Abstract
We conducted a large-scale genome-wide association study in UK Caucasians to identify susceptibility alleles for lung cancer, analyzing 1529 cases and 2707 controls. To increase the likelihood of identifying disease-causing alleles, we genotyped 1476 nonsynonymous single nucleotide polymorphisms (nsSNPs) in 871 candidate cancer genes, biasing SNP selection toward those predicted to be deleterious. Statistically significant associations were identified for 64 nsSNPs, generating a genome-wide significance level of P = 0.002. Eleven of the 64 SNPs mapped to genes encoding pivotal components of the growth hormone/insulin-like growth factor (GH-IGF) pathway, including CAMKK1 E375G (OR = 1.37, P = 5.4 × 10−5), AKAP9 M463I (OR = 1.32, P = 1.0 × 10−4) and GHR P495T (OR = 12.98, P = 0.0019). Significant associations were also detected for SNPs within genes in the DNA damage-response pathway, including BRCA2 K3326X (OR = 1.72, P = 0.0075) and XRCC4 I137T (OR = 1.31, P = 0.0205). Our study provides evidence that inherited predisposition to lung cancer is in part mediated through low-penetrance alleles and specifically identifies variants in GH-IGF and DNA damage-response pathways with risk of lung cancer.
Lung cancer is the most common cancer in the world and represents a major public health problem, accounting for ∼1.2 million cancer-related deaths worldwide each year (Parkin et al. 2005). Tobacco smoking is acknowledged to be the major risk factor for lung cancer, contributing to a 10-fold increase in risk in long-term smokers compared with nonsmokers (Doll and Peto 1981). Other environmental risk factors include exposure to radiation, asbestos, heavy metals, polycyclic aromatic hydrocarbons, and chloromethyl ethers (IARC 1986).
Lung cancer is frequently cited as a malignancy solely attributable to environmental exposure. However, it has long been postulated that individuals may differ in their susceptibility and there is increasing evidence from epidemiological studies for a familial risk (Matakidou et al. 2005). Direct evidence for a genetic predisposition is provided by the increased risk of lung cancer associated with a number of rare Mendelian cancer syndromes, such as carriers of constitutional tumor protein p53 (TP53) (Hwang et al. 2003) and retinoblastoma (Sanders et al. 1989) gene mutations, as well as in patients with Bloom’s (Takemiya et al. 1987) and Werner’s syndromes (Yamanaka et al. 1997).
The genetic basis of inherited susceptibility to lung cancer outside the context of the rare Mendelian cancer predisposition syndromes is at present undefined, but a model in which dominantly acting, high-risk alleles account for all of the excess familial risk seems unlikely. An alternative hypothesis about the allelic architecture of lung cancer susceptibility proposes that most of the genetic risk is caused by low-penetrance alleles. This hypothesis implies that testing for allelic association should be a powerful strategy for identifying lung cancer predisposition alleles.
We sought to identify novel low-penetrance susceptibility alleles to lung cancer by genotyping SNPs across 871 genes with relevance to cancer biology. To increase the likelihood of identifying disease-causing alleles, we biased selection of nsSNPs to those likely to have functionally deleterious consequences. Genotyping 1529 lung cancer cases and 2707 controls from the UK population across 1476 nsSNPs provided strong evidence that low-penetrance alleles in genes involved in the hormone/insulin-like growth factor (GH-IGF) and DNA damage-response pathways are associated with lung cancer susceptibility
Results
Genotypes were obtained for 1526 cases (99.8%) and 2695 controls (99.6%). Of the 1476 SNPs submitted for analysis, 1221 SNPs had sample call rates >95%. Of these, 180 were fixed, leaving 1041 SNPs for which genotype data were informative (Supplemental Table 1). Implementing the genomic control method indicated no evidence of population stratification in our data as a cause of false-positive results, as the 95% confidence interval for the stratification parameter  (0.92–1.31) encompassed unity. As deviates from Hardy-Weinberg equilibrium followed the expected distribution, we concluded that genotyping error is unlikely to have impacted on the statistics generated.
(0.92–1.31) encompassed unity. As deviates from Hardy-Weinberg equilibrium followed the expected distribution, we concluded that genotyping error is unlikely to have impacted on the statistics generated.
Significant associations with risk of lung cancer were identified for 64 of 1041 nsSNPs at the 5% level. The over-representation of associations between SNPs and lung cancer risk was confirmed by a joint analysis of their combined effect using the set-association approach (smallest global significance level of P = 4.2 × 10−4). After further adjustment for the number of terms in the set being a priori, unknown, the genome-wide significance was P = 0.002.
