T.G. Kinzy for the nonsense suppression plasmids, and J.R. Warner for antibodies to RPL3. We thank Elizabeth M. Link, David W. Powell, R. Vincent Gerbasi, and Adam Farley for critical comments in the preparation of this manuscript. This project was supported by NIH grant GM64779. T.C.F. is supported by NIH post-doctoral training grant T32 AI49824 and ACS post-doctoral fellowship PF-05-148-01-MBC. C.M.W. and J.L.J are supported by NIH grants GM64779 and HL68744. K.J.M. is supported by NIH grants ES11993, GM64779, and GM68900. A.J.L is supported by NIH grants GM64779, HL68744, ES11993, and CA098131. In addition, this project was funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN266200400079C/N01-AI-40079.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1422006
References
- Albrecht M., Lengauer T. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 2004;569:18–26. doi: 10.1016/j.febslet.2004.03.126. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anand M., Chakraburtty K., Marton M.J., Hinnebusch A.G., Kinzy T.G. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 2003;278:6985–6991. doi: 10.1074/jbc.M209224200. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M., Vazquez D. (3H)anisomycin binding to eukaryotic ribosomes. J. Mol. Biol. 1974;84:603–623. doi: 10.1016/0022-2836(74)90119-3. [DOI] [PubMed] [Google Scholar]
- Beggs J.D. Lsm proteins and RNA processing. Biochem. Soc. Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- Botstein D., Chervitz S.A., Cherry J.M. Yeast as a model organism. Science. 1997;277:1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven C.F., Muller C., Leutz A. Translational control of gene expression and disease. Trends Mol. Med. 2002;8:577–583. doi: 10.1016/s1471-4914(02)02424-3. [DOI] [PubMed] [Google Scholar]
- Carr-Schmid A., Valente L., Loik V.I., Williams T., Starita L.M., Kinzy T.G. Mutations in elongation factor 1β, a guanine nucleotide exchange factor, enhance translational fidelity. Mol. Cell. Biol. 1999;19:5257–5266. doi: 10.1128/mcb.19.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.K., Lee J.H., Zoll W.L., Merrick W.C., Dever T.E. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- Costanzo M.C., Hogan J.D., Cusick M.E., Davis B.P., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., et al. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): Comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierov J., Prosniak M., Gallia G., Gartenhaus R.B. Increased G1 cyclin/cdk activity in cells overexpressing the candidate oncogene, MCT-1. J. Cell. Biochem. 1999;74:544–550. [PubMed] [Google Scholar]
- Dinman J.D., Kinzy T.G. Translational misreading: Mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- Donahue T.F. Genetic approaches to translation initiation in Saccharomyces cerevisiae. In: Sonenberg N., editor. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 487–502. [Google Scholar]
- Dua K., Williams T.M., Beretta L. Translational control of the proteome: Relevance to cancer. Proteomics. 2001;1:1191–1199. doi: 10.1002/1615-9861(200110)1:10<1191::AID-PROT1191>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Funk M., Niedenthal R., Mumberg D., Brinkmann K., Ronicke V., Henkel T. Vector systems for heterologous expression of proteins in Saccharomyces cerevisiae. Methods Enzymol. 2002;350:248–257. doi: 10.1016/s0076-6879(02)50967-8. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gerbasi V.R., Weaver C.M., Hill S., Friedman D.B., Link A.J. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 2004;24:8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Granneman S., Baserga S.J. Probing the yeast proteome for RNA-processing factors. Genome Biol. 2003;4:229. doi: 10.1186/gb-2003-4-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S., Baserga S.J. Ribosome biogenesis: Of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Grossman L., Moldave K. Nucleic acids and protein synthesis, part G. Academic Press; New York: 1979. [Google Scholar]
- Hansen J.L., Moore P.B., Steitz T.A. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N., editor. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 185–243. [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Holland E.C., Sonenberg N., Pandolfi P.P., Thomas G. Signaling control of mRNA translation in cancer pathogenesis. Oncogene. 2004;23:3138–3144. doi: 10.1038/sj.onc.1207590. [DOI] [PubMed] [Google Scholar]
- Hong E.L., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Livestone M.S., Nash R. 2005. ‘Saccharomyces genome database.’ http://www.yeastgenome.org. [Google Scholar]
- Hsu H.L., Shi B., Gartenhaus R.B. The MCT-1 oncogene product impairs cell cycle checkpoint control and transforms human mammary epithelial cells. Oncogene. 2005;24:4956–4964. doi: 10.1038/sj.onc.1208680. [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K.M., Lanier J., Du M., Salas-Marco J., Gao L., Kaenjak-Angeletti A., Bedwell D.M. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R.E., Chen C.A., Okayama H. Calcium phosphate transfection. In: Ausubel F.M., et al., editors. Current protocols in molecular biology. John Wiley & Sons; New York: 1996. pp. 9.1.4–9.1.11. [Google Scholar]
