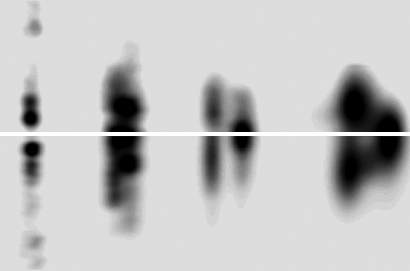Abstract
The fibrous collagens are ubiquitous in animals and form the structural basis of all mammalian connective tissues, including those of the heart, vasculature, skin, cornea, bones, and tendons. However, in comparison with what is known of their production, turnover and physiological structure, very little is understood regarding the three-dimensional arrangement of collagen molecules in naturally occurring fibrils. This knowledge may provide insight into key biological processes such as fibrillo-genesis and tissue remodeling and into diseases such as heart disease and cancer. Here we present a crystallographic determination of the collagen type I supermolecular structure, where the molecular conformation of each collagen segment found within the naturally occurring crystallographic unit cell has been defined (P1, a ≈ 40.0 Å, b ≈ 27.0 Å, c ≈ 678 Å, α ≈ 89.2°, β ≈ 94.6°, γ ≈ 105.6°; reflections: 414, overlapping, 232, and nonoverlapping, 182; resolution, 5.16 Å axial and 11.1 Å equatorial). This structure shows that the molecular packing topology of the collagen molecule is such that packing neighbors are arranged to form a supertwisted (discontinuous) right-handed microfibril that interdigitates with neighboring microfibrils. This interdigitation establishes the crystallographic superlattice, which is formed of quasihexagonally packed collagen molecules. In addition, the molecular packing structure of collagen shown here provides information concerning the potential modes of action of two prominent molecules involved in human health and disease: decorin and the Matrix Metallo-Proteinase (MMP) collagenase.
Keywords: x-ray, fiber, crystallography, fibril, extracellular matrix
Although the general features of the structure of type I collagen have been known for a long time, the specific packing arrangement of collagen molecules in situ has remained difficult to define, despite a great deal of effort by many investigators (1–16) (the general organization of type I collagen is summarized in Fig. 5, which is published as supporting information on the PNAS web site). Recently, we approached this difficult structural problem by employing conventional crystallographic techniques in x-ray fiber diffraction experiments (13, 17, 18), culminating in an initial electron density map (13, 19) that allowed a crude look at some aspects of the supermolecular arrangement of collagen molecules in situ. Unfortunately, the high degree of disorder observed in the gap region of the electron density map precluded the fitting of a molecular model to the electron density; the gap region was largely uninterpretable. Without the structure of the gap region, it was impossible to determine the overall molecular arrangement of collagen molecules in situ, and therefore its potential for improving our understanding of the structural, developmental, and pathological function of the collagen fibril at the molecular level remained unrealized. We have subsequently integrated additional (nonoverlapping) intensity data into the structural determination process (by multiple isomorphous replacement), which, along with improved scaling of the intensity data and, hence, better phase estimates, produced a much improved and now clearly interpretable electron density map of collagen molecules in situ. A model that included all of the amino acid residues of the collagen molecule then was fitted to this experimentally determined electron density map, which verified the correct solution of the phase problem and aided in the interpretation of the native map. Collectively, this work presents a molecular resolution structure of whole collagen molecules in situthat has not been described previously.
Results and Discussion
Summary of Findings.
The new electron density map now clearly shows all of the molecular segments within a single unit cell [of axial length D, where D is one 670-Å repeat as defined by the axial Hodge–Petruska scheme (20)], including the four molecular segments within the previously unresolved gap region. This map not only allows the full path of each collagen chain to be visualized and so determine the nature of the molecular packing topology (described here as interdigitated microfibril), but provides specific spatial data relevant to potential molecular interactions within the mammalian extracellular matrix (ECM).
An accompanying model that describes this structure was fitted to the electron density in a straightforward manner and has an overall R factor of 9.55% (Fig. 1; see also Table 1, which is published as supporting information on the PNAS web site). Although previous, high-resolution crystallographic studies accurately describe the triple-helical and potential electrostatic features within the molecule, and have been very important in these respects, (15, 16), this work provides a visualization of the overall, native, molecular structure and its packing arrangement.
Fig. 1.