Two of the 64 SNPs identified through the set association procedure, rs2602141 (K1136Q) and rs560191 (D353E), map to the tumor protein p53 binding protein 1 (TP53BP1) and are in strong linkage disequilibrium (LD). A further group of three SNPs in the MHC region spaced within 100 kb; rs1052486 (S625P) in HLA-B-associated transcript 3 (BAT3), rs3130618 (R41L) in HLA-B-associated transcript 4 (BAT4), and rs16900023 (P786S) in mutS homolog 5 (MSH5) also formed a cluster of high LD. Although the permutation procedure implemented in the set-association strategy allows for such substructure in the data when estimating significance levels, it may not be desirable to include highly correlated SNPs in the analysis. A total of 262 SNPs displayed high LD with an adjacent SNP. High LD was found to occur primarily within the same gene, but there were 80 instances where strong LD was observed between SNPs in different genes. We repeated our analysis by omitting markers in LD, retaining one SNP per LD set on the basis of maximum GenCall score or call rate, yielding almost identical sum statistics (P = 0.005) with inclusion of 70 SNPs.
Sixty-seven SNPs displayed significant association at the 5% level with familial lung cancer, but only 52 when the analysis was restricted to sporadic cases. After permutation, the overall significance level attained from the set-association analysis for the familial cases was P = 0.015 compared with P = 0.076 for sporadic cases. Familial cases contributed significantly to overall study findings with 13 SNPs contributing to the 20 associated at the 1% level in the overall data set (Table 1). Stratification of cases by cancer histology (small cell and non-small cell, global P-values 0.11 and 0.06, respectively), age at diagnosis (<60 and ≥60; global P-values 0.17 and 0.18, respectively) and sex (male and female; global P-values 0.19 and 0.08, respectively) did not impact significantly on study findings. Furthermore, limiting our analysis to the 93.7% of cases who were smokers indicated that there was no evidence of confounding due to smoking.
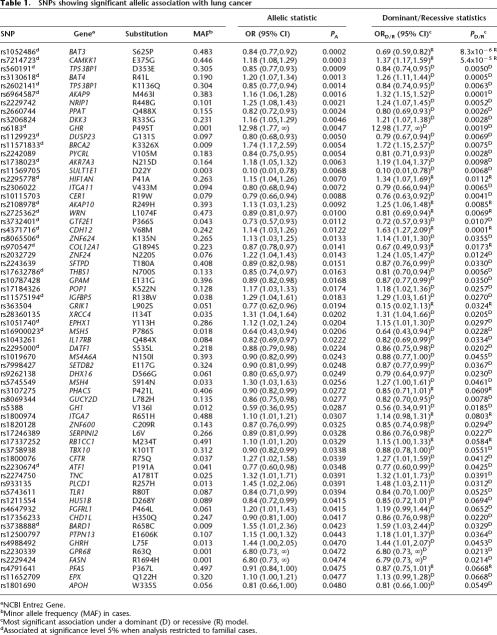
Table 1.
SNPs showing significant allelic association with lung cancer
aNCBI Entrez Gene.
bMinor allele frequency (MAF) in cases.
cMost significant association under a dominant (D) or recessive (R) model.
dAssociated at significance level 5% when analysis restricted to familial cases.
The SNP showing the most significant allelic association with lung cancer was rs1052486 (S625P) in BAT3, a nuclear protein implicated in the control of apoptosis, with strongest association under a recessive model (ORR = 0.69, 95% CI: 0.59–0.82, PR = 8.3 × 10−6) (Table 1). Two additional SNPs, rs7214723 (E375G) in calcium/calmodulin-dependent protein kinase kinase 1 α (CAMKK1), belonging to the Serine/Threonine protein kinase family (ORR = 1.37, 95% CI: 1.17–1.59, PR = 5.4 × 10−5) and rs6964587 (M463I) in A kinase anchor protein 9 (AKAP9), a key component of signal transduction (ORD = 1.32, 95% CI: 1.15–1.52, PD = 7.6 × 10−5), also showed highly significant nominal association under recessive and dominant models, respectively. Empirical limits for genome-wide significance for individual TA, TD, and TR statistics were established at 16.12, 16.23, and 15.66, respectively. Hence, BAT3 S625P and CAMKK1 E375G were both significantly associated with lung cancer with adjusted P-values of 0.006 and 0.036, respectively, with AKAP9 M463I showing borderline significance with adjusted P = 0.066.