- Klann E., Dever T.E. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat. Rev. Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Peng W.T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Pestova T.V., Shin B.S., Cao C., Choi S.K., Dever T.E. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. 2002;99:16689–16694. doi: 10.1073/pnas.262569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson A.S., Thurn K.E., Simons L.A., Veliceasa D., Jarrett J., Osipo C., Jordan V.C., Volpert O.V., Satcher, R.L. , Gartenhaus R.B. MCT-1 oncogene contributes to increased in vivo tumorigenicity of MCF7 cells by promotion of angiogenesis and inhibition of apoptosis. Cancer Res. 2005;65:10651–10656. doi: 10.1158/0008-5472.CAN-05-0845. [DOI] [PubMed] [Google Scholar]
- Lewis P.W., Beall E.L., Fleischer T.C., Georlette D., Link A.J., Botchan M.R. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes & Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Trueb B. DRG represents a family of two closely related GTP-binding proteins. Biochim. Biophys. Acta. 2000;1491:196–204. doi: 10.1016/s0167-4781(00)00025-7. [DOI] [PubMed] [Google Scholar]
- Link A.J., Eng J., Schieltz D.M., Carmack E., Mize G.J., Morris D.R., Garvik B.M., Yates III J.R. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Link A.J., Jennings J.L., Washburn M.P. Analysis of protein composition using multidimensional chromatography and mass spectrometry. In: Coligan J.E., et al., editors. Current protocols in protein science. John Wiley and Sons; New York: 2003. pp. 23.21.21–23.21.25. [DOI] [PubMed] [Google Scholar]
- Link A.J., Fleischer T.C., Weaver C.M., Gerbasi V.R., Jennings J.L. Purifying protein complexes for mass spectrometry: Applications to protein translation. Methods. 2005;35:274–290. doi: 10.1016/j.ymeth.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Amrani N., Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D.A., Smith M.M., McSweeney J.M., Jacobson A. Identification of factors regulating poly(A) tail synthesis and maturation. Mol. Cell. Biol. 2004;24:4196–4206. doi: 10.1128/MCB.24.10.4196-4206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric F., Hunt K.K. Translation initiation in cancer: A novel target for therapy. Mol. Cancer Ther. 2002;1:971–979. [PubMed] [Google Scholar]
- Milkereit P., Kuhn H., Gas N., Tschochner H. The pre-ribosomal network. Nucleic Acids Res. 2003;31:799–804. doi: 10.1093/nar/gkg165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- Pe’ery T., Mathews M.B. Viral translation strategies and host defense mechanisms. In: Sonnenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 371–424. [Google Scholar]
- Powell D.W., Weaver C.M., Jennings J.L., McAfee K.J., He Y., Weil P.A., Link A.J. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosniak M., Dierov J., Okami K., Tilton B., Jameson B., Sawaya B.E., Gartenhaus R.B. A novel candidate oncogene, MCT-1, is involved in cell cycle progression. Cancer Res. 1998;58:4233–4237. [PubMed] [Google Scholar]
- Rajesekhar V.K., Holland E.C. Postgenomic global analysis of translational control induced by oncogenic signaling. Oncogene. 2004;23:3248–3264. doi: 10.1038/sj.onc.1207546. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Sonenberg N. The Akt of translational control. Oncogene. 2005;24:7426–7434. doi: 10.1038/sj.onc.1209098. [DOI] [PubMed] [Google Scholar]
- Rutter J., Probst B.L., McKnight S.L. Coordinate regulation of sugar flux and translation by PAS kinase. Cell. 2002;111:17–28. doi: 10.1016/s0092-8674(02)00974-1. [DOI] [PubMed] [Google Scholar]
- Schneider R.J., Mohr I. Translation initiation and viral tricks. Trends Biochem. Sci. 2003;28:130–136. doi: 10.1016/S0968-0004(03)00029-X. [DOI] [PubMed] [Google Scholar]
- Shi B., Hsu H.L., Evens A.M., Gordon L.I., Gartenhaus R.B. Expression of the candidate MCT-1 oncogene in B- and T-cell lymphoid malignancies. Blood. 2003;102:297–302. doi: 10.1182/blood-2002-11-3486. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M., Prescott C.D. Throwing a spanner in the works: Antibiotics and the translation apparatus. J. Mol. Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- Tadauchi T., Inada T., Matsumoto K., Irie K. Posttranscriptional regulation of HO expression by the Mkt1–Pbp1 complex. Mol. Cell. Biol. 2004;24:3670–3681. doi: 10.1128/MCB.24.9.3670-3681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Yanagida M., Fujiyama S., Hayano T., Isobe T. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 2003;22:287–317. doi: 10.1002/mas.10057. [DOI] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A.E., Lennertz P., Beggs J.D., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vanderklish P.W., Edelman G.M. Differential translation and fragile X syndrome. Genes Brain Behav. 2005;4:360–384. doi: 10.1111/j.1601-183X.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- Vilardell J., Warner J.R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Czaplinski K., Rao Y., Peltz S.W. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiaegenome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]