Abstract
Early in vertebrate development, endodermal signals act on mesoderm to induce cardiogenesis. The F-type SOXs SOX7 and SOX18β are expressed in the cardiogenic region of the early Xenopus embryo. Injection of RNAs encoding SOX7 or SOX18β, but not the related F-type SOX, SOX17, leads to the nodal-dependent expression of markers of cardiogenesis in animal cap explants. Injection of morpholinos directed against either SOX7 or SOX18 mRNAs lead to a partial inhibition of cardiogenesis in vivo, while co-injection of SOX7 and SOX18 morpholinos strongly inhibited cardiogenesis. SOX7 RNA rescued the effects of the SOX18 morpholino and visa versa, indicating that the proteins have redundant functions. In animal cap explants, it appears that SOX7 and SOX18 act indirectly through Xnr2 to induce mesodermal (Eomesodermin, Snail, Wnt11), organizer (Cerberus) and endodermal (endodermin, Hex) tissues, which then interact to initiate cardiogenesis. Versions of SOX7 and SOX18 with their C-terminal, β-catenin interaction domains replaced by a transcriptional activator domain failed to antagonize β-catenin activation of Siamois, but still induced cardiogenesis . These observations identify SOX7 and SOX18 as important, and previously unsuspected, regulators of cardiogenesis in Xenopus.
Keywords: cardiogenesis, SOX7, SOX18, β-catenin signaling, nodal-signaling
Introduction
The heart is the first functional organ to form during vertebrate development (Foley and Mercola, 2004; Mohun et al., 2003). Generated primarily from mesodermal tissues, in Xenopus the cardiogenic domain is specified during gastrulation (Jacobson and Sater, 1988; Sater and Jacobson, 1989; Sater and Jacobson, 1990) by signals that arise within the organizer and anterior endoderm (Foley and Mercola, 2005; Nascone and Mercola, 1995; Sater and Jacobson, 1990; Schultheiss et al., 1995; Sugi and Lough, 1994). Dorsal lateral domains are brought together through the morphogenic movements of gastrulation and neurulation to form the heart tube. The regulatory interactions involved in cardiogenesis appear to be conserved among both invertebrates and vertebrates (Cripps and Olson, 2002; Davidson and Levine, 2003). The homeobox-containing transcription factor Nkx2.5, and related proteins (Newman and Krieg, 1998), sit atop a regulatory cascade that leads to the differentiation of mesodermal cells into cardiomyocytes.
Wnts are a phylogenetically ancient group of secreted signaling proteins (Jockusch and Ober, 2000; Kusserow et al., 2005; Prud’homme et al., 2002). Wnts can activate a number of downstream signaling pathways, which in turn modulate target gene activity, cell morphology, motility, and polarity (Logan and Nusse, 2004; Miller, 2002; Nelson and Nusse, 2004; Walston et al., 2004). One of the best studied of these Wnt-activated pathways involves stabilization of the cytosolic form of the cell adhesion protein β-catenin. Originally identified through its ability to interact with cadherins and to mediate the anchorage of cytoskeletal systems to cell adhesion sites, cytosolic β-catenin is normally rapidly degraded. In the presence of a “canonical” Wnt signal, cytosolic β-catenin is stabilized, enters the nucleus and interacts with LEF/TCF-type HMG box transcription factors, altering their effects on gene expression.
Studies in Xenopus, chick and mouse suggest the inhibition of Wntmediated, β-catenin-regulated gene expression plays a conserved role in the induction of cardiogenic mesoderm (Lickert et al., 2002; Marvin et al., 2001; Schneider and Mercola, 2001; Tzahor and Lassar, 2001). In X. laevis, inhibitors of β-cateninregulating Wnt signaling act on endoderm, leading to the expression of Hex, which encodes a homeobox-containing transcription factor. Hex expression, in turn, leads to the generation of inductive signals that act on adjacent mesoderm (Foley and Mercola, 2005; Schneider and Mercola, 2001). That this mechanism is conserved in mice is suggested by the phenotype of Hex mutant mice, and the observation that mice carrying a conditional null mutation of β-catenin in the mesoderm produce multiple hearts (Foley and Mercola, 2005; Lickert et al., 2002).
A second “non-canonical” Wnt signaling pathway plays an important role in cardiogenesis. In the chick and in X. laevis, Wnt11 induces cardiogenesis through activation RhoA and Jun-N-terminal kinase (JNK)(Eisenberg and Eisenberg, 1999; Eisenberg et al., 1997; Pandur et al., 2002a; Pandur et al., 2002b). Wnt11 also acts as an antagonist of β-catenin-regulating Wnt signaling (Maye et al., 2004; Pandur et al., 2002a). Wnt 11 is expressed in the developing heart field of the mouse and the chick (Eisenberg et al., 1997; Kispert et al., 1996; Terami et al., 2004) and its expression is elevated as mouse P19 carcinoma and embryonic stem cells differentiate into cardiomyocytes within embryoid bodies (Pandur et al., 2002a; Terami et al., 2004). Circulating endothelial progenitor cells can be induced to differentiate into cardiomyocytes by exposure to Wnt11 (Koyanagi et al., 2005)
A number of observations led us to examine the role of the F-type SOX transcription factors, SOX7, SOX17 and SOX18, in cardiogenesis. The first was the observation that both SOX7 and SOX17 can act as antagonists of β-catenin-mediated Wnt signaling (Takash et al., 2001; Zorn et al., 1999). Next, there was their the pattern of expression. In the mouse and human, SOX7, SOX17 and SOX18 are expressed in the adult heart (Katoh, 2002; Saitoh and Katoh, 2002; Takash et al., 2001; Taniguchi et al., 1999). During mouse development, SOX7 is expressed in the parietal endoderm, but is apparently absent from the definitive endoderm and embryonic gut (Kanai-Azuma et al., 2002). SOX7 appears to be essential for endodermal differentiation in F9 embryonal carcinoma cells, acting up-stream of GATA factors (Futaki et al., 2004; Murakami et al., 2004; Niimi et al., 2004). By embryonic day 7.5, mouse SOX18 is expressed in the extraembryonic allantois and yolk-sac blood islands; shortly after, it is expressed in foregut endothelial and presumptive endocardial cells and in the dorsal aorta and the heart (Pennisi et al., 2000b). SOX17 expression is present at day 6 in the extraembryonic endoderm and later within the definitive endoderm (Kanai-Azuma et al., 2002).
Next, there was the observation that in mice, semi-dominant mutations in SOX18 lead to severe cardiovascular defects, whereas mice homozygous for a null mutation in SOX18 display only a mild coat-color phenotype (Downes and Koopman, 2001; Pennisi et al., 2000a; Pennisi et al., 2000b). This suggests that the mutant protein interferes with the activity of other proteins, perhaps other SOXs, and that one or more related SOXs may compensate for the absence of SOX18 during cardiovascular development.
Finally, our attention was drawn to a possible role for SOX7 and SOX18 in cardiogenesis when, in the course of studies on foregut differentiation, we observed that the injection of SOX7 mRNA induced the expression of markers of cardiogenesis in Xenopus animal cap explants. While we had previously found only mild gross morphological defects following SOX7 morpholino injection (Zhang et al., 2005), the ability of SOX7 (and SOX18) to induce cardiogenic markers in animal caps lead us to more thoroughly examination of the role of these proteins in heart formation. Using a combination of in vivo and explant studies, we find that both SOX7 and SOX18 are expressed in the early cardiogenic region of the Xenopus embryo, and are required for, and capable of inducing, cardiogenesis in a process that depends upon nodal signaling.
RESULTS
Three F-type SOXs have been described in tetrapods, SOX7, SOX17 and SOX18. In X. laevis SOX7 RNA is supplied maternally and is present in the aortic arch, the procardiac tube, within cells of the developing embryonic vasculature, in the notochord, and within the hindbrain (Fawcett and Klymkowsky, 2004). SOX17 α and SOX17 β are expressed zygotically in early embryonic endoderm (Hudson et al., 1997). As in the case of SOX17, there are two SOX18 genes in X. laevis, α and β (Hasegawa et al., 2002). Expression of SOX18β begins following the on-set of zygotic transcription at the mid-blastula transition (Hasegawa et al., 2002); SOX18β RNA is detectable by RT-PCR in both animal and vegetal hemispheres of the embryo at this early stage (FIG. 1A). Expression of SOX18α begins at stage 32 (Hasegawa et al., 2002).
Figure 1. Expression and activity of SOX18β.
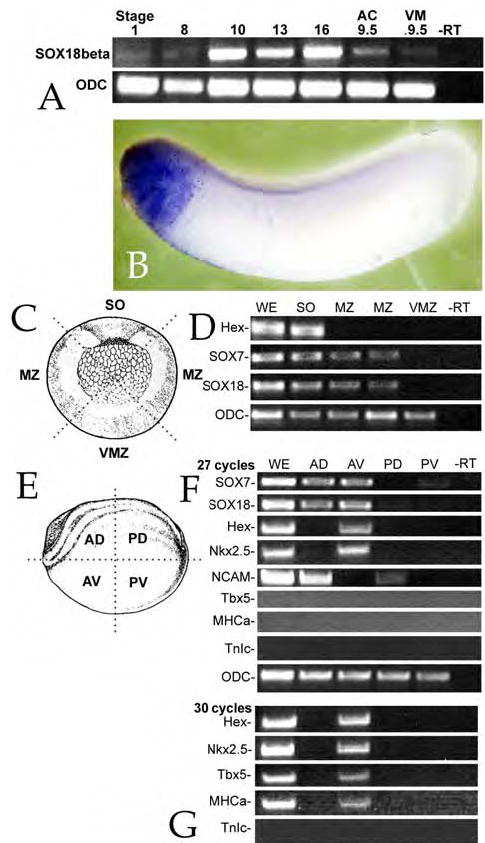
A: Fertilized eggs were analyzed by RT-PCR for SOX18 RNA at various stages. A weak signal was observed in pre-MBT embryos, but strong expression was detected by stage 10. When animal caps and vegetal masses were analyzed at stage 9.5, SOX18β mRNA appeared to be present at similar levels in both regions. Ornithine decarboxylase (ODC) was used as a control for RNA quality throughout. B: In situ hybridization staining with an anti-sense SOX18β probe revealed expression primarily in the anterior region of stage 30 embryos – at earlier stages no consistent, regionspecific staining was observed; no staining was observed using a sense probe (data not shown). C: To define the regional distribution of SOX7 and SOX18 in the gastrula (stage 10/11) embryos, such embryos were dissected into four regions: Spemann Organizer/Dorsal marginal zone (“SO”), lateral marginal zones (“MZ”), and ventral marginal zone (“VZ”). The schematic shows these regions. D: RNA was isolated from the various regions and used for RT-PCR analysis (30 cycles); SOX7 and SOX18 RNAs were found in the SO and MZ, but not in the VZ region; Hex RNA was restricted to the SO region. ODC was found to be uniformly distributed in all four regions. E: A similar analysis was carried out with neurula stage embryos (stage 16/17); these embryos were dissected into anterior dorsal (“AD”), anterior ventral (“AV”), posterior dorsal (“PD”) and posterior ventral (“PV”) regions and RT-PCR analysis was performed. F: Using 27 cycles of amplification, we found SOX7 and SOX18 RNA in both anterior regions, but little if any in the posterior regions. Hex and Nkx2.5 RNAs were found in the AV region only, while NCAM RNA (a marker of neural specification) was found in the two dorsal regions (AD and PD) but not in ventral regions. G: When 30 cycles of PCR were used, Tbx5 and MHCa, but not TnIc RNAs could be detected in the anterior ventral region of the embryo. Images in parts C and E are modified from Nieuwkoop & Faber (1976).