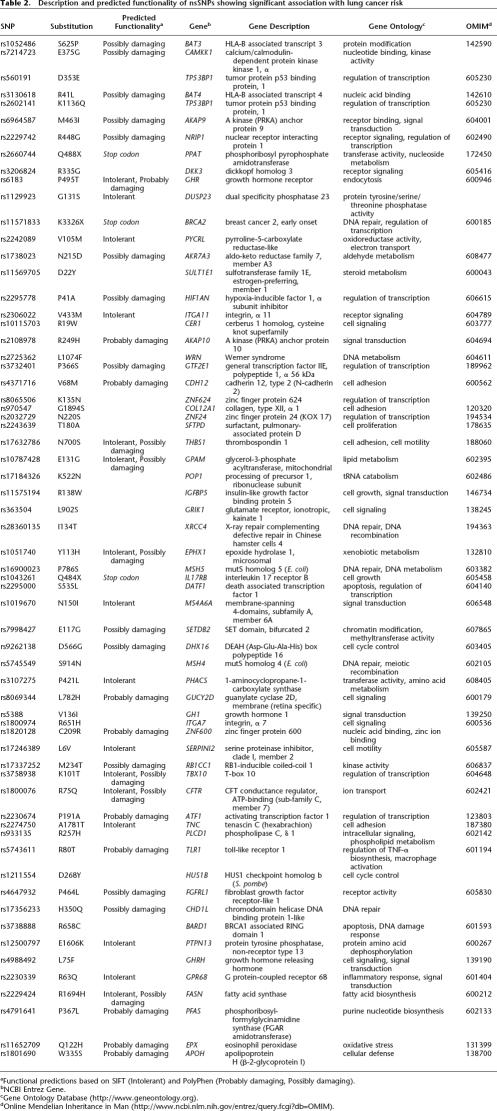
Of the 64 SNPs identified, two SNPs have been documented to be functional, i.e., K3326X in breast cancer 2 early onset (BRCA2) and N700S in thrombospondin 1 (THBS1), and a further 37 SNPs are predicted in silico to deleteriously impact on the expressed proteins (Table 2).
Table 2.
Description and predicted functionality of nsSNPs showing significant association with lung cancer risk
Through interrogation of the Pathway Assist program (Stratagene), 11 of the 64 SNPs associated with risk of lung cancer were located within individual genes encoding pivotal components of the extended GH-IGF pathway, including CAMKK1 E375G and AKAP9 M463I (both of which were globally significant), growth hormone receptor (GHR) P495T (ORD = 12.98, PD = 0.0019), A kinase anchor protein 10 (AKAP10) R249H (ORR = 1.25, PR = 0.0085), and insulin-like growth-factor binding protein 5 (IGFBP5) R138W (ORD = 1.29, PD = 0.027) (Table 1). A further five SNPs were located in genes directly involved in the DNA damage-response pathway, including the functional BRCA2 SNP K3326X (ORD = 1.72, PD = 0.0075), X-ray repair complementing defective repair in Chinese hamster cells 4 (XRCC4) I134T (ORD = 1.31, PD = 0.0205), mutS homolog 5 (MSH5) P786S (ORD = 0.64, PD = 0.0228), mutS homolog 4 (MSH4) S914N (ORD = 1.27, PD = 0.0461), and BRCA1-associated RING domain 1 (BARD1) R658C (ORD = 1.59, PD = 0.0329) (Table 1).
Haplotype frequencies defined by the two sets of SNPs displaying high LD, TP53BP1 K1136Q and D353E, and BAT3 S625P, BAT4 R41L, and MSH5 P786S were significantly different in cases and controls (adjusted P-values, 0.01 and 0.01, respectively, after permutation testing).
We examined for potential interactive effects between the 64 SNPs significantly associated with lung cancer risk (PA < 0.05) by fitting full logistic regression models for each pair, generating 2016 models, and comparing these with the main effects model. Ninety-six pairs of SNPs showed nominally significant interaction at the 5% level. The largest interactive effect identified was between 1-aminocyclopropane-1-carboxylate synthase (PHACS) P421L and toll-like receptor 1 (TLR1) R80T (P = 3.3 × 10−4), albeit nonsignificant after correction for multiple testing.
Discussion
To date, the only evidence for a major locus for lung cancer susceptibility is provided by the linkage scan conducted by Bailey-Wilson et al. (2004) which reported linkage of the disease to chromosome 6q23–25, and a model based on involvement of multiple low-penetrance alleles is eminently plausible.
Previous association studies aimed at identifying low-penetrance alleles for lung cancer susceptibility