Background subtracted off-meridian diffraction pattern of rat tail tendon (Upper) and simulated diffraction pattern from model-derived intensities (Lower). Rsim (R factor between model generated and observed diffraction pattern) was determined to be 16.7%, whereas the R factor for the integrated structure factors is 9.55% (Rw being 21.73%).
Although of low resolution, the electron density map allows assignment of the amino acid sequence to the molecular coordinates of each chain (Fig. 2), because the N and C termini can be identified by the heavy atom labeling (13). Identification of the remaining molecular segments (2–4) was achieved by following a single collagen molecule from the N terminus through several successive unit cells to the C terminus (Fig. 3), the results of which were found to agree with the lattice positions determined from difference Fourier maps (13).
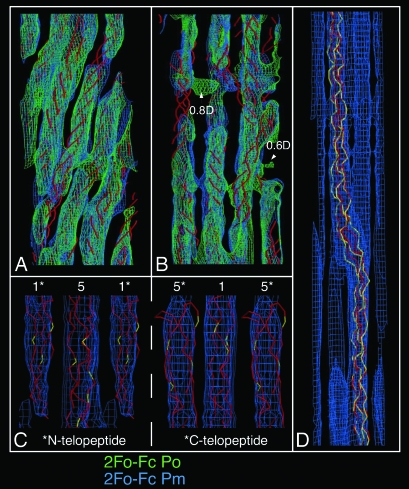
Fig. 2.
Electron density maps. Map elements in A and B have been compressed five times along the direction parallel to the c-axis for clarity. (A) Overlap region of 1D repeat, showing the common tilt of the five collagen segments within the overlap region. The triple-helical backbone of the prerefined model is shown in red as a stylized Cα trace. (B) Expanded view of the gap region showing the different paths of the gap region molecules to that of those in the overlap and to each other in the gap. Small patches of unassigned electron density are seen at 0.6D and 0.8D of the unit cell in the observed/experimental phase (Po) maps.‖ Two large cavities are also found at these locations. The triple-helical backbone is shown in red as a stylized Cα trace, and the calculated/model phase map is shown as 2Fo − Fc Pm (phases averaged between calculated and observed to reduce possible phase bias). (C) The electron density at the N and C telopeptide levels of the D-periodic unit cell and conformation of the telopeptides. The amino acid residue Cα trace is shown in red except for lysine/hydroxy-lysine residues, which are colored yellow. The N-telopeptide-containing segment (segment 1; Left) and C-telopeptide-containing segment (segment 5; Right) are labeled. (D) An ≈225-Å-long section from the native cell (original aspect ratio preserved, i.e., not compressed along the c-axis), showing the section of the bent collagen helix just below 0.6D of the axial unit cell. The electron density of neighboring chain segments can be seen along the side-chain traced segment. The rigid-body refined model is shown in red; the final relaxed model is in yellow.
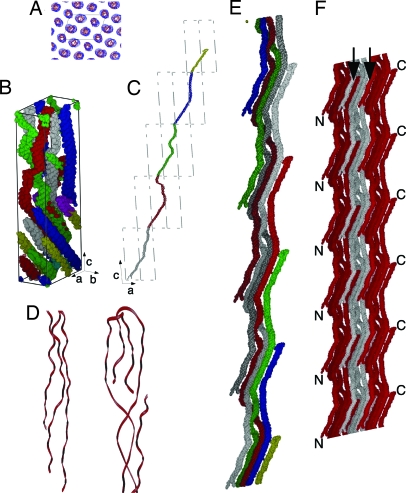
Fig. 3.
