Summary
The probability to predict correctly a protein structure can be enhanced through introduction of spatial constraints - either from NMR experiments or from homologous structures. However, the additional constraints lead often to new local energy minima and worse sampling efficiency in simulations. In this work we present a new parallel tempering variant that alleviates the energy barriers resulting from spatial constraints and therefore yields to an enhanced sampling in structure prediction simulations.
Keywords: CABS model, Monte Carlo sampling, protein folding, protein structure prediction, replica method
1 Introduction
Proteins are only functional if they assume specific shapes. For this reason, it is not sufficient to know their chemical composition (the sequence of amino acids as specified in the genome), understanding the working of proteins requires in addition knowledge of their three dimensional structure. However, the number of experimentally solved structures is much smaller than the number of known sequences, and the recent completion of the Human Genome Project has only widened this gap. Consequently, one of the most pressing tasks in computational biology and biochemistry is is the development of techniques for prediction of a protein structure from its amino acid sequence.
Computer experiments attempt to forecast the biologically active structure of a protein through modeling. Such simulations require the use of a suitable model that describes sufficiently accurate the interactions within a protein and with its environment (most noticeable the surrounding water molecules). Unfortunately, current energy functions do not always distinguish between native-like conformations and other low-energy conformers [1], and their use in structure prediction calculations is therefore limited.
In many cases we know more than just the sequence of a target protein. Searching structural databases one often finds a homologous protein, whose structure is known. Such a homolog is termed a template. If the sequence similarity between target and template is high (40% and more) the two proteins are evolutionary related and will share a common fold. Hence, distance constraints derived from template structures can be employed to force a simulation into the correct fold as they favor the correct topology, decrease the energy of a native-like fold and destabilize other topologies. Thus, the probability that native-like configurations form the basin of attraction of the global minimum becomes higher. However, if the sequence similarity is lower than 40% and the differences between the structures become more pronounced, one often needs several templates to guide the simulation into the correct fold. Such an “guided simulation” approach was successfully used in the CASP6 (Critical Assessment of Techniques for Protein Structure Prediction) competition. For instance, the Kolinski-Bujnicki group, using the CABS (CAlpha,CBeta,Side chain) model with distance constraints, presented very good (in some cases the best) solutions for a large number of targets [8].
Adding constraints to an energy function is not without problems. Often distance constraints are derived from several templates. While this increases the chance to find the correct topology, it also introduces additional local minima if the constraints are inconsistent or competing with the force field. The resulting larger roughness can lead to a slower sampling of the energy landscape rendering it difficult or even impossible to find the correct structure (even if that structure has now become the global minimum of the (constraint) energy function).
Slow convergence of simulations due to a rough energy landscapes is a well-known problem in computational protein science. Various numerical techniques have been developed to overcome this multiple minima problem [2]. For instance, in parallel tempering (PT) [3], which was first introduced to protein studies in Ref. [4], one considers an artificial system built up of N non-interacting copies of a molecule, each at a different temperature Ti. In addition to standard Monte Carlo or molecular dynamics moves that affect only one copy, parallel tempering allows also with a certain probability the exchange of conformations between two copies i and j = i + 1. The resulting random walk in temperature enables configurations to cross energy barriers and move out of local minima leading in this way to an enhanced sampling of low-energy structures. In this paper we present a new variant of PT, where the replicas are all at the same temperature, but differ in the strength with that constraints contribute to the total energy. Hence, our approach is similar to the Hamilton Exchange Method [5] and “model hopping” [6] where the various copies also differ not by their temperature but in their energy function. Our new approach is tested for a target from the last CASP competition (CASP6). We find that tempering in constraint space leads to faster and broader sampling of the conformational space than regular parallel tempering and requires less human input.