Hasegawa et al., (2002) were unable to localize SOX18β RNA by in situ hybridization; we were also unable to visualize a clear pattern of SOX18β expression in the early embryo via in situ hybridization (data not shown). By stage 30, however, in situ staining for SOX18β RNA was found in the rostral region of the embryo (FIG. 1B).
To better define the pattern of SOX7 and SOX18β expression, gastrula and neurula stage embryos were dissected into distinct regions. In gastrula stage embryos (FIG. 1C,D), both SOX7 and SOX18β RNAs were found in the region of the dorsal and lateral marginal zones; RNA for the anterior endodermal marker Hex was restricted to the dorsal marginal zone. At neurula stage 16/17 (FIG. 1E-G), SOX7 and SOX18 RNAs were found in both anterior dorsal and ventral regions of the embryo, while Hex RNA was restricted to the anterior ventral region. Expression of Nkx2.5, an early marker of the cardiogenic domain (Raffin et al., 2000; Tonissen et al., 1994), was also restricted to the anterior ventral region, whereas NCAM, a marker of neurogenic regions (Jacobson and Rutishauser, 1986; Kintner and Melton, 1987), was restricted to the dorsal regions of the embryo. Markers of cardiogenic differentiation (Tbx5 and MHCα, but not TnIc - see below) could be detected by RT-PCR in stage 16/17 embryos when 30 cycles of amplification were used (FIG. 1G). This contrasts to the previous report that MHCα expression first begins as stage 29/30 (Logan and Mohun, 1993); we attribute this to differences in the sensitivity of the techniques used, that is, RT-PCR versus RNA protection assays.
SOX7 and SOX17 share similar DNA-binding HMG boxes, but activate distinct sets of target genes (Sinner et al., 2004; Zhang et al., 2005). In animal cap explants SOX7 activates expression of the nodal-related protein-encoding genes Xnr1, Xnr2, Xnr4, Xnr5 and Xnr6 in a nodalsignaling independent manner (Zhang et al., 2005). Of these, Xnr4, Xnr5 and Xnr6 appear to be direct targets (see below). In contrast, SOX17β induces the expression of Xnr4 alone (Sinner et al., 2004; Zhang et al., 2005). To determine which Xnr genes were regulated by SOX18, animal caps derived from embryos injected with either SOX7- GFP, mt-SOX17-GFP or SOX18β-GFP RNAs were analyzed by RT-PCR. SOX18β induced the expression Xnr2 and Xnr4, and Xnr1 weakly (FIG. 2). Both SOX7 and SOX18β, but not SOX17, induce the expression of Mixer (FIG. 2), a homeodomain-containing transcription factor involved in mesendodermal differentiation (Henry and Melton, 1998; Kofron et al., 2004). All three SOXs induce expression of the pan-endodermal marker endodermin (Edd)(FIG. 2)(Hudson et al., 1997; Sinner et al., 2004; Zhang et al., 2005).
Figure 2. SOX18 induction of Nodal-related genes in animal caps.
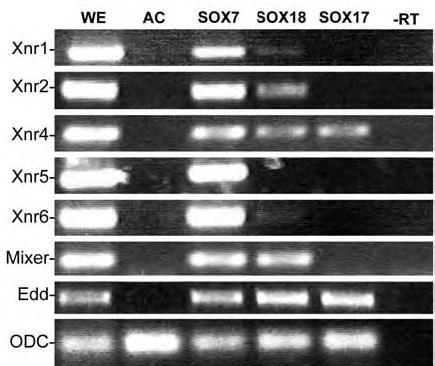
To compare the activity of SOX18 to SOX7 and SOX17 in the animal cap system, fertilized eggs were injected with RNAs encoding SOX7-GFP, mtSOX17β-GFP or SOX18β- GFP (0.5 ng/embryo). At stage 8, animal caps were prepared, cultured for approximately 4 hours (when control embryos had reached stage 10/11) and then analyzed by RT-PCR (Unless otherwise noted, this is the “standard” animal cap assay used throughout this work). As reported previously, SOX7-GFP induced the expression of the five mesoderminducing nodal-related genes Xnr1, Xnr2, Xnr4, Xnr5 and Xnr6, as well as Mixer and Endodermin (Edd), while SOX17β-GFP induce the expression of Xnr4 and Edd, but not Mixer. SOX18β-GFP induced weak expression of Xnr1, and stronger expression of Xnr2, Xnr4, Mixer and Edd.
Redundant roles for SOX7 and SOX18 in cardiogenesis
There are two common methods for studying gene function in Xenopus – the injection of RNA encoding dominant negative polypeptides or the injection of translation-blocking morpholinos (MO). Given the observation that SOX3 and SOX7 share at least partial DNA binding site selectivity (Zhang et al., 2005), there is a real possible of “crossinhibition” when using DNA-binding forms of either SOX7 or SOX18 to study their role in early embryos, where multiple SOXs (e.g. SOXD, SOX3, SOX7, SOX11, SOX17α and β and SOX18β) are expressed (St Amand and Klymkowsky, 2001). We therefore designed morpholinos to specifically target SOX7 (MO7) and SOX18 (MO18) RNAs (FIG. 3A). The SOX7 morpholino (MO7) has previously been shown to block the translation of a SOX7 RNA in embryos and its effects can be rescued by injection of altSOX7-GFP RNA, which contains silent mutations that reduce its affinity for MO7 (Zhang et al., 2005). MO7 has 16 mismatches with the SOX18β RNA, 14 mismatches with SOX17α RNA, and 12 mismatches with the SOX17α RNA. The MO18 is a perfect match to SOX18β and has one mismatch with the SOX18α sequence; it has 16 mismatches to SOX7, and 17 mismatches with SOX17α and SOX17β RNAs. The SOX18MO differs from the SOX18b sequence at 4 sites, although it still blocks translation of the SOX18b RNA.
Figure 3. SOX7 and SOX18 morpholinos.
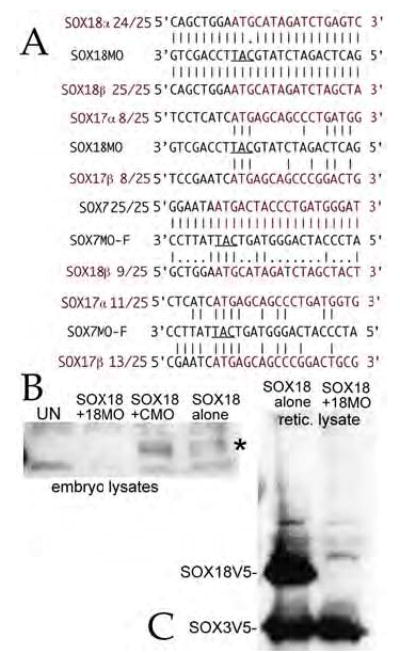
A: The sequence of the morpholinos against SOX7 and SOX18, their alignment with their target sequences, and the analogous regions of the other F-type SOX mRNAs are shown. The ability of the SOX7 morpholino to inhibit SOX7 RNA translation has been established previously (Zhang et al., 2005). B: To test the specificity of the SOX18 morpholino, fertilized eggs where injected with 0.5 ng/embryo utr-SOX18β-V5 RNA either alone or in the presence of control (CMO) or SOX18 (18MO) morpholinos (15 ng/embryo). At stage 8/9 embryonic lysates were generated and analyzed by SDS-PAGE/immunoblot using a monoclonal antiV5 antibody. The accumulation of SOX18β-V5 polypeptides (marked “*”) was blocked by the SOX18 morpholino, but unaffected by the control morpholino. C: Rabbit reticulocyte extracts (25 μL final volume) were programmed with 2 μg each of utr-SOX18β-V5 and SOX3-V5 RNAs either alone or together with 80ng SOX18 morpholino. The reactions were then analyzed by SDS-PAGE and immunoblot using the monoclonal antiV5 antibody. The SOX18 morpholino completely inhibited the accumulation of SOX18-V5 polypeptide, but had no effect on SOX3V5 polypeptide accumulation.
To characterize the ability of the SOX18 morpholino to inhibit the translation of SOX18 RNA, we generated a version of the pCS2 plasmid in which the 5’ UTR region of β-globin was replaced by the 123 base pair region of the SOX18β cDNA characterized Hasegawa et al (Hasegawa et al., 2002); MO18 perfectly matches the utrSOX18V5 RNA (FIG. 3A). We injected utrSOX18V5 RNA (0.5 ngs/embryo) into fertilized eggs either alone, or together with control or SOX18 morpholinos (15 ngs/embryo); embryos were analyzed at stage 9/10 by immunoblot using a monoclonal antiV5 epitope antibody. MO18 blocked the accumulation of the SOX18V5 polypeptide, while the control morpholino had no apparent effect (FIG. 3B). We also tested the specificity of the MO18 morpholino using a rabbit translation reticulocyte in vitro assay programmed with both SOX3V5 and utrSOX18V5 RNAs; addition of MO18 inhibited SOX18V5 accumulation, but had no discernable effect on SOX3V5 accumulation (FIG. 3C).
We injected either MO7 and MO18 (total final concentration 30 ng/embryo) into animal and vegetal sites of fertilized eggs and examined the phenotypes at larval stages. Gastrulation appeared normal and, as described previously for SOX7, the larvae displayed only mild gross morphological phenotypes (FIG. 4)(Zhang et al., 2005). When such embryos were stained in situ for either myosin heavy chain α (MHCα) RNA, a specific marker of cardiomyocyte differentiation (Logan and Mohun, 1993)(FIG. 4B-D) or Nkx2.5 RNA (FIG. 4F), both MO7 (FIG. 4B,E) and MO18 (FIG. 4C,E) produced dramatic decreases in the extent and intensity of staining; no effect on these markers was observed when control morpholino was injected (data not shown). When the MO7 and MO18 were injected together (each 15ng/embryo), the complete loss of expression of Nkx2.5 (FIG 4F) and MHCα (FIG. 4D) was more frequent (Table 2). It is worth noting that while some morpholino-injected embryos displayed aberrant head morphologies, others appeared quite normal, even while the expression of cardiac markers was extinguished (see FIG. 4D,F).
Figure 4. Inhibition of cardiogenesis by SOX7 and SOX18 morpholinos.