Collagen organization and structure. The collagen segments are labeled as follows for B, C, and E: 1, gray; 2, red; 3, green; 4, blue; 5, yellow. Part of segment 1 is colored cyan (the N terminus), and part of segment 5 is colored magenta (the C terminus) to allow easier identification in B. The c-axis has been compressed five times for B, C, and E. (A) Electron density and model showing the quasihexagonal packing of the molecular segments. The approximate outline of the unit cell (a and b sides) is marked with black lines. (B) Cα carbons rendered as line spheroids showing the conformation of the D-staggered collagen segments within a single unit cell (cell axis shown). (C) Molecular path of a collagen molecule through successive unit cells in the a–c plane. (D) Enlarged view of the telopeptides of type I collagen, showing N-telopeptide (left and bottom of C) and C-telopeptide (right and top of C). Both have been rotated with respect to C for clarity of display. (E) Taking several 1D staggered collagen molecules from the collagen packing structure (single molecule shown in C), it is possible to represent the collagen microfibril. The collagen molecules progress from bottom to top (N to C terminus) and are colored as previously (except that chains starting in successive D-periods are darker equivalent colors). A clear right-handed twist can be seen, particularly between segments 2 and 3 (which is roughly at the midpoint of each collagen molecule). The noncrystallographic symmetry that relates the collagen molecules within the microfibrillar structure is a simple fractional translational function (Nu, 0v, Nw) where N is an integer. Five successive D-repeats of the microfibril can be visualized with nine copies of the coordinates (A–E1) of a single collagen molecule by applying the following translations. (A) 0, 0, 0. (B) −1, 0, −1. (C) −2, 0, −2. (D) −3, 0, −3. (E) −4, 0, −4. (B1) 1, 0, 1. (C1) 2, 0, 2. (D1) 3, 0, 3. (E1) 4, 0, 4. (F) Three microfibrils are shown side by side to indicate the probable binding relationship. The N-terminal segment of each collagen molecule is bound to two other collagen molecules (one inter- and one intramicrofibrillar) and a single crosslinked partnership at the C-terminal telopeptide (one intermicrofibrillar link). N- and C-terminal areas are marked (see also Fig. 2C). Note the positions (arrows) where the molecules belonging to one microfibril interdigitate with that of its neighbors.
The extent of the ordered regions (the crystallites) within collagen fibrils has been measured from the Bragg peak broadening in the lateral plane of the tendon diffraction pattern, after the Williamson–Hall method (21) (see Fig. 6, which is published as supporting information on the PNAS web site). The crystallite size is shown to be 429 Å, a value that corresponds well to the value inferred by Hulmes et al. (22) of ≈450 Å.
Hulmes et al. (22) proposed a liquid-crystal model of fibrils composed of well ordered crystallite domains randomly organized within each collagen fibril. The size of the crystallite suggests that there is an average of 10 or 11 unit cells per organized domain, and given the normal range of fibril size being ≈3,000–5,000 Å, there would be ≈10 crystallite domains per fibril. The Williamson–Hall method also provides an estimate of lattice distortion, which in this case is calculated to be 1.55 Å (Fig. 6). This result can be taken to imply an overall average temperature factor of ≈190 Å2 for the collagen molecule, although this value is not uniform because the gap region is relatively more disordered than the overlap (approximately two times; average q factor for overlap is 0.89 and 0.49 for the gap region).
Features of the Structure and Their Interpretation.
Inspection of the electron density map and fitted model shows that each collagen molecule is arranged within a quasihexagonal lattice throughout the overlap and interface regions (the axial level of the telopeptides) and are observed to be tilted through the overlap region (11, 13) (Figs. 2 and 3). An unexpected finding is that this quasihexagonal arrangement (Fig. 3A) is seen to continue into and throughout the gap region, despite the absence of one collagen molecule per unit cell and the fact that, in this region, each of the four molecular segments adopts a unique conformation (Fig. 2D), whereas in the overlap region the segments have virtually the same conformation (Fig. 2A). In addition, there is a general change of direction between overlap and gap regions, giving rise to a pleated arrangement of the 1D staggered collagen molecules (Fig. 3 E and F).
The inflection point for each “pleat” is found within both telopeptide regions, the sites where intermolecular attachment is believed to occur via lysine and hydroxy-lysine crosslinks. This structure supports this assumption, although the resolution of the study does not allow a specific delineation of the long lysine/hydroxy-lysine side chains. Several clusters of lysine/hydroxy-lysine residues found within the telopeptides are found to be within plausible bonding distances between the telopeptides and their helical neighbors (Fig. 2C; see also Table 2, which is published as supporting information on the PNAS web site).
Collagen Molecular Packing and the Microfibrillar Structure.