2 Methods
A necessary ingredient in protein simulations is a model that approximates the interactions within a protein and between the protein and surrounding water. Such models differ by the degree of coarse graining - from all-atom models up to such that describe each amino acid by a single sphere - and the way interactions are described. For structure prediction purposes the CABS model is very successful as it combines accuracy with very fast calculation of energies. CABS is a high-resolution lattice model. Each amino acid is represented by four interaction centers C α, C β, the center of mass of the side chain and the center of the peptide bond. The positions of C α-atoms are restricted to a simple cubic lattice with lattice spacing equal to 0.61Å. The resulting 800 possible C α - C α vector orientations diminish adverse lattice effects from anisotropy. The C β atoms are located off-lattice and the position of a central C β is fixed by three consecutive C α atoms. The location of the center of mass of the side chain depends on the local secondary structure. An additional atom is placed in the center of the virtual C α - C α bond if a hydrogen bond needs to be defined.
Various forms of interactions are considered in the CABS model. Short range sequence dependent potential terms derived from the PDB data base control the distances ri,i +2, ri,i +3 and ri,i +4 between C α atoms. They reduce the conformational space and make the model chain behave more like a protein then a polymer. Complex multibody effects are accounted for indirectly through sequence dependent pairwise interactions between the side-groups. These long range interactions are context dependent and take into account the identity of interacting groups, their spatial separation, mutual orientation and the geometry of corresponding fragments of the main chain. A detailed description of the interactions considered in the CABS model is given elsewhere [7].
CABS and other energy functions are often not accurate enough to guarantee that the native structure of a protein is indeed the global minimum configurations. This is one of the main obstacle in structure prediction simulations. Distance constraints derived from template structures can be employed to steer the simulation into the correct fold as they decrease the energy of a native-like fold and destabilize other topologies. For this purpose one writes down an energy function
| (1) |
Here, ECABS(c) is the energy of a configuration c in the CABS model and
| (2) |
a term that describes the “distance” of a configuration c to the template structure t. The strength of this constraint term is tuned with the parameter a. Constraints are derived from structures of homologous proteins as found by the Genesilico.pl [11] metaserver (http://www.genesilico.pl). The metaserver sends a target sequence to many threading servers, secondary structure predictors and other fully automated prediction tools, and calculates a consensus based on these predictions. Based on the so-obtained coarse-grained structures a more refined set of full-atom models was built using FRankensteins monster approach (fold recognition with fragment assembly approach)[9]. These models are evaluated using Verify3D [10] which assesses protein models with three-dimensional profiles. Spatial restraints are derived from the high scoring regions.
Addition of EConstraints to the CABS energy ECABS ensures that for a suitable choice of templates the unknown structure of the target protein is the global minimum state in the total energy ETot. However, for proteins with low sequence similarity one has often to use several templates. This can lead to inconsistent constraints or such that compete with the CABS energy. As a consequence, additional minima and barriers are introduced into the energy landscape of the protein. The increased roughness can then lead to a much slower sampling of the energy landscape rendering it difficult or even impossible to find the correct structure.
One way to overcome this so-called multiple minima problem is parallel tempering [3]. In this method one considers an artificial system built up of N non-interacting copies of a molecule, each at a different temperature Ti. In addition to standard Monte Carlo or molecular dynamics moves that affect only one copy, parallel tempering allows also the exchange of conformations between two copies i and j = i + 1 with probability
| (3) |
The resulting random walk in temperature enables configurations to cross energy barriers and move out of local minima leading in this way to an enhanced sampling of low-energy structures.
In the present paper we consider a variant of parallel tempering where all copies are simulated at the same temperature T but differ instead in the parameter ai that describes how strongly the constraint energy EConstraint is coupled to ECABS in replica i:
| (4) |
In each replica, configurations evolve through standard Monte Carlo or Molecular Dynamics moves but are also exchanged between two adjacent copies with probability
Here, Δa = aj - ai and ΔEConstraints = EConstraints(Cj) - EConstraints(Ci). Due to this exchange move configurations perform a random walk on a ladder of models with a 1 > a 2 > a 3 > .... > aN. This random walk works in two ways. The replica which is the closet to the the “physical” system (smallest a) is “fed” with configurations biased toward the correct structure from the replicas with non-vanishing contributions of EConstraints. On the other hand, configurations at the replica where the constraints are fully employed (the highest value of a) can escape out of local minima resulting from the constraints by walking into replicas with diminishing contributions of EConstraints. In this way, sampling of low-energy configurations will be enhanced and the probability increases to find the correct structure.