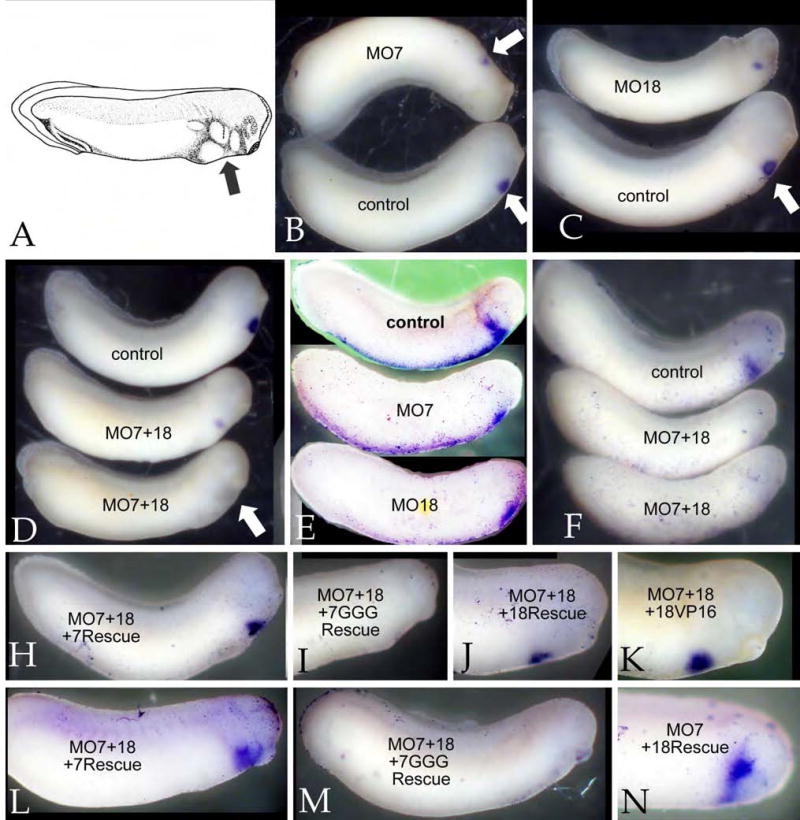
Fertilized eggs were injected at animal and vegetal sites with a total of 30 ngs of morpholino and examined at stage ~30. A: A schematic of a stage 30 embryo showing the location of the cardiogenic region (“arrow”)(Image from Nieuwkoop & Faber, 1976). In situ hybridization using antisense RNAs directed against either MHCα (B-D) or Nkx2.5 (E,F). Both the SOX7 morpholino (MO7)(B,E) and the SOX18 morpholino (MO18) (C,E) reduced the size and intensity of the MHCα and Nkx2.5 staining domains. A combination of both morpholinos (15+15 ng/embryo)(D,F) produced a more complete suppression of MHCα and Nkx2.5 staining (see also Table 2). Rescue studies: To confirm the specificity of the morpholino effects, MO7+MO18-injected embryos were injected with RNAs (0.5 ng/embryo) encoding either altSOX7-GFP (H,L), SOX7GGG-GFP (I,M), SOX18β-GFP (J,N), or mtSOX18βΔC-VP16 (18VP16)(K); at stage 30 the embryos were stained in situ for MHCα (H-K) or Nkx2.5 (L-N). SOX7, SOX18, and mtSOX18βΔC-VP16 rescued the effects of the combined morpholinos, whereas SOX7GGG-GFP did not.
TABLE 2.
| normal | reduced | absent | ||||
|---|---|---|---|---|---|---|
| treatment | MHCα | Nkx2.5 | MHCα | Nkx2.5 | MHCα | Nkx2.5 |
| Control (uninjected) | 16/16 (100%) | 0/16 (0%) | 0/16 (0%) | |||
| Control MO (20 ng/embro) | 21/24 (87.5%) | 11/13 (85%) | 3/24 (12.5%) | 2/13 (15%) | 0/24 (0%) | 0/13 (0%) |
| SOX7 MO 30 ng/embryo | 3/25 (12%) 3/19 (16%) | 4/15 (27%) | 14/25 (56%) 10/19 (53%) | 5/15 (33%) | 8/25 (32%) 6/19 (32%) | 6/15 (40%) |
| SOX7 MO + 0.5 ng/embryo altSOX7-GFP RNA | 51/74 (69%) | 17/22 (77%) | 19/74 (26%) | 3/12 (25%) | 4/74 (5%) | 2/12 (17%) |
| SOX7 MO + 0.5 ng/embryo SOX18β-GFPRNA | 7/12 (58%) | 3/12 (25%) | 2/12 | |||
| SOX7 MO + 0.5 ng/embryo mtSOX18βΔC-VP6 RNA | 6/10 (60%) | 3/10 (30%) | 1/10 (10%) | |||
| SOX18MO (30 ng/embryo) | 2/15 (13%) 3/13 (23%) | 3/12 (25%) | 7/15 (47%) 8/13 (62%) | 6/12 (50%) | 6/15 (40%) 2/13 (15%) | 3/12 (25%) |
| SOX18MO + 0.5 ng/embryo altSOX7-GFP RNA | 28/41 (68%) | 13/15 (87%) | 9/41 (22%) | 2/15 (13%) | 4/41 (10%) | 0/15 (0%) |
| SOX18MO + 0.5 ng/embryo mtSOX7βΔC-VP6 RNA | 7/11(64%) | 3/11(27%) | 1/11(9%) | |||
| SOX7+SOX18 MO (15+15 ngs/embryo) | 1/23 (4%) 3/22 (14%) | 0/13 (0%) | 2/23 (9%) 8/22 (36%) | 9/13 (69%) | 20/23 (87%) 11/22(50%) | 4/13 (31%) |
| SOX7+SOX18 MO + 0.5 ng/embryo altSOX7-GFP RNA | 14/17 (82%) | 11/15 (73%) | 2/17 (12%) | 2/15 (13%) | 1/17 (6%) | 4/13 (31%) |
| SOX7+SOX18 MO +0.5 ng/embryo SOX7GGG-GFP RNA | 1/24 (4%) | 2/12 (17%) | 12/24 (50%) | 8/12 (75%) | 11/24(46%) | 2/12 (17%) |
The expression of both MHCα and Nkx2.5 in MO18 injected embryos could be rescued by injection of RNA encoding altSOX7-GFP (0.5ng/embryo)(data not shown), while the injection of SOX18β-GFP (0.5ng/embryo) rescued the effects of MO7 (Fig. 4N and data not shown). Similarly, the loss of Nkx2.5 and MHCα expression observed in MO7+MO18 injected embryo was efficiently rescued by either altSOX7-GFP RNA or SOX18β-GFP RNA injection (FIG. 4H, J, L). In contrast, little or no rescue was observed using a mutated form of SOX7, SOX7GGG-GFP (FIG. 4I,M; see below). Together these observations indicate that SOX7 and SOX18 are required for normal cardiogenesis and have largely redundant activities.
SOX7, SOX18 and the cardiogenic gene regulatory network
It is possible that SOX7 and SOX18 are not necessary for cardiogenesis per se, but are required for developmental processes upon which cardiogenesis depends. Previous studies indicate that animal ectodermal explants, animal caps, form cardiac tissue and even beating hearts following activin treatment (Ariizumi et al., 2003; Logan and Mohun, 1993; Okabayashi and Asashima, 2003). In contrast, naïve animal caps differentiate into atypical epidermis, and do not express endodermal, mesodermal or cardiac markers (Sive et al., 2000). If SOX7 and SOX18 are actively involved in cardiogenesis, they might be able to induce expression of markers of cardiogenesis when introduced into animal caps. Animal caps were generated at stage 8 from embryos injected with SOX7-GFP RNA (0.1–0.5 ng/embryo) and cultured until controls reached stage 30 and then stained in situ for Nkx2.5 (FIG. 5A) or MHCα (FIG. 5B) – in each case, a single distinct domain of staining was observed; no staining was observed in uninjected animal caps (data not shown). Markers of cardiogenesis were examined by RT-PCR, when control embryo had reached stage 25, we obtained a similar result (FIG. 5C). In addition to Nkx2.5 and MHCα, both SOX7-GFP and SOX18β-GFP induced expression of the heart specific T-box transcription factor Tbx5 (Horb and Thomsen, 1997), the cardiac-specific structural protein troponin c inhibitory chain (TnIc)(Drysdale et al., 1994), as well as the endodermal marker endodermin. It is worth noting that, as in the case of injection of Hex RNA, which also induces the expression of cardiac markers in animal caps (Foley and Mercola, 2005), we did not observe obvious beating tissue in SOX7 or SOX18 RNA injected animal caps (data not shown). When SOX7-GFP or SOX18β-GFP RNAs were injected into fertilized eggs and intact embryos were examined by in situ hybridization; the extent and intensity of the Nkx2.5 and MHCα expression domains appeared to be increased compared to uninjected controls (data not shown; see supplemental figure). In contrast to the effects of SOX7 and SOX18, injection of RNA encoding mtSOX17β-GFP induced endodermin but none of the cardiac markers (FIG. 5C).
Figure 5. Induction of cardiogenesis in animal caps.
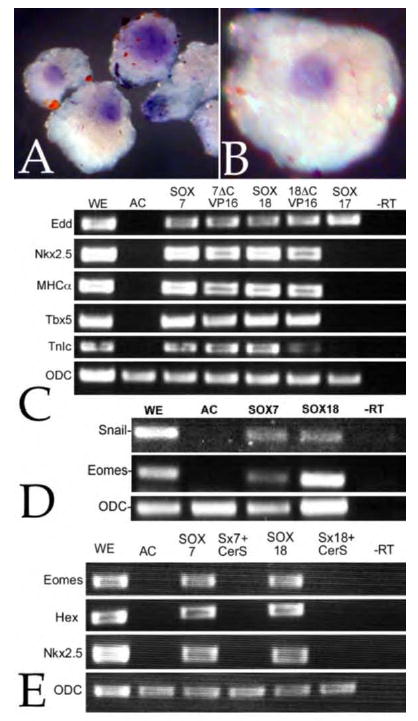
To determine whether SOX7 induced markers of cardiogenesis, fertilized eggs were injected with RNA encoding SOX7-GFP (0.5ng/embryo); animal caps were prepared at stage 8 and analyzed by in situ hybridization when control embryo had reached stage 30. Both Nkx2.5 (A) or MHCα (B) were expressed in discrete domains, one per animal cap; no expression of either gene was observed in cap derived from uninjected embryos (data not shown). C: A similar analysis was carried using RT-PCR. Fertilized eggs were injected with RNA (0.5 ng/embryo) encoding either SOX7-GFP (SOX7), mtSOX7ΔC-VP16 (7ΔCVP16), SOX18β-GFP (SOX18), mtSOX18ΔC-VP16 (18ΔCVP16), or mtSOX17β-GFP (SOX17). Animal caps were prepared and analyzed when controls reached stage 25. Each of the injected RNAs induced the expression of the endodermal marker Edd, but only SOX7 or SOX18 constructs induced markers of cardiogenesis, Nkx2.5, MHCα Tbx5 and TnIc. D: In a similar study, SOX7-GFP and SOX18β-GFP induced expression of the mesodermal markers Snail and Eomesodermin (Eomes). E: The SOX7-GFP and SOX18β-GFP induction of Eomes, Hex and Nkx2.5 expression was blocked by the coinjection of RNA encoding the nodal inhibitor CerS (0.2 ng/embryo).