A “microfibril” is thought to be the basic building block of the collagen fibril. The specific arrangement of collagen molecules that constitute the putative microfibril has long been sought, and numerous models have been proposed describing its possible structure (see below and Figs. 7 and 8, which are published as supporting information on the PNAS web site). We can identify the microfibrillar structure in our multiple isomorphous replacement determination of the in situ packing structure of collagen by tracing the full path of a single collagen molecule through several successive unit cells and observing the central axis that it and its neighbors rotate around (Fig. 3 B and E). It can be seen that collagen molecules from four (lateral plane) neighboring unit cells comprise a self-contained, rope-like repeating unit (Fig. 3E). Of the various, previously hypothesized, structural arrangements, the one that most closely resembles this observed structure is the compressed microfibril (7, 9, 11, 13). However, this theoretical model is seen to be slightly simplistic, because one cannot obtain a self-contained pentameric unit according to previous, cyclic definitions (7, 9, 11, 13) (because the molecular progression, although of a cyclic nature, would be better described as spiral) without the addition of at least one “extra” collagen molecule to fill unoccupied positions within the lattice (see Fig. 8 and also Fig. 9, which is published as supporting information on the PNAS web site). In contrast, the repeating arrangement of pentameric collagen molecules presented here is consistently self-satisfying with respect to the collagen-packing lattice and can be accurately described as a microfibril, consisting of five 1D staggered, pleated collagen molecules, with a right-handed supertwist.
Neighboring microfibrils are interdigitated with one another (Figs. 3F, 6D, 8F, and 9D). Specifically, the quasihexagonal packing of the collagen molecules is continuous and uninterrupted because neighboring N- and C-terminal-containing molecular segments are contained within neighboring microfibrils, rather than being internally directed within the microfibril. This arrangement indicates why microfibril-like structures have not yet been extracted from tissue samples (23) because, although they can be identified as topological entities, they are not separable structural units in the mature form. Each microfibril contains at least two to three intermicrofibrillar crosslinkage (7, 24) but also one intramicrofibrillar linkage; hence, the disruption of the N- and C-terminal bonds during collagen extraction disrupts not only the fibrillar but also the microfibrillar structure of the sample. This feature also may help explain the ability of fibrillar tissues to absorb and transmit mechanical force, i.e., the collagen structure can be considered to be a networked rope where each element of the array transmits force to the rest of the array via the lysine-hydroxy-lysine mediated crosslinks (Figs. 2C and 3F), while the microfibrillar elements themselves maintain structural stability through the right-handed supertwist (Fig. 3). This supertwist (which appears greatly exaggerated in these figures because of the compression along the c-axis, approximately parallel to the collagen triple-helix; see Fig. 10, which is published as supporting information on the PNAS web site, for noncompressed view of a single collagen molecule) may allow the structure to absorb torsion effects without interfering with the superhelix of the collagen molecule itself. The torsion on the supertwist would be converted into a force that is transmitted three-dimensionally through the crosslinked molecular array to neighboring collagen molecules in neighboring microfibrillar cords. These crosslinks are found at both the (axially contracted) N-telopeptide and the (folded) C-terminal telopeptide, which is crosslinked with the neighboring 1D staggered C- and N-terminal collagen segments, respectively (7, 25) (Figs. 2C, 3D, and 4).
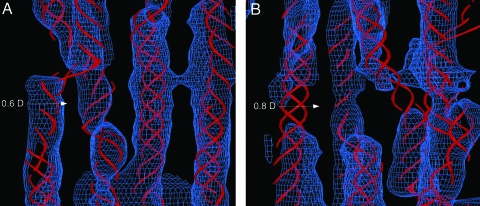
Fig. 4.
Electron density maps. Map elements have been compressed five times in the direction parallel to the c-axis. The collagen triple helix is displayed as a stylized Cα trace. (A) The electron density (2Fo − Fc Pm) map and model structure of the gap region at ≈0.6D. (B) The electron density map and model structure of the gap region at ≈0.8D. Note that density patches at 0.6D and 0.8D seen in Fig. 2 are not seen here because the displayed density is calculated from the average of the experimental and model phases (Pm); the model structure does not contain or account for these sites. Electron density is displayed at a lower threshold than that in Fig. 2 because of the average lower density values in the gap region [because of higher disorder relative to that in the overlap region (11, 22)].
Implications for Molecular Interactions Within the ECM.
The packing structure of collagen as presented here offers insight into the binding of important macromolecules such as the archetypical Small Leucine-Rich Proteoglycan (SLLP) decorin with type I collagen. Decorin has been found to be a potent regulator of collagen fibrillar-genesis, apparently by inhibiting the lateral association of collagen molecules by its prior binding (26–28). In an animal disease model where the decorin gene was disrupted, otherwise viable mice possessed fragile skin with reduced tensile strength, and abnormally large and irregularly shaped collagen fibrils were observed (29), similar traits to those seen in the human connective tissue disease Ehlers–Danlos syndrome. These data have been accepted as a clear demonstration of the importance of the collagen–decorin interaction to the development and maintenance of healthy connective tissue, and any insight offered into the nature of the decorin–collagen interaction in situ is likely to be valuable in understanding the pathology of such connective tissue diseases.