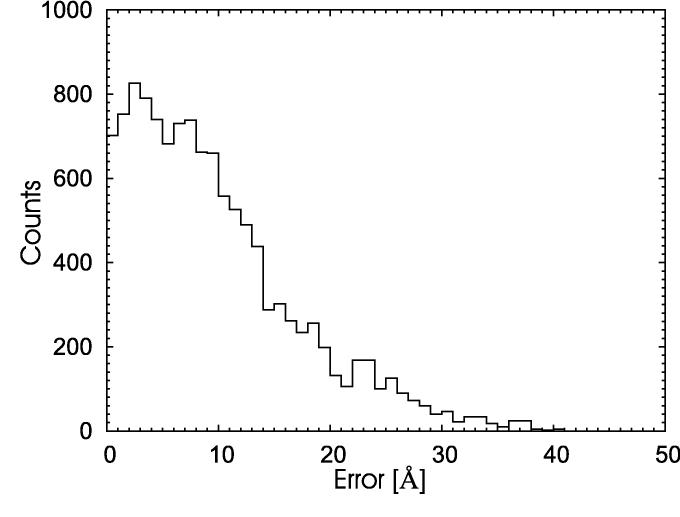
We have selected the target T0206 from last CASP competition to test this assumption. The sequence provided by the organizers has 220 amino acids. However, the CASP organizer provided the additional information that the N-terminal has a collagen-like structure rather than a globular one. That region can be annotated as ‘collagen-like’ by many homology tools rendering the prediction trivial. For this reason, we have in the present work (as during CASP6) simulated only only the 142 C-terminal residues. We have chosen this sequence because it is short enough to allow for sufficient long sampling of the conformational space within our model, but requires at the same time to select the constraints from four different protein domains: 1pk6A, 1pk6B, 1pk6C and 1gr3A. This renders our test more difficult as it introduces frustration and additional local energy minima into the system. This can be seen from the broad and rugged histogram on Figure 1. It shows the distribution of deviations between the true distances, as calculated from the published native structure, and the distances calculated from homology models. A high fraction of distances has been predicted correctly however the distribution has a broad peak between 0 and 10 Å.
Fig. 1.
Distribution of deviations between the true distances, as calculated from the published native structure, and the distances calculated from homology models.
3 Results
In order to test how “tempering” in constraints compares with such in temperature we performed long simulations with both methods. Our simulations use 17 replicas, with 42000 steps for each replica and target. Replicas are exchanged every 30 steps. Each step consists of N two-atom moves, N three-atom moves, 10 N single atom moves and N moves of a larger part of a chain (4-16 atoms) The regular parallel tempering simulations were performed for a fixed (constraint) scaling parameter a = 0.1 with the set of inverse temperatures: 0.526, 0.500, 0.476, 0.455, 0.435, 0.417, 0.400, 0.385, 0.370, 0.357, 0.345, 0.339, 0.333, 0.328, 0.323, 0.317, 0.312. The temperatures are selected around the phase transition point and manually optimized to achieve equal replica swapping ratio for each pair of adjacent copies. In the case of our new approach we choose as (fixed) inverse temperature β = 0.33 and the set of constrains’ scaling parameters: 0.290, 0.280, 0.270, 0.260, 0.240, 0.220, 0.200, 0.180, 0.160, 0.140, 0.120, 0.100, 0.090, 0.080, 0.070, 0.060, 0.050.