Supplemental figure.
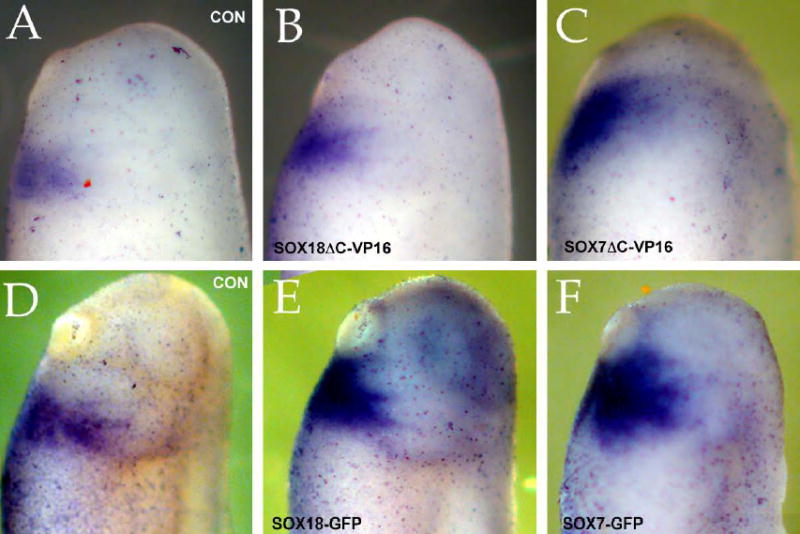
Fertilized eggs were injected with either SOX18βΔC-VP16myc (B), SOX7ΔC-VP16myc (C), SOX18β-GFP (E) or SOX7-GFP (F) RNAs (0.5 ng/embryo). At stages between 25 (A-C) and 30 (D-F) the embryos were stained in situ for Nkx2.5. Compared to uninjected controls (A,D), both GFP and ΔCVP16myc forms of SOX7 and SOX18 induced an apparent increase in the extent and intensity of the Nkx2.5 expression domain.
Cardiogenesis involves mesodermal tissues. Both SOX7 and SOX18β, but not SOX17, induce expression of Xnr2 which in turn has been reported to activate expression of Eomesodermin (Eomes)(Loose and Patient, 2004), a T-box containing transcription factor expressed very early in mesodermal specification and known to play a role in cardiac development (Ryan et al., 1996; Ryan et al., 2004; Sousa-Nunes et al., 2003). Both SOX7 and SOX18 induced animal cap expression of Eomes and Snail, which encodes a Zn+-finger transcription factor expressed in the embryonic mesoderm (Essex et al., 1993)(FIG. 5D). SOX7 and SOX18β induction of animal cap Eomes and Snail expression was inhibited by the co-injection of RNA encoding the nodal-specific antagonist CerS (FIG. 5E), indicating that this induction is indirect.
To further characterize how SOX7 (and presumably SOX18) acts to induce cardiogenesis, we generated a plasmid encoding a dexamethasone-regulated version of the polypeptide, GR-SOX7-GFP. In the absence of dexamethasone, GRSOX7-GFP RNA is translated but the polypeptide is expected to be inactive due to interactions with hsp90 (Sive et al., 2000). When animal caps expressing GR-SOX7-GFP were analyzed in the absence of dexamethasone, there was no detected activation of known SOX7 targets; upon addition of dexamethasone, however, there was clear expression of the mesoderminducing nodal-related protein encoding genes Xnr1, Xnr2, Xnr4, Xnr5 and Xnr6 (FIG. 6). When animal caps were pretreated with the protein synthesis inhibitor emetine (commonly used in studies of cell cycle proteins) prior to, and during dexamethasone activation, expression of Xnr4, Xnr5 and Xnr6 remained robust, suggesting that these are immediate-early, i.e., direct targets of SOX7 regulation. The expression of Eomes was blocked by emetine treatment, confirming that it is an indirect target of SOX7 regulation. Emetine-treatment alone did not lead to the expression of any of the genes examined (FIG. 6).
Figure 6. Direct versus indirect targets of SOX7 regulation.
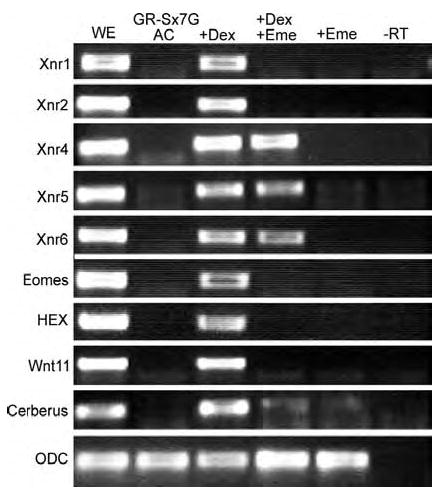
Fertilized eggs were injected with RNA encoding GR-SOX7-GFP (0.5 ng/embryo). Animal caps were treated in four different ways: either left in standard media (AC) with 1% ethanol (the carrier for the dexamethasone), incubated with 20 μM dexamethasone (+DEX), pretreated with 100 μg/ml emetine for 30 minutes and then incubated with 20mM dexamethasone and 100 μg/ml emetine (+DEX +Eme), or incubated with 100 μg/ml emetine alone (+Eme). In contrast to cycloheximide (see Sinner et al 2004), emetine treatment had no apparent effect on the expression of any of the genes examined in the absence of dexamethasone. In the presence of dexamethasone, the Xnrs were induced, along with Eomes, Hex, Wnt11 and Cerberus; of these genes, Xnr4, Xnr5 and Xnr6 were expressed in the presence of both dexamethasone and emetine, indicating that they are direct targets of SOX7 regulation.
Formation of the heart depends upon the release of inhibitors of canonical Wnt signaling from the organizer that act on the endoderm (Foley and Mercola, 2005; Schneider and Mercola, 2001). In addition, Wnt11 both inhibits canonical Wnt signaling and acts through the protein kinase C (PKC) and jun kinase (JnK) to induce cardiogenesis (Pandur et al., 2002a). Cerberus encodes a secreted Wnt/BMP/Nodal inhibitor first expressed in the deep region of the organizer (Bouwmeester et al., 1996; Piccolo et al., 1999; Schneider and Mercola, 1999), while Hex encodes a homeobox-containing protein expressed in the anterior endoderm in response to the inhibition of Wnt signaling in the endoderm (Foley and Mercola, 2005; Jones et al., 1999; Smithers and Jones, 2002). Both SOX7 and SOX18β induced expression of Cerberus (FIG. 6 & 7C), Hex (FIG. 5E & 6), and Wnt11 (FIG. 6 & data not shown) in animal caps. Two lines of evidence indicate that SOX7/SOX18 activation of Cerberus, Hex and Wnt11 is indirect: GR-SOX7-GFP induction of Cerberus, Hex, and Wnt11 was blocked by the protein synthesis inhibitor emetine (FIG. 6), and the SOX7/18-induced expression of Eomes, Hex and Nkx2.5 was blocked by injection of RNA encoding the nodal-inhibitor, CerS (200 pg/embryo)(FIG. 5E & data not shown)
Figure 7. SOX7 antagonism of b-catenin signaling and cardiogenic ability.
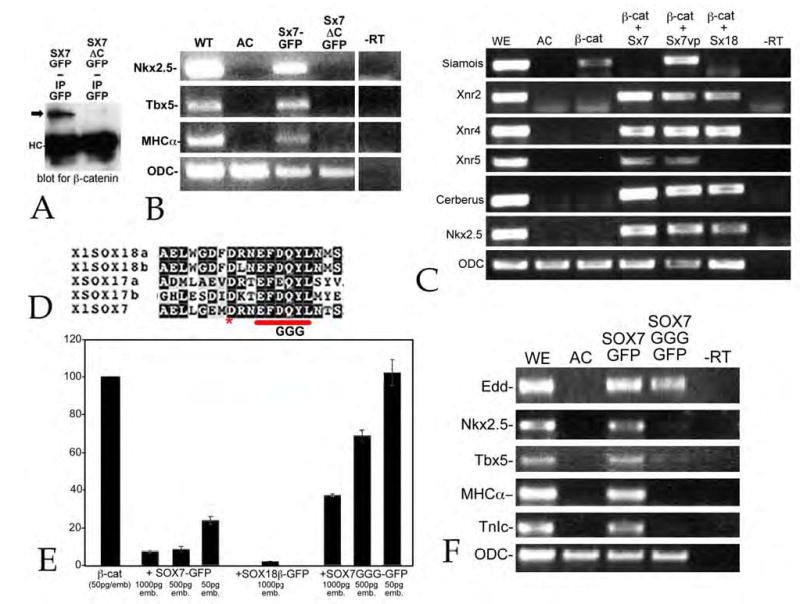
A: Fertilized eggs were injected with RNA encoding either SOX7-GFP (Sx7GFP) or SOX7ΔC-GFP (Sx7ΔC-GFP)(0.5 ng/embryo); at stage 9/10 embryo lysates were prepared and immunoprecipitated with a rabbit antiGFP antibody. Immunoprecipitates where analyzed by SDS-PAGE and immunoblot using a rabbit anti-β-catenin antibody. β-catenin was coprecipitated with SOX7-GFP (arrow), but little or no β-catenin co-precipitated with SOX7ΔC-GFP (immunoglobulin heavy chain marked “HC”). Immunoblot with antiGFP antibody of embryonic lysates indicated that both SOX7-GFP and SOX7ΔC-GFP polypeptides accumulated to similar extents (data not shown). B: In an animal cap assay, injection of RNA (0.5 ng/embryo) encoding SOX7ΔC-GFP failed to induce Nkx2.5, Tbx5 or MHCα expression (animal caps analyzed when control embryos reached stage 25). C: Injection of RNA encoding a mutationally stabilized form of β-catenin (mt-ΔG-β-catenin) induces the expression of Siamois in animal caps. Fertilized eggs were injected with RNA encoding mt-ΔG-β-catenin (50 pg/embryo) alone or together with RNAs encoding SOX7-GFP, mt-SOX7ΔC-VP16 or SOX18β-GFP (500 pg/embryo); animal caps were prepared at stage 8 and analyzed when control embryos reached stage 10/11. β-catenin induced expression of Siamois; both SOX7-GFP and SOX18β-GFP suppressed β-catenin-induced Siamois expression, and induced expression of Xnr2, Xnr4, Cerberus and Nkx2.5; SOX7-GFP also induced Xnr5 expression. SOX7ΔC-VP16 failed to suppress β-catenin-induced Siamois expression, but induced Xnr2, Xnr4, Xnr5, Cerberus and Nkx2.5 expression. D: The SOX7GGG-GFP construct is analogous to the SOX17G3 construct generated and characterized by Sinner et al (2004). In animal caps assayed at stage 25, SOX7GGG-GFP (0.5 ng/embryo) induced the expression of Edd, but not Nkx2.5, Tbx5, MHCα or TnIc. E: To examine the ability of SOX7-GFP, SOX7GGG-GFP and SOX18β-GFP to inhibit β-catenin activation of the OT reporter, fertilized eggs were injected with OT and pTK-Renilla DNAs (20 pgs each/embryo) together with RNAs encoding mt-ΔG-β-catenin (50 pg/embryo) and varying amounts of SOX RNAs; animal caps were analyzed when control embryos reached stage 10/11. SOX7-GFP and SOX18β-GFP RNAs inhibited of β-catenin activation of OT; SOX7GGG-GFP was a much less active antagonist of β-catenin in this assay.