In the electron density map, two collagen segments (molecular segments 2 and 3 of our model) make relatively sharp turns in the gap region, vacating two large molecular spaces in the packing arrangement at ≈0.6D and ≈0.8D (Figs. 2B and 4). These segments contain two of the several sites that have been implicated for decorin binding, at 0.8D and 0.6D (and 1.6D) of collagens’ molecular length (30, 31) (also equivalent to 0.8D and 0.6D within the 1Dstaggered packing system). If collagen molecules on the surface of the fibril are similarly structured, then these locations are clearly suited to accommodate the large decorin protein core, whether in monomeric (32) or dimeric (25, 33) form, although this finding certainly does not preclude the viability of other proposed binding sites along the collagen fibril’s surface. The significance of the 0.6D and 0.8D sites revealed by the electron density map and model presented here is that each could provide a “niche” (shallow hole) in the growing fibril surface that would provide a larger protein-to-protein contact area, and therefore the potentially stronger association between decorin and collagen, than at other possible (and flatter) decorin binding sites, while still allowing decorin to “stick out” of the fibril surface sufficiently to be effective in regulating fibril diameter.
Two small patches of electron density were initially seen at the 0.6D and 0.8D sites in the experimentally determined electron density map (Fig. 2B). Neither of the patches described constitute any part of the path of the collagen molecules, but both are large enough to accommodate two or more leucine-rich repeats (LRRs) of the decorin protein core. Two LRRs, IV–VI, have been previously implicated in collagen binding (34). However, there is no known direct evidence of decorin being found within the collagen fibril, and this scenario seems unlikely because it would require some small but significant rearrangement of the packing arrangement in at least 4% of the unit cells within each collagen fibril [there are ≈0.043 decorin molecules per collagen monomer (35)]. Either this scenario would disrupt the crystallineity of the system or would form part of the mechanism that limits fibril diameter during normal growth. It is also possible that the density at 0.6D and 0.8D represents some other molecule bound at low occupancy.
Collagenase (MMP1) is active in the homeostatic regulation of the ECM and is strongly implicated in the pathology of heart disease and cancer (metastasis) and a host of other diseases. The mechanism of cleavage site recognition by collagenase is not fully understood, but it is thought to rely on aspects of the supermolecular arrangement of the substrate, i.e., native collagen, in addition to recognizing the tripeptide cleavage sequence (36). Its expression also is up-regulated by the presence of native fibrillar collagen (37). The substrate cleavage site is located within the overlap region at 0.316D, but there is insufficient space for the enzyme to enter this dense packing matrix in the interior of the collagen fibril. Depending on the nature of the trimolecular binding complex (catalytic and hemopexin domains of MMP1 and the collagen molecule substrate), access to the site also may be restricted to the enzyme on the exterior of the collagen fibril because of the proximity of the bulbous C-terminal telopeptide (18, 38) to the cleavage site. A prior cleavage of the folded C-telopeptide (Figs. 2C and 3D) would reduce this constriction, although the collagen molecules would still need to be moved aside at least 10 Å for the catalytic domain and 20 Å for the hemopexin-like domain to accommodate the enzyme in the interior of the fibril. Alternatively, it seems entirely possible that the mode of action of the enzyme is to proteolyse collagen chains exclusively from the outside of the fibril inwards. In both cases, the 3D structure of the collagen arrangement would still determine the substrate-binding and recognition site for the enzyme as it proteolyses from the outside to inside of the fibril.
In conclusion, we have presented here a previously undescribed, objectively determined structure showing the packing of the entire collagen molecule in situ. This structure provides strong evidence for a specific microfibrillar substructure for the collagen fibril and important insights regarding the binding of collagen by other ECM macromolecules such as decorin and collagenase. The availability of this experimentally determined model should be very useful to researchers investigating the organization and molecular biology of the ECM.
Methods
Data Collection, Intensity Estimation, and Validation.