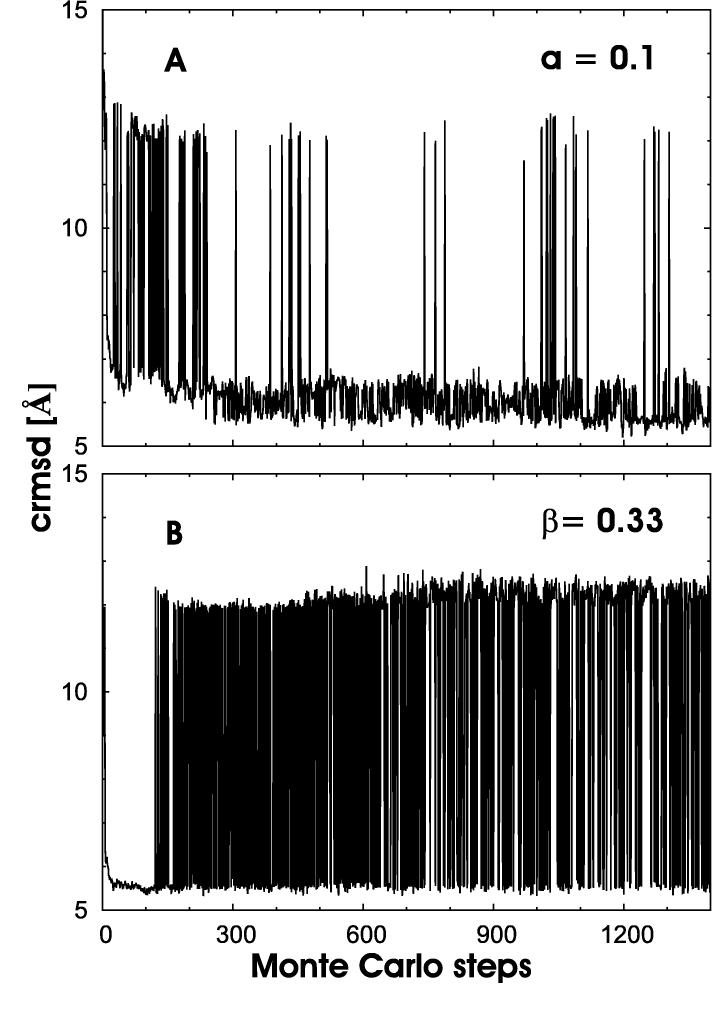
Trajectories of the two runs are shown in Figure 2a (regular parallel tempering) and Figure 2b (tempering in constraint space), respectively. Shown are here the crmsd (root-mean-square deviation between Cα-atoms) as a function of Monte Carlo time. Only data for the lowest temperature (highest β) (regular parallel tempering) or highest constraint scaling factor ai (tempering in constraint space) are shown. Comparing the two graphs one can clearly see the faster sampling in conformation space. In regular parallel tempering the simulation moves 134 times between a region with Rcrmsd > 10 Å and one with Rcrmsd < 6Å. On the other hand, our new variant moves 654 times up and down between configurations with rcrmsd ≤ 6 and rcrmsd 10Å. Consequently, the average time to go from a conformation with rcrmsd ≤ 6 to one with rcrmsd ≥ 10 is 10.5 sweeps for regular parallel tempering, but only t = 2.1 sweeps for our new algorithm.
Fig. 2.
Trajectories of (a) a regular parallel tempering run (in temperature) and (b) a tempering-in-constraints run.
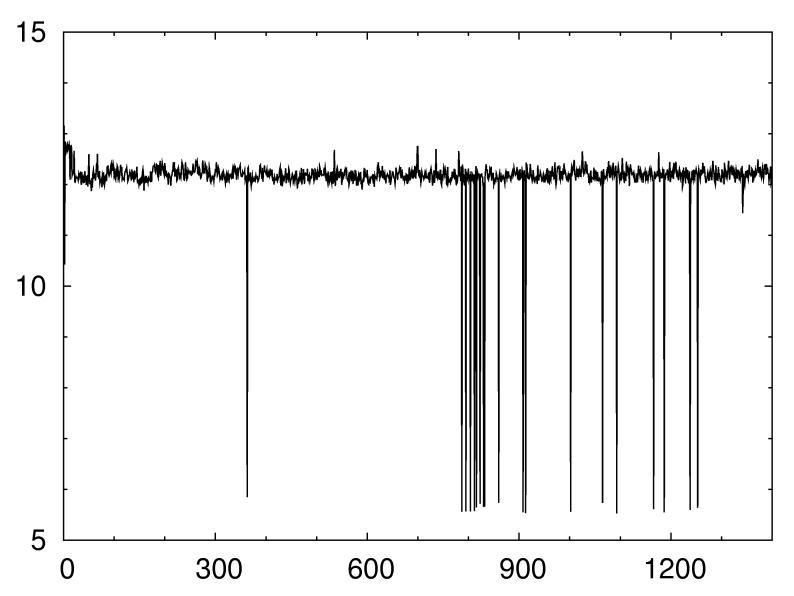
Besides the enhanced sampling our new approach offers also the advantage that in our context it is easier to tune than regular parallel tempering. This is because the efficiency of parallel tempering runs with constraint energy functions does only depend on the temperature distribution but also on the specific value of the scaling parameter a. Figure 3 shows the trajectory of a regular parallel tempering run with the same temperatures as in Figure 2a but slightly different choice of a. The number of independent visits to configurations with Rcrmsd ≤ 6 Å is reduced from 134 (in Figure 2a) to only 38. This strong dependence on the scaling parameter is a serious problem as there is no simple relation to determine the optimal value of a. Its value depends on the number of constraints, their quality and the induced frustration, all quantities that are difficult to estimate if the native structure is not known. On the other hand, in our approach, the user need not to provide a specific value for the scaling parameter as the replicas cover a wide range of values. Instead he has to choose an “optimal” temperature. This is a much easier task as transition temperatures depend mainly on the length of a protein chain and vary little. In CABS it is located in almost all cases between 1.0 and 2.0 kT. Hence, our new approach with its tempering of spatial constraints is much easier to tune. This property could prove important in the development of truly automatic protein structure prediction algorithms.