SOX7 and SOX18 are β-catenin-signaling antagonists
SOX7 and SOX17 have previously been identified as antagonists of β-catenin activation of the β-catenin/TCF regulated OT reporter (Takash et al., 2001; Zorn et al., 1999). The β-catenin binding domain of SOX17 has been localized to its C-terminus (Sinner et al., 2004; Zorn et al., 1999). We generated a version of SOX7, SOX7ΔC-GFP, which lacks this C-terminal domain, beginning 20 residues downstream of the HMG box. In immunoprecipitation studies, SOX7-GFP co-precipitated endogenous β-catenin, while SOX7ΔC-GFP failed to co-precipitate β-catenin (FIG. 7A). In animal cap studies, SOX7ΔC-GFP failed to induce the expression of Nkx2.5, Tbx5 or MHCα (FIG. 7B), although it was able to bind to DNA (data not shown). Replacing the C-terminal domains of SOX7 and SOX18βwith a VP16 transcriptional activation domain (Sadowski et al., 1988) restores the ability of these proteins to induce the cardiogenic markers Nkx2.5, Tbx5, MHCα and TnIc (FIG. 5C).
In animal caps, stabilized β-catenin induces the expression of the TCFregulated gene Siamois (Brannon and Kimelman, 1996; Fagotto et al., 1997). Coinjection of either SOX7-GFP or SOX18β-GFP RNAs suppressed β-catenin-mediated activation of Siamois in this assay, while mtSOX7ΔC-VP16 did not (FIG. 7C). Both mtSOX7ΔC-VP16 and mtSOX18βΔC-VP16 RNAs retained the ability, when injected into fertilized eggs, of increasing the extent and intensity of the Nkx2.5 expression domains, as determined by in situ hybridization (data not shown - supplemental figure 1), and both can rescue the loss of MCHα staining produced by SOX7/18 morpholino injection (FIG. 2K; Table 2).
Sinner et al (2004) identified a conserved sequence motif in the tail domain of F-type SOXs as a critical β-catenin binding element in SOX17 (FIG. 7D). We generated a version of SOX7, SOX7GGG-GFP, analogous to their SOX17-3G construct. We compared the ability of SOX7-GFP, SOX18β-GFP and SOX7GGGGFP to inhibit β-catenin activation of the OT reporter in animal caps (FIG. 7E). Fertilized eggs were injected with linearized OT and pTK-Renilla luciferase plasmids, and RNAs encoding a mutationally stabilized form of β-catenin and either SOX7-GFP, SOX18β GFP or SOX7GGG-GFP. Both SOX7 and SOX18β were potent inhibitors of OT activation, whereas SOX7GGG-GFP was reproducibly less effective (FIG. 7E). In animal caps, SOX7GGG-GFP retained the ability to induce the expression of the endodermal marker Edd but failed to induce expression of the markers of cardiogenesis (FIG. 7F). In the embryo, injection of RNA encoding SOX7GGG-GFP failed to rescue the effects of SOX7 and SOX18 morpholinos (FIG. 4I,M; TABLE 2).
Discussion
There are three main conclusions from the studies presented here. The first is the demonstration, based on the effects of targeted morpholinos, RNA rescue, and animal cap studies, that the F-type SOXs, SOX7 and SOX18 play an unexpected, essential and overlapping role in cardiogenesis in Xenopus. The second is that while SOX7, SOX17 and SOX18 can each inhibit β-catenin-mediated gene/reporter activation, this activity is not sufficient for them to induce cardiogenesis; while both SOX7 and SOX18 are cardiogenic, SOX17 is not. Moreover, a version of SOX7 in which the C-terminal β-catenin binding domain is replaced by a generic transcription activation domain still induces the expression of cardiogenic markers. Finally, based on an analysis of their activity in animal caps, the cardiogenic effects of SOX7 and SOX18 appear to depend upon the indirect activation of Xnr2 in particular and nodal signaling in general.
Overlapping functions of SOX7 and SOX18 in cardiogenesis
A role for SOX18 in cardiovascular development was first suggested by the observation that semidominant mutations in SOX18 lead to cardiovascular defects (Pennisi et al., 2000b). On the other hand, the observation that mice homozygous for a SOX18 null mutation have no such defect (Pennisi et al., 2000a) implies that the mutant SOX18 proteins must disrupt the functions of genes other than SOX18 to produce the cardiovascular phenotype observed. In a previous study, we found that a “dominantnegative” version of SOX7 produces a much more severe gross morphological phenotype that did the SOX7 morpholino, presumably because of their ability to interfere with the activity of other SOX proteins (Zhang et al., 2005). This lead us to avoid the use of “dominant-negative” constructs in this study.
Both SOX7 and SOX18β RNAs are present in the embryo during the period of cardiogenic specification (Fawcett and Klymkowsky, 2004; Hasegawa et al., 2002; Shiozawa et al., 1996)(FIG. 1) and the coinjection of SOX7 and SOX18 morpholinos produces a more complete suppression of cardiac marker expression than does either morpholino alone (FIG. 3; Table 2) supporting the conclusion that both proteins are normally involved in cardiogenesis. At the same time, the observation that either SOX7 or SOX18 RNA can rescue the loss of cardiac markers produced by SOX7 or SOX18 morpholinos, when used alone or together, indicates that the two proteins are functionally similar, at least at this level of resolution (FIG. 2; Table 2). It is possible that the expression of both genes is required to produce a level of protein activity necessary to insure that cardiogenesis occurs normally. It remains to be determined whether the combination of SOX7 and SOX18 are required for cardiogenesis in other organisms, such as the mouse or human, or what roles they may play in different tissues.
Wnt signaling and the cardiogenic activities of SOX7 and SOX18
The inhibition of canonical Wnt signaling in the endoderm, by Wnt inhibitors secreted by the organizer and Wnt11, expressed in the mesoderm, plays a key role in the induction of mesodermal cardiogenesis (Foley and Mercola, 2004; Foley and Mercola, 2005; Mohun et al., 2003; Pandur et al., 2002a; Sater and Jacobson, 1989; Sater and Jacobson, 1990). SOX7, SOX17 and SOX18 can each induce endodermal differentiation, as revealed by their ability to induce expression of Endodermin. They also each antagonize β-catenin-regulated gene expression (FIG. 7)(Sinner et al., 2004; Takash et al., 2001; Zorn et al., 1999). That said, it is clear that the ability to induce endodermal differentiation and to antagonize β-catenin signaling is not in and of itself the reason SOX7 and SOX18 are cardiogenic, since SOX17 does not induce cardiogenic markers in the animal cap system.
To further define the role, if any, of the anti-β-catenin activity of SOX7 and SOX18 in cardiogenesis, we replaced the Cterminal β-catenin binding domain of SOX7 or SOX18 with a virally-derived, transcriptional activation domain. The resulting “VP16” polypeptides retained the ability to induce cardiac markers in animal caps and to rescue the effects of SOX7 and SOX18 morpholinos on heart markers in vivo, but did not suppress β-catenininduction of Siamois. On the other hand, a mutant form of SOX7, SOX7GGG-GFP, reduced, but did not eliminate, the anti-β-catenin activity of SOX7 in the OT assay, failed to suppress the β-catenin activation of Siamois expression, and, while clearly retaining some transcriptional activity, was unable to either induce the expression of cardiogenic markers in animal cap explants or to rescue the effects of SOX7 and SOX18 morpholinos on cardiogenesis in vivo. Our results support the conclusions of Sinner et al (2004), working with an analogous mutation in SOX17, that this conserved region of the SOX7/SOX17 polypeptides is clearly critical for some, but not all of the transcriptional activities of these proteins. One possibility is that the proteins differ quantitatively rather than qualitatively, and that threshold effects are involved in the differences in gene expression patterns they provoke. A detailed quantitative analysis that examines direct and indirect targets should be able to identify qualitative differences associated with the GGG mutation if they exist, and is currently planned.
SOX7 and SOX18 regulation of nodal expression
Given the apparent role of SOX7 and SOX18 in cardiogenesis, the next question we sought to answer was, where in the cardiogenic pathway do these proteins act? To place SOX7 and SOX18 in the cardiogenic pathway, we examined their effects in animal caps. Although the F-type SOXs have similar DNA binding domains, they differ in the genes they activate. SOX17 activates only Xnr4, SOX18 activates Xnr2 and Xnr4, while SOX7 activates Xnr1, Xnr2, Xnr4, Xnr5 and Xnr6 (Sinner et al., 2004; Zhang et al., 2005)(this work). Given that the pattern of Xnr activation is similar between full length and ΔC-VP16 versions of SOX7 and SOX18, it appears that this specificity resides in the N-terminal and HMG box regions of the proteins.
Based on the use of hormoneregulated forms of SOX7 (this work) and SOX17 (Sinner et al., 2004), Xnr4 appears to be a direct target of both proteins, but whether they bind to common or distinct sites is not yet known. We expect, but have yet directly determined, that SOX18 also directly regulates Xnr4 and, like SOX7, indirectly regulates Xnr2. Since SOX7 induces Xnr2 expression in the presence of the nodal inhibitor CerS (Zhang et al., 2005), it appears that SOX7 acts through an as yet unidentified intermediate to activate Xnr2 expression. It should be possible to use the GR-SOX7-GFP construct to identify this intermediate factor, particularly as Xenopus DNA microarrays become more complete.
Nodals and cardiogenesis
Although there may well be other targets involved in the regulation of cardiogenesis by SOX7 and SOX18, their common activation of Xnr2, together with the failure of SOX17 to activate Xnr2, and the ability of CerS to block the expression of cardiac markers by SOX7 and SOX18, suggests that Xnr2 plays a key upstream role in the induction of cardiogenesis (FIG. 8). A number of previous studies demonstrate that, with the exception of Xnr3, all of the Xnrs induce mesendodermal specification (Schier, 2003). The requirement for nodals in mesoderm formation in general is based on the inhibition of mesoderm formation by the nodal-inhibitor CerS (Agius et al., 2000; Piccolo et al., 1999) and the defects in mesoderm formation observed in mice null for Nodal (Conlon et al., 1994).
Figure 8. SOX7, SOX18 and the cardiogenic pathway.