The methods used for sample preparation, derivative labeling, and x-ray diffraction protocols have been described (11, 13, 18), as have those for background subtraction and intensity estimation (13, 19). Briefly, x-ray fiber diffraction data were obtained from whole, intact rat tail tendons at room temperature, and the tendons were maintained in a well hydrated state within a sealed sample cell. Data were collected from both native samples and those derivatized using gold chloride and potassium iodide. Because of a significant number of overlapping reflections, individual reflection intensities were estimated by fitting a simulated diffraction pattern to the observed background-subtracted diffraction pattern by using a simulated annealing procedure (13, 19). Because the 677.9-Å c-axis of the 3D unit cell is approximately parallel with the fiber axis (see Table 1), both the meridional and equatorial intensities from the derivative tendons contain complementary axial information regarding the separation between heavy atom labeling sites in the N- and C-telopeptide regions. A previous study (18) using only meridional data (001 indices) produced a detailed 1D structure of collagen’s packing structure via multiple isomorphous replacement and gave the specific axial separation of the heavy atom labeling sites (a fractional unit cell distance of w = 0.33–0.41). Therefore, the vectorial contribution to the Patterson map corresponding to the c-axis direction, estimated from the near-equatorial intensity data, should correlate with vectors observed from the meridional series. This finding helped verify that the indexing and intensity fitting procedures produced an accurate estimate of the data, and not just one that fortuitously produced a low rms residual value.
Initial Phases.
Initial phases were obtained by searching through a sparse matrix of possible heavy atom labeling sites, based on the 3D difference Patterson and knowledge of the 1D structure. Two-dimensional difference Patterson maps (13) were generated at the axial level of the heavy atom attachment sites determined from the 1D structure (18). These were compared with the native Patterson maps of the same axial length scales and, used together, provided a matrix of possible labeling positions in three dimensions. Two principal vectors were used in determining the location of the heavy atoms: the iodide vector (u = 0.2, v = 0.4) and the gold vector (u = 0.05, v = 0.25), which represented the 2D distance (uv plane) between the heavy atom labeled nonhelical telopeptides. The heavy atom labeling positions remained consistent after several rounds of phase calculations: iodide being centered at the N- and C-telopeptide locations (u = 0.23, v = 0.51, w = 0.04; and u = 0.45, v = 0.93, w = 0.45, segments 1 and 5, respectively) and gold at essentially the opposite locations (u = 0.37, v = 0.09, w = 0.04; and u = 0.31, v = 0.31, w = 0.45, segments 5 and 1, respectively).
Additional details of model building, refinement, and validation are described in Supporting Methods, which is published as supporting information on the PNAS web site (see also Figs. 11 and 12 and Table 3, which are published as supporting information on the PNAS web site).
Model Validation.
The R factor for the integrated amplitudes and final model amplitudes was found to be 9.55% and 16.7% respectively by comparison of the observed and simulated diffraction patterns (R factor and Rsim; see Fig. 1; Rw was 21.73%). The model is clearly consistent with the experimentally determined electron density map (Figs. 2 and 4), and the experimental and model phases differ by ≈62° [equivalent to a figure of merit (FOM) of 0.47].
The coordinates for the monomeric decorin model (39), the recombinant decorin dimer [Protein Data Bank (PDB) ID codes 1XKU, 1XEC, and 1XCD] (25), and fibroblast collagenase model (PDB ID code 1FBL) (40) were used to perform initial studies of collagen–decorin and collagen–collagenase interactions within the context of the structure presented here. All model structures were viewed by using spock (http://quorum.tamu.edu) (41) and x-fit (42). Both collagen models (steps 3 and 4) communicated to the PDB have been deposited as Cα traces as displayed in Figs. 2–4 because of the relatively low resolution of this study (5.16/11.1 Å). The observed structure factors accompany the entry for PDB ID code 1Y0F.
Supplementary Material
Acknowledgments
We thank Drs. Reed and Iozzo for providing a copy of the coordinates for their model of decorin, Olga Antipova for critical sequence checking, the staff of the Biophysical Collaborative Access Team (BioCAT) (a National Institutes of Health-supported Research Center RR08630), and the staff of the Structural Biology Center Collaborative Access Team (SBC-CAT) (supported by Department of Energy Grant W-31-109-ENG-38) and Southeast Regional Collaborative Access Team (SER-CAT) (supporting institutions may be found at www.ser-cat.org/members.html) beamlines for their assistance in the development of this project. This work was supported by American Heart Association Greater Midwest Affiliate Grant 0435339Z (to J.P.R.O.O.). A.M was supported by a Leverhulme Emeritus Research Fellowship. T.J.W. was supported by Biotechnology and Biological Sciences Research Council Grant BBS/B/09643. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research, under Contract W-31-109-ENG-38.
Abbreviation
- ECM
extracellular matrix.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1Y0F and 1YGV).