Fig. 3.
Parallel tempering (in temperature) run at a slightly different value of constraint coupling parameter a.
4 Conclusion
We have presented a variant of the parallel tempering approach for simulation of energy functions with additional constraints. In this variant, the random walk in temperature is replaced by one in the strength of the constraints. In the context of structure prediction simulations that use distance constraints derived from homologous structures the new approach not only leads to a faster and broader sampling but is also easier to tune. It therefore may become useful in the development of reliable automatic prediction server.
Acknowledgments:
Support by a research grant from the National Institutes of Health (GM062838) and a grant from KBN (3 T09A 087 028) is gratefully acknowledged.
Contributor Information
Dominik Gront, Department of Physics, Michigan Technological University, Houghton, MI 49931-1295, USA, Faculty of Chemistry, Warsaw University, Pasteura 1 02-093 Warsaw .
Andrzej Kolinski, Faculty of Chemistry, Warsaw University, Pasteura 1 02-093 Warsaw.
Ulrich H.E. Hansmann, Department of Physics, Michigan Technological University, Houghton, MI 49931-1295, USA .
References
- 1.Lin C-Y, Hu C-K, Hansmann UHE. Proteins: Structure, Functions and Genetics. 2003;52:436. doi: 10.1002/prot.10351. [DOI] [PubMed] [Google Scholar]
- 2.Hansmann UHE. In: New Directions in Statistical Physics - Econophysics, Bioinformatics and Pattern Recognition. Wille LT, editor. Springer Verlag; Berlin: 2004. p. 173. [Google Scholar]
- 3.Hukushima K, Nemoto K. J. Phys. Soc. Jpn. 1996;65:1604. Geyer, G.J., Stat. Sci., 1992; 7:437. [Google Scholar]
- 4.Hansmann UHE. Chem. Phys. Lett. 1997;281:140. [Google Scholar]
- 5.Fukunishi H, Watanabe O, Takada S. J. Chem. Phys. 2004;116:9058. [Google Scholar]
- 6.Kwak W, Hansmann UHE. submitted for publication.
- 7.Kolinski A. Acta. Biochim. Pol. 2004;51:349. [PubMed] [Google Scholar]
- 8.Kolinski A, Bujnicki JM. submitted for publication.
- 9.Kosinski J, et al. Proteins. 2003;53(Suppl 6):369. doi: 10.1002/prot.10545. [DOI] [PubMed] [Google Scholar]
- 10.Eisenberg D, Luthy R, Bowie JU. Methods Enzymol. 1997;277:396. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 11.Kurowski MA, Bujnicki JM. Nucleic Acids Res. 2003;31:3305. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]