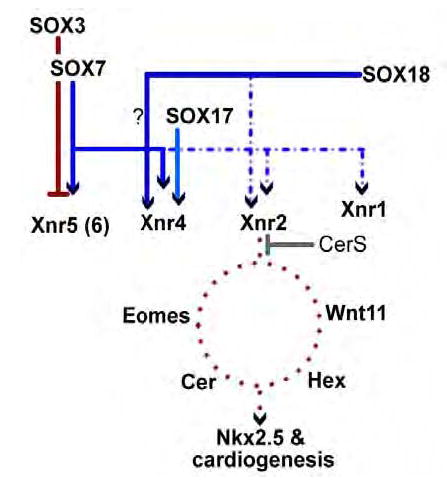
In this diagram, direct interactions are indicated by solid lines, indirect interactions by dotted lines. SOX17 induces Xnr4, but fails to induce cardiogenesis in animal caps, whereas SOX7 and SOX18 induce both Xnr2 and Xnr4 and induce cardiogenesis. The presence of the nodal inhibitor CerS inhibits SOX7 and SOX18-induced cardiogenesis. How other direct and indirect targets of SOX7 and SOX18 interact with “downstream” targets of Xnr-regulation (e.g. Eomes, Wnt11, Cerberus and Hex) remains unclear - nevertheless, the end result in the activation of Nkx2.5 and other cardiac markers by SOX7 and SOX18.
Identifying the specific roles of the specific Xnrs in the patterning of the early Xenopus embryo in general, and cardiogenesis in particular is, for a number of reasons, not completely straightforward. Aside from the rather extravagant number of nodal genes in X. laevis (6 compared to 3 in the zebrafish and 1 in the mouse)(Schier, 2003; Weng and Stemple, 2003; Whitman, 2001), there is the added complexity that nodals, like other members of the TGFβ-superfamily, normally act as dimers and can heterodimerize with other TGFβ polypeptides, such as Derrierre and BMPs (Eimon and Harland, 2002; Yeo and Whitman, 2001). Commonly used dominant “cleavage mutant” forms of Xnrs inhibit the activity of both nodals and other TGFβ family members (Onuma et al., 2002; Takahashi et al., 2000), but may not inhibit all nodal functions (Eimon and Harland, 2002). At the same time, nodals regulate each other’s expression in a dose dependent fashion, e.g., low levels of Xnr5 and Xnr6 induce the expression of Derriere, while higher levels lead to Xnr1, Xnr2 and Xnr4 expression (Takahashi et al., 2000). Nodals also can diffuse some distance from the cells that secrete them (Williams et al., 2004). Finally, nodals can use the same receptors as other TGFβ family members (Chen et al., 2004; Kumar et al., 2001; Reissmann et al., 2001).
That the details of nodal signaling remain to be fully understood is illustrated by experiments on SOX regulation of nodal activity. In animal caps, SOX7 induces the expression of Xnr1, Xnr2, Xnr4, Xnr5 and Xnr6; each of these nodals has been found to induce expression of prospective posterior mesodermal marker Xbra in animal caps (Joseph and Melton, 1997; Osada and Wright, 1999; Takahashi et al., 2000), yet SOX7 does not induce Xbra expression in animal caps (Zhang et al., 2005). Similarly, SOX17 induces expression of Xnr4, which can induce Xbra, but SOX17 does not induce Xbra expression (Joseph and Melton, 1997; Sinner et al., 2004). Finally, both Xnr5 (Takahashi et al., 2000) and Xnr2 (data not shown) induce the cardiac marker Nkx2.5 in animal caps. SOX7 and SOX7GGG both induce Xnr2 and Xnr5 expression (data not shown), but SOX7GGG-GFP does not induce Nkx2.5. This suggests that genes, differentially regulated by SOX7 and SOX7GGG, play an important role in the cardiogenic cascade. Since replacing the C-terminal domain of SOX7 with a transcriptional activation domain restores the ability to induce cardiogenesis, it may be this effect is due simply to quantitative, rather than qualitative changes in protein activity. In either case, such data suggest that interactions between Xnr2 and other SOX7/SOX18 target genes may be important in regulating cardiogenesis in vivo.
Methods and Materials
Generation and manipulation of embryos and animal caps
Fertilized eggs were generated, injected at 1 to 4 cell stages, and cultured at 16 ºC or room temperature (22–24 ºC) in 20% Marc’s modified Ringers (MMR), following established laboratory protocols (Karnovsky and Klymkowsky, 1995; Sive et al., 2000). Embryos were staged according to Nieuwkoop & Faber (Nieuwkoop and Faber, 1967). Animal caps were prepared from embryos at stage 8/9 using a Gastromaster (Xenotek, Inc.) as described previously (Sive et al., 2000; Zhang et al., 2003). For experiments involving hormone activation of chimeric polypeptide, animal caps were first treated for 30 minutes with the protein synthesis inhibitor emetine (100 μg/ml)(Sigma)(Entner and Grollman, 1973; Roy et al., 1991), and then with dexamethasone (20 μM)(diluted 1:100 from a 2mM solution in 100% ethanol (Sigma). Control caps were incubated in 1% ethanol.
Plasmids, RNAs and morpholinos
The pCS2.SOX3-V5H, pCS2.SOX7-GFP, pCS2.altSOX7-GFP, pCS2.mt-SOX17β-GFP, pCS2.SOX7ΔC-EnRmyc, pCS2mycΔG-β-catenin, and pCS2.CerS plasmids have been described previously (Merriam et al., 1997; Piccolo et al., 1999; Zhang et al., 2005; Zhang et al., 2003). For this work, we generated new forms of SOX7 and SOX18β. In pCS2.SOX7ΔC-GFP, the region of SOX7 encoding amino acids 161-362, which begins 20 amino acids downstream of the DNA-binding HMG box, was deleted. In pCS2mt-SOX7ΔC-VP16, the SOX7ΔC region was subcloned into the pCS2mt-SOX3ΔCVP16 plasmid, replacing the SOX3ΔC sequence. A plasmid containing the Xenopus SOX18β cDNA was supplied by Sadakazu Aiso (Keio University, Japan)(Hasegawa et al., 2002) and the full length and “ΔC” (amino acids 206-361 removed) regions were amplified by PCR and subcloned to form pGEM.SOX18β, pCS2-SOX18β-GFP, pCS2mt-SOX18βΔCVP16. In SOX7GGG-GFP, amino acids 310–312 of the original SOX7 coding sequence were changed, using a QuickChange Kit (Stratagene), from DQL to GGG; the change was confirmed by DNA sequencing. For testing morpholino inhibition of SOX18β mRNA translation, we generated the pCS2utrSOX18β-V5H plasmid in which the 5’ globin-derived UTR of the pCS2 plasmid was replaced by the 123 base pair region of the 5’ UTR region of the SOX18β cDNA (Hasegawa et al., 2002). The utrSOX18β-V5H RNA is a perfect match to the SOX18 morpholino. Finally, the hormone-binding region of the human glucocorticoid receptor (originally supplied by H. Sive, MIT) was amplified by PCR and subcloned into pCS2.altSOX7-GFP to form pCS2.GRaltSOX7-GFP. Plasmids were linearized and capped RNAs were synthesized using a mMessage mMachine kit (Ambion) following manufacturer’s instructions. Modified antisense DNA oligonucleotides (morpholinos) were synthesized by Gene Tools, Inc. and are directed against the 5’ UTR and coding sequence of the SOX7 and SOX18β cDNAs. The morpholinos were (7MO: 5′-ATCCCATCAGGGTAGTCATTATTCCFL3′; 18MO: 5′ GACTCAGATCTATGCATTCCAGCTG-3′)(FIG. 3A). As a control, we used the standard control morpholino sold by Gene Tools. In vitro translation reactions were carried out using Promega Rabbit Reticulocyte Lysate, following manufacturer’s instructions.
In situ hybridization, histochemical staining imaging and quantitative RT-PCR
Digoxygenin-labeled antisense RNA probes against Myosin Heavy Chain-α (MHCα) (Logan and Mohun, 1993), troponin c inhibitory chain (TnIc) (Drysdale et al., 1994), Nkx2.5 (Tonissen et al., 1994), Tbx5 (Horb and Thomsen, 1997) and SOX18β (Hasegawa et al., 2002) mRNAs were prepared and in situ hybridization was carried out following standard laboratory protocols (Sive et al., 2000). In the case of SOX18β, the pGEM.SOX18β plasmid was linearized with Not I and antisense RNA was synthesized using SP6 RNA polymerase. All photographs were taken using a Nikon CoolPix 995 Camera on an Inverted Leica M400 Photomicroskop. Images were manipulated using Macromedia Fireworks Software using the “auto levels” and “curves” functions only. Primers used for RT-PCR analyses are listed in TABLE I.
Table 1.
PCR primer sets for RT-PCR
| Endodermin | F, 5’-AGC AGA AAA TGG CAA ACA CAC-3’
R, 5’-GGT CTT TTA ATG GCA ACA GGT-3’ |
(Sasai et al., 1996) |
| Eomes | F, 5 ’-TGG TCC TCA AGG TCA AGT CC-3’
R, 5’-GGG GAG TTT TCA TTG CTT GA-3’ |
(Ryan et al., 1996) |
| Snail | F, 5’-GCA CAA TGG ACT CCT TAA ATT CCT G-3’
R, 5’-GTG ACC GGG TGC TCA TTG TG-3’ |
(Aybar et al., 2003) |
| MHCα | F, 5’-GCC AAC GCG AAC CTC TCC AAG TTC CG-3’
R, 5’-GGT CAC ATT TTA TTT CAT GCT GGT TAA CAG G-3’ |
(Schneider and Mercola, 2001) |
| Nkx2.5 | F.5’-GAG CTA CAG TTG GGT GTG TGT GGT-3’
R, 5’-GTG AAG CGA CTA GGT ATG TGT TCA-3’ |
(Schneider and Mercola, 2001) |
| Tbx5 | F,5’-GGC GGA CAC AGA GGA GGC TTA T-3’
R, 5’-GTG GCT GGT GAA TCT GGG TGA AC-3’ |
(Schneider and Mercola, 2001) |
| TnIc | F, 5’-CTG ATG AGG AAG AGG TAA CC-3’
R, 5’-CCT CAC GTT CCA TTT CTG CC-3’ |
(Schneider and Mercola, 2001) |
| SOX17β | F, 5 ’-CAG GTG AAG AGG ATG AAG AG-3’
R, 5’-CAT TGA GTT GTG GCC CTC AA-3’ |
(Engleka et al., 2001) |
| SOX18β | F, 5’-CTT TTC CCA CAT CCT CAT-3’
R, 5’-TGC AGG AAG CTG AAT GCC-3’ |
This work |
| Cerberus | F, 5’ CCT TGC CCT TCA CTC AG-3’
R, 5’ TGG CAG ACA GTC CTT T-3’ |
(Foley and Mercola, 2005) |
| Hex | F, 5’ GTG GCT ACT TAC CGG AC-3’
R, 5’ CCT TTC CGC TTG TGC A-3’ |
(Foley and Mercola, 2005) |
| Xnr1 | F, 5’-TGG CCA GAT AGA GTA GAG-3’
R, 5’-TCC AAC GGT TCT CAC TTT-3’ |
(Kofron et al., 1999) |
| Xnr2 | F, 5’-ATC TGA TGC CGT TCT AAG CC-3’
R, 5’-GAC CTT CTT CAA CCT CAG CC-3’ |
(Takahashi et al., 2000) |
| Xnr4 | F, 5 ’-TTA CAA GAT GCT GCA CAC TCC-3’
R, 5’-AAC TCT GCA TGT ATG CGT GG-3 |
(Takahashi et al., 2000) |
| Xnr5 | F, 5’-TCA CAA TCC TTT CAC TAG GGC-3’
R,5’ -GGA ACC TCT GAA AGG AAG GC-3’ |
(Yang et al., 2002) |
| Xnr6 | F, 5’-TCC AGT ATG ATC CAT CTG TTG C-3’
R, 5’-TTC TCG TTC CTC TTG TGC CTT-3’ |
(Takahashi et al., 2000) |
| NCAM | F, 5’-CAC AGT TCC ACC AAA TGC-3’ R 5’-GGA ATC AAG CGG TAC AGA-3’ | (XenBase) |
| Siamois | F, 5’-CTC CAG CCA CCA GTA CCA GAT C-3’ R 5’ GGG GAG AGT GGA TAG AAA CAG T-3’ | (Brannon and Kimelman, 1996) |
| ODC | F, 5 ’-CAG CTA GCT GTG GTG TGG-3’
R, 5’-CAA CAT GGA AAC TCA CAC-3’ |
(Agius et al., 2000) |
Reporter assays
Fertilized eggs were injected with linearized pOT-firefly luciferase (Korinek et al., 1998) and pTKRenilla luciferase reporter plasmids (Promega)(20 pg/embryo each), together with 50 pg/embryo RNA encoding the mutationally stabilized form of β-catenin, pCS2mtΔGβ-catenin (Merriam et al., 1997), and various amounts of SOX7-GFP, SOX7GGG-GFP, or SOX18β-GFP RNAs. At stage 8, animal caps were prepared and analyzed when unmanipulated embryos reached stage 10/11 using the Promega dual luciferase assay system. Alternatively, fertilized eggs were injected with 50 pgs/embryo of mtΔGβ-catenin RNA and SOX7-GFP, SOX7GGG-GFP, mtSOX7ΔC-VP16, SOX18β-GFP or mt-SOX18β ΔC-VP16 RNAs (500pg/embryo); animal caps were prepared at stage 8, and analyzed by RT-PCR for expression of Siamois, a direct target of β-catenin regulation (Brannon and Kimelman, 1996), when unmanipulated embryos reached stage 10/11.