These two isolated patches of density at 0.6D and 0.8D were observed in the initial electron density map (Fo Po) and did not form any part of the collagen molecules. However, attempts to build in this density (using two leucine-rich proteins from decorin model structure as a generic model for this density; data not shown) resulted in a significant deterioration of the R factor when modeled within normal occupancy ranges, and the diminishing effects of significantly lower molecular occupancies made it difficult to assess the accuracy of the modeled leucine-rich repeat’s relative position and orientation within the collagen packing matrix. These patches of density are diminished in size in the 2 Fo − Fc map.
References
- 1.Hulmes D. J., Miller A. Nature. 1979;282:878–880. doi: 10.1038/282878a0. [DOI] [PubMed] [Google Scholar]
- 2.North A. C., Cowan P. M., Randall J. T. Nature. 1954;174:1142–1143. doi: 10.1038/1741142a0. [DOI] [PubMed] [Google Scholar]
- 3.Smith J. W. Nature. 1968;219:157–158. doi: 10.1038/219157a0. [DOI] [PubMed] [Google Scholar]
- 4.Miller A. Biochemistry of Collagen. New York: Plenum; 1976. [Google Scholar]
- 5.Bornstein P., Traub W. The Proteins. London: Academic; 1979. [Google Scholar]
- 6.Fraser R. D. B., MacRae T. P. Int. J. Biol. Macromol. 1981;3:193–200. [Google Scholar]
- 7.Piez K. A., Trus B. L. Biosci. Rep. 1981;1:801–810. doi: 10.1007/BF01114803. [DOI] [PubMed] [Google Scholar]
- 8.Fraser R. D., MacRae T. P., Miller A., Suzuki E. J. Mol. Biol. 1983;167:497–521. doi: 10.1016/s0022-2836(83)80347-7. [DOI] [PubMed] [Google Scholar]
- 9.Fraser R. D., MacRae T. P., Miller A. J. Mol. Biol. 1987;193:115–125. doi: 10.1016/0022-2836(87)90631-0. [DOI] [PubMed] [Google Scholar]
- 10.Wess T. J., Hammersley A., Wess L., Miller A. J. Mol. Biol. 1995;248:487–493. doi: 10.1016/s0022-2836(95)80065-4. [DOI] [PubMed] [Google Scholar]
- 11.Wess T. J., Hammersley A. P., Wess L., Miller A. J. Mol. Biol. 1998;275:255–267. doi: 10.1006/jmbi.1997.1449. [DOI] [PubMed] [Google Scholar]
- 12.Holmes D. F., Gilpin C. J., Baldock C., Ziese U., Koster A. J., Kadler K. E. Proc. Natl. Acad. Sci. USA. 2001;98:7307–7312. doi: 10.1073/pnas.111150598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orgel J. P., Miller A., Irving T. C., Fischetti R. F., Hammersley A. P., Wess T. J. Structure (London) 2001;9:1061–1069. doi: 10.1016/s0969-2126(01)00669-4. [DOI] [PubMed] [Google Scholar]
- 14.Venturoni M., Gutsmann T., Fantner G. E., Kindt J. H., Hansma P. K. Biochem. Biophys. Res. Commun. 2003;303:508–513. doi: 10.1016/s0006-291x(03)00390-5. [DOI] [PubMed] [Google Scholar]
- 15.Bella J., Eaton M., Brodsky B., Berman H. M. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 16.Kramer R. Z., Bella J., Brodsky B., Berman H. M. J. Mol. Biol. 2001;311:131–147. doi: 10.1006/jmbi.2001.4849. [DOI] [PubMed] [Google Scholar]
- 17.Bradshaw J. P., Miller A., Wess T. J. J. Mol. Biol. 1989;205:685–694. doi: 10.1016/0022-2836(89)90314-8. [DOI] [PubMed] [Google Scholar]
- 18.Orgel J. P., Wess T. J., Miller A. Struct. Folding Des. 2000;8:137–142. doi: 10.1016/s0969-2126(00)00089-7. [DOI] [PubMed] [Google Scholar]
- 19.Orgel J. P. R. O., Miller A., Irving T. C., Wess T. J. Fibre Diffraction Rev. 2002;10:40–50. [Google Scholar]
- 20.Petruska J. A., Hodge A. J. Proc. Natl. Acad. Sci. USA. 1964;51:871–876. doi: 10.1073/pnas.51.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson G. K., Hall W. H. Acta Metall. 1953;1:22–31. [Google Scholar]
- 22.Hulmes D. J., Wess T. J., Prockop D. J., Fratzl P. Biophys. J. 1995;68:1661–1670. doi: 10.1016/S0006-3495(95)80391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulmes D. J. J. Struct. Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura I. Int. J. Biol. Macromol. 1987;8:281–290. [Google Scholar]
- 25.Scott P. G., McEwan P. A., Dodd C. M., Bergmann E. M., Bishop P. N., Bella J. Proc. Natl. Acad. Sci. USA. 2004;101:15633–15638. doi: 10.1073/pnas.0402976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel K. G., Trotter J. A. Collagen Relat. Res. 1987;7:105–114. doi: 10.1016/s0174-173x(87)80002-x. [DOI] [PubMed] [Google Scholar]
- 27.Birk D. E., Nurminskaya M. V., Zycband E. I. Dev. Dyn. 1995;202:229–243. doi: 10.1002/aja.1002020303. [DOI] [PubMed] [Google Scholar]
- 28.Birk D. E., Hahn R. A., Linsenmayer C. Y., Zycband E. I. Matrix Biol. 1996;15:111–118. doi: 10.1016/s0945-053x(96)90152-3. [DOI] [PubMed] [Google Scholar]
- 29.Danielson K. G., Baribault H., Holmes D. F., Graham H., Kadler K. E., Iozzo R. V. J. Cell Biol. 1997;136:729–743. doi: 10.1083/jcb.136.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan Y., Cummings C., Sheehan J. K., Kadler K. E., Holmes D. F., Chapman J. A. Dermatan Sulphate Proteoglycans. London: Portland; 1993. [Google Scholar]
- 31.Tenni R., Viola M., Welser F., Sini P., Giudici C., Rossi A., Tira M. E. Eur. J. Biochem. 2002;269:1428–1437. doi: 10.1046/j.1432-1033.2002.02784.x. [DOI] [PubMed] [Google Scholar]
- 32.Goldoni S., Owens R. T., McQuillan D. J., Shriver Z., Sasisekharan R., Birk D. E., Campbell S., Iozzo R. V. J. Biol. Chem. 2004;279:6606–6612. doi: 10.1074/jbc.M310342200. [DOI] [PubMed] [Google Scholar]
- 33.Scott P. G., Grossmann J. G., Dodd C. M., Sheehan J. K., Bishop P. N. J. Biol. Chem. 2003;278:18353–18359. doi: 10.1074/jbc.M211936200. [DOI] [PubMed] [Google Scholar]
- 34.Svensson L., Heinegard D., Oldberg A. J. Biol. Chem. 1995;270:20712–20716. doi: 10.1074/jbc.270.35.20712. [DOI] [PubMed] [Google Scholar]
- 35.Nareyeck G., Seidler D. G., Troyer D., Rauterberg J., Kresse H., Schonherr E. Eur. J. Biochem. 2004;271:3389–3398. doi: 10.1111/j.1432-1033.2004.04273.x. [DOI] [PubMed] [Google Scholar]
- 36.Brandstetter H., Grams F., Glitz D., Lang A., Huber R., Bode W., Krell H. W., Engh R. A. J. Biol. Chem. 2001;276:17405–17412. doi: 10.1074/jbc.M007475200. [DOI] [PubMed] [Google Scholar]
- 37.Nakamuta M., Kotoh K., Enjoji M., Nawata H. World J. Gastroenterol. 2005;11:2264–2268. doi: 10.3748/wjg.v11.i15.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bozec L., Horton M. Biophys. J. 2005;88:4223–4231. doi: 10.1529/biophysj.104.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber I. T., Harrison R. W., Iozzo R. V. J. Biol. Chem. 1996;271:31767–31770. doi: 10.1074/jbc.271.50.31767. [DOI] [PubMed] [Google Scholar]
- 40.Li J., Brick P., O’Hare M. C., Skarzynski T., Lloyd L. F., Curry V. A., Clark I. M., Bigg H. F., Hazleman B. L., Cawston T. E., et al. Structure (London) 1995;3:541–549. doi: 10.1016/s0969-2126(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 41.Christopher J. A., Swanson R., Baldwin T. O. Comput. Chem. 1996;20:339–345. doi: 10.1016/0097-8485(95)00075-5. [DOI] [PubMed] [Google Scholar]
- 42.McRee D. E. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.