Immunoprecipitation
Thes studies were carried out using streptavidin-protein A beads (Sigma) as described previously (Zhang et al., 2003). A rabbit anti-β-catenin antibody was used to visualize endogenous β-catenin, and immunoprecipitation was performed using an anti-GFP antibody, while V5 tagged SOX18 was visualized using a monoclonal antiV5 antibody; antiGFP and V5 antibodies were purchased from Invitrogen.
Acknowledgments
We thank Mark Mercola and Ann Foley for helpful discussions, Sadakazu Aiso, Paul Krieg, Doug Melton, Hazel Sive and Eddy DeRobertis for various plasmids; the Leinwand lab for use of their luminometer, the Pace lab for use of their real-time PCR machine. This work was funded by NIH grant GM54001 to MWK.
Footnotes
This manuscript is dedicated to the memory of Spencer I. Browne, whose friendship over the years has meant a great deal.
References
- Agius E, Oelgeschlager M, Wessely O, Kemp C, De Robertis EM. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127:1173–83. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Kinoshita M, Yokota C, Takano K, Fukuda K, Moriyama N, Malacinski GM, Asashima M. Amphibian in vitro heart induction: a simple and reliable model for the study of vertebrate cardiac development. Int J Dev Biol. 2003;47:405–10. [PubMed] [Google Scholar]
- Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130:483–94. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann’s organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- Brannon M, Kimelman D. Activation of Siamois by the Wnt pathway. Dev Biol. 1996;180:344–7. doi: 10.1006/dbio.1996.0306. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mironova E, Whitaker LL, Edwards L, Yost HJ, Ramsdell AF. ALK4 functions as a receptor for multiple TGF beta-related ligands to regulate left-right axis determination and mesoderm induction in Xenopus. Dev Biol. 2004;268:280–94. doi: 10.1016/j.ydbio.2003.12.035. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–28. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci U S A. 2003;100:11469–73. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M, Koopman P. SOX18 and the transcriptional regulation of blood vessel development. Trends Cardiovasc Med. 2001;11:318–24. doi: 10.1016/s1050-1738(01)00131-1. [DOI] [PubMed] [Google Scholar]
- Drysdale TA, Tonissen KF, Patterson KD, Crawford MJ, Krieg PA. Cardiac troponin I is a heart-specific marker in the Xenopus embryo: expression during abnormal heart morphogenesis. Dev Biol. 1994;165:432–41. doi: 10.1006/dbio.1994.1265. [DOI] [PubMed] [Google Scholar]
- Eimon PM, Harland RM. Effects of heterodimerization and proteolytic processing on Derriere and Nodal activity: implications for mesoderm induction in Xenopus. Development. 2002;129:3089–103. doi: 10.1242/dev.129.13.3089. [DOI] [PubMed] [Google Scholar]
- Eisenberg CA, Eisenberg LM. WNT11 promotes cardiac tissue formation of early mesoderm. Dev Dyn. 1999;216:45–58. doi: 10.1002/(SICI)1097-0177(199909)216:1<45::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Eisenberg CA, Gourdie RG, Eisenberg LM. Wnt-11 is expressed in early avian mesoderm and required for the differentiation of the quail mesoderm cell line QCE-6. Development. 1997;124:525–36. doi: 10.1242/dev.124.2.525. [DOI] [PubMed] [Google Scholar]
- Engleka MJ, Craig EJ, Kessler DS. VegT activation of Sox17 at the midblastula transition alters the response to nodal signals in the vegetal endoderm domain. Dev Biol. 2001;237:159–72. doi: 10.1006/dbio.2001.0366. [DOI] [PubMed] [Google Scholar]
- Entner N, Grollman AP. Inhibition of protein synthesis: a mechanism of amebicide action of emetine and other structurally related compounds. J Protozool. 1973;20:160–3. doi: 10.1111/j.1550-7408.1973.tb06025.x. [DOI] [PubMed] [Google Scholar]
- Essex LJ, Mayor R, Sargent MG. Expression of Xenopus snail in mesoderm and prospective neural fold ectoderm. Dev Dyn. 1993;198:108–22. doi: 10.1002/aja.1001980205. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Guger K, Gumbiner BM. Induction of the primary dorsalizing center in Xenopus by the Wnt/GSK/betacatenin signaling pathway, but not by Vg1, Activin or Noggin. Development. 1997;124:453–60. doi: 10.1242/dev.124.2.453. [DOI] [PubMed] [Google Scholar]
- Fawcett SR, Klymkowsky MW. Embryonic expression of Xenopus laevis SOX7. Gene Expr Patterns. 2004;4:29–33. doi: 10.1016/j.modgep.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Foley A, Mercola M. Heart induction: embryology to cardiomyocyte regeneration. Trends Cardiovasc Med. 2004;14:121–5. doi: 10.1016/j.tcm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Foley A, Mercola M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Gene and Development. 2005;19:387–96. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futaki S, Hayashi Y, Emoto T, Weber CN, Sekiguchi K. Sox7 plays crucial roles in parietal endoderm differentiation in F9 embryonal carcinoma cells through regulating Gata-4 and Gata-6 expression. Mol Cell Biol. 2004;24:10492–503. doi: 10.1128/MCB.24.23.10492-10503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Hiraoka Y, Hagiuda J, Ogawa M, Aiso S. Expression and characterization of Xenopus laevis SRY-related cDNAs, xSox17alpha1, xSox17alpha2, xSox18alpha and xSox18beta. Gene. 2002;290:163–72. doi: 10.1016/s0378-1119(02)00554-1. [DOI] [PubMed] [Google Scholar]
- Henry GL, Melton DA. Mixer, a homeobox gene required for endoderm development. Science. 1998;281:91–6. doi: 10.1126/science.281.5373.91. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. A vegetally localized T-box transcription factor in Xenopus eggs specifies mesoderm and endoderm and is essential for embryonic mesoderm formation. Development. 1997;124:1689–98. doi: 10.1242/dev.124.9.1689. [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17α and -β mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Jacobson AG, Sater AK. Features of embryonic induction. Development. 1988;104:341–59. doi: 10.1242/dev.104.3.341. [DOI] [PubMed] [Google Scholar]
- Jacobson M, Rutishauser U. Induction of neural cell adhesion molecule (NCAM) in Xenopus embryos. Dev Biol. 1986;116:524–31. doi: 10.1016/0012-1606(86)90153-3. [DOI] [PubMed] [Google Scholar]
- Jockusch EL, Ober KA. Phylogenetic analysis of the Wnt gene family and discovery of an arthropod wnt-10 orthologue. J Exp Zool. 2000;288:105–19. [PubMed] [Google Scholar]
- Jones CM, Broadbent J, Thomas PQ, Smith JC, Beddington RS. An anterior signalling centre in Xenopus revealed by the homeobox gene XHex. Curr Biol. 1999;9:946–54. doi: 10.1016/s0960-9822(99)80421-7. [DOI] [PubMed] [Google Scholar]
- Joseph EM, Melton DA. Xnr4: a Xenopus nodal-related gene expressed in the Spemann organizer. Dev Biol. 1997;184:367–72. doi: 10.1006/dbio.1997.8510. [DOI] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–79. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- Karnovsky A, Klymkowsky MW. Anterior axis duplication in Xenopus induced by the over-expression of the cadherin-binding protein plakoglobin. Proc Natl Acad Sci U S A. 1995;92:4522–6. doi: 10.1073/pnas.92.10.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Molecular cloning and characterization of human SOX17. Int J Mol Med. 2002;9:153–7. [PubMed] [Google Scholar]
- Kintner CR, Melton DA. Expression of Xenopus N-CAM RNA in ectoderm is an early response to neural induction. Development. 1987;99:311–25. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- Kispert A, Vainio S, Shen L, Rowitch DH, McMahon AP. Proteoglycans are required for maintenance of Wnt-11 expression in the ureter tips. Development. 1996;122:3627–37. doi: 10.1242/dev.122.11.3627. [DOI] [PubMed] [Google Scholar]
- Kofron M, Demel T, Xanthos J, Lohr J, Sun B, Sive H, Osada S, Wright C, Wylie C, Heasman J. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors. Development. 1999;126:5759–70. doi: 10.1242/dev.126.24.5759. [DOI] [PubMed] [Google Scholar]
- Kofron M, Wylie C, Heasman J. The role of Mixer in patterning the early Xenopus embryo. Development. 2004;131:2431–41. doi: 10.1242/dev.01132. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/betacatenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–56. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi, M., Haendeler, J., Badorff, C., Brandes, R. P., Hoffmann, J., Pandur, P., Zeiher, A. M., Kuhl, M. and Dimmeler, S. (2005). Non-canonical Wnt signaling enhances differentiation of human circulating progenitor cells to cardiomyogenic cells. J Biol Chem [DOI] [PubMed]
- Kumar A, Novoselov V, Celeste AJ, Wolfman NM, ten Dijke P, Kuehn MR. Nodal signaling uses activin and transforming growth factor-beta receptor-regulated Smads. J Biol Chem. 2001;276:656–61. doi: 10.1074/jbc.M004649200. [DOI] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–60. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo MM, Kemler R. Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev Cell. 2002;3:171–81. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- Logan, C. Y. and Nusse, R. (2004). The Wnt Signaling Pathway in Development and Disease. Annu Rev Cell Dev Biol [DOI] [PubMed]
- Logan M, Mohun T. Induction of cardiac muscle differentiation in isolated animal pole explants of Xenopus laevis embryos. Development. 1993;118:865–75. doi: 10.1242/dev.118.3.865. [DOI] [PubMed] [Google Scholar]
- Loose M, Patient RK. A genetic regulatory network for Xenopus mesendodermal formation. Devel Biol. 2004;271:467–78. doi: 10.1016/j.ydbio.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–27. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maye P, Zheng J, Li L, Wu D. Multiple mechanisms for Wnt11-mediated repression of the canonical Wnt signaling pathway. J Biol Chem. 2004;279:24659–65. doi: 10.1074/jbc.M311724200. [DOI] [PubMed] [Google Scholar]
- Merriam JM, Rubenstein AB, Klymkowsky MW. Cytoplasmically anchored plakoglobin induces a WNT-like phenotype in Xenopus. Dev Biol. 1997;185:67–81. doi: 10.1006/dbio.1997.8550. [DOI] [PubMed] [Google Scholar]
- Miller JR. The Wnts. Genome Biol. 2002;3:REVIEWS3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohun T, Orford R, Shang C. The origins of cardiac tissue in the amphibian, Xenopus laevis. Trends Cardiovasc Med. 2003;13:244–8. doi: 10.1016/s1050-1738(03)00102-6. [DOI] [PubMed] [Google Scholar]
- Murakami A, Shen H, Ishida S, Dickson C. SOX7 and GATA-4 are competitive activators of Fgf-3 transcription. J Biol Chem. 2004;279:28564–73. doi: 10.1074/jbc.M313814200. [DOI] [PubMed] [Google Scholar]
- Nascone N, Mercola M. An inductive role for the endoderm in Xenopus cardiogenesis. Development. 1995;121:515–23. doi: 10.1242/dev.121.2.515. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman CS, Krieg PA. tinman-related genes expressed during heart development in Xenopus. Dev Genet. 1998;22:230–8. doi: 10.1002/(SICI)1520-6408(1998)22:3<230::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop, P. D. and Faber, J. (1967). Normal Table of Xenopus laevis (Daudin). Amsterdam: North-Holland Publishing Company.
- Niimi T, Hayashi Y, Futaki S, Sekiguchi K. SOX7 and SOX17 regulate the parietal endoderm-specific enhancer activity of mouse laminin alpha1 gene. J Biol Chem. 2004;279:38055–61. doi: 10.1074/jbc.M403724200. [DOI] [PubMed] [Google Scholar]
- Okabayashi K, Asashima M. Tissue generation from amphibian animal caps. Curr Opin Genet Dev. 2003;13:502–7. doi: 10.1016/s0959-437x(03)00111-4. [DOI] [PubMed] [Google Scholar]
- Onuma Y, Takahashi S, Yokota C, Asashima M. Multiple nodalrelated genes act coordinately in Xenopus embryogenesis. Dev Biol. 2002;241:94–105. doi: 10.1006/dbio.2001.0493. [DOI] [PubMed] [Google Scholar]
- Osada SI, Wright CV. Xenopus nodal-related signaling is essential for mesendodermal patterning during early embryogenesis. Development. 1999;126:3229–40. doi: 10.1242/dev.126.14.3229. [DOI] [PubMed] [Google Scholar]
- Pandur P, Lasche M, Eisenberg LM, Kuhl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002a;418:636–41. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- Pandur P, Maurus D, Kuhl M. Increasingly complex: new players enter the Wnt signaling network. Bioessays. 2002b;24:881–4. doi: 10.1002/bies.10164. [DOI] [PubMed] [Google Scholar]
- Pennisi D, Bowles J, Nagy A, Muscat G, Koopman P. Mice null for sox18 are viable and display a mild coat defect. Mol Cell Biol. 2000a;20:9331–6. doi: 10.1128/mcb.20.24.9331-9336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D, Gardner J, Chambers D, Hosking B, Peters J, Muscat G, Abbott C, Koopman P. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet. 2000b;24:434–7. doi: 10.1038/74301. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De RE. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–10. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B, Lartillot N, Balavoine G, Adoutte A, Vervoort M. Phylogenetic analysis of the Wnt gene family. Insights from lophotrochozoan members. Curr Biol. 2002;12:1395. doi: 10.1016/s0960-9822(02)01068-0. [DOI] [PubMed] [Google Scholar]
- Raffin M, Leong LM, Rones MS, Sparrow D, Mohun T, Mercola M. Subdivision of the cardiac Nkx2.5 expression domain into myogenic and nonmyogenic compartments. Dev Biol. 2000;218:326–40. doi: 10.1006/dbio.1999.9579. [DOI] [PubMed] [Google Scholar]
- Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH. The orphan receptor ALK7 and the Activin receptor ALK4 mediate signaling by Nodal proteins during vertebrate development. Genes Dev. 2001;15:2010–22. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy LM, Swenson KI, Walker DH, Gabrielli BG, Li RS, Piwnica-Worms H, Maller JL. Activation of p34cdc2 kinase by cyclin A. J Cell Biol. 1991;113:507–14. doi: 10.1083/jcb.113.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Mitchell A, Gurdon JB. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell. 1996;87:989–1000. doi: 10.1016/s0092-8674(00)81794-8. [DOI] [PubMed] [Google Scholar]
- Ryan K, Russ AP, Levy RJ, Wehr DJ, You J, Easterday MC. Modulation of eomes activity alters the size of the developing heart: implications for in utero cardiac gene therapy. Hum Gene Ther. 2004;15:842–55. doi: 10.1089/hum.2004.15.842. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–4. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Katoh M. Expression of human SOX18 in normal tissues and tumors. Int J Mol Med. 2002;10:339–44. [PubMed] [Google Scholar]
- Sasai Y, Lu B, Piccolo S, De Robertis EM. Endoderm induction by the organizer-secreted factors chordin and noggin in Xenopus animal caps. Embo J. 1996;15:4547–55. [PMC free article] [PubMed] [Google Scholar]
- Sater AK, Jacobson AG. The specification of heart mesoderm occurs during gastrulation in Xenopus laevis. Development. 1989;105:821–30. doi: 10.1242/dev.105.4.821. [DOI] [PubMed] [Google Scholar]
- Sater AK, Jacobson AG. The role of the dorsal lip in the induction of heart mesoderm in Xenopus laevis. Development. 1990;108:461–70. doi: 10.1242/dev.108.3.461. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schneider VA, Mercola M. Spatially distinct head and heart inducers within the Xenopus organizer region. Curr Biol. 1999;9:800–809. doi: 10.1016/s0960-9822(99)80363-7. [DOI] [PubMed] [Google Scholar]
- Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–15. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss TM, Xydas S, Lassar AB. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121:4203–14. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- Shiozawa M, Hiraoka Y, Komatsu N, Ogawa M, Sakai Y, Aiso S. Cloning and characterization of Xenopus laevis xSox7 cDNA. Biochim Biophys Acta. 1996;1309:73–6. doi: 10.1016/s0167-4781(96)00145-5. [DOI] [PubMed] [Google Scholar]
- Sinner D, Rankin S, Lee M, Zorn AM. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004;131:3069–3080. doi: 10.1242/dev.01176. [DOI] [PubMed] [Google Scholar]
- Sive, H. L., Grainger, R. M. and Harland, R. M. (2000). Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- Smithers LE, Jones CM. Xhex-expressing endodermal tissues are essential for anterior patterning in Xenopus. Mech Dev. 2002;119:191–200. doi: 10.1016/s0925-4773(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Rana AA, Kettleborough R, Brickman JM, Clements M, Forrest A, Grimmond S, Avner P, Smith JC, Dunwoodie SL, et al. Characterizing embryonic gene expression patterns in the mouse using nonredundant sequence-based selection. Genome Res. 2003;13:2609–20. doi: 10.1101/gr.1362303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Amand AL, Klymkowsky MW. Cadherins and catenins, Wnts and SOXs: embryonic patterning in Xenopus. Int Rev Cytol. 2001;203:291–355. doi: 10.1016/s0074-7696(01)03010-8. [DOI] [PubMed] [Google Scholar]
- Sugi Y, Lough J. Anterior endoderm is a specific effector of terminal cardiac myocyte differentiation of cells from the embryonic heart forming region. Dev Dyn. 1994;200:155–62. doi: 10.1002/aja.1002000207. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yokota C, Takano K, Tanegashima K, Onuma Y, Goto J, Asashima M. Two novel nodal-related genes initiate early inductive events in Xenopus Nieuwkoop center. Development. 2000;127:5319–29. doi: 10.1242/dev.127.24.5319. [DOI] [PubMed] [Google Scholar]
- Takash W, Canizares J, Bonneaud N, Poulat F, Mattei MG, Jay P, Berta P. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–83. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Hiraoka Y, Ogawa M, Sakai Y, Kido S, Aiso S. Isolation and characterization of a mouse SRY-related cDNA, mSox7. Biochim Biophys Acta. 1999;1445:225–31. doi: 10.1016/s0167-4781(99)00047-0. [DOI] [PubMed] [Google Scholar]
- Terami H, Hidaka K, Katsumata T, Iio A, Morisaki T. Wnt11 facilitates embryonic stem cell differentiation to Nkx2.5-positive cardiomyocytes. Biochem Biophys Res Commun. 2004;325:968–75. doi: 10.1016/j.bbrc.2004.10.103. [DOI] [PubMed] [Google Scholar]
- Tonissen KF, Drysdale TA, Lints TJ, Harvey RP, Krieg PA. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–8. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- Tzahor E, Lassar AB. Wnt signals from the neural tube block ectopic cardiogenesis. Genes Dev. 2001;15:255–60. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walston T, Tuskey C, Edgar L, Hawkins N, Ellis G, Bowerman B, Wood W, Hardin J. Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev Cell. 2004;7:831–41. doi: 10.1016/j.devcel.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Weng W, Stemple DL. Nodal signaling and vertebrate germ layer formation. Birth Defects Res Part C Embryo Today. 2003;69:325–32. doi: 10.1002/bdrc.10027. [DOI] [PubMed] [Google Scholar]
- Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell. 2001;1:605–17. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- Williams PH, Hagemann A, Gonzalez-Gaitan M, Smith JC. Visualizing long-range movement of the morphogen Xnr2 in the Xenopus embryo. Curr Biol. 2004;14:1916–23. doi: 10.1016/j.cub.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Betacatenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129:5743–52. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Yeo C, Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell. 2001;7:949–57. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Fawcett SR, Klymkowsky MW. SOX7 is an immediate-early target of VegT and regulates Nodal expression in Xenopus. Dev Biol. 2005;278:526–41. doi: 10.1016/j.ydbio.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Jensen ED, Klymkowsky MW. The betacatenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development. 2003;130:5609–24. doi: 10.1242/dev.00798. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol Cell. 1999;4:487–98. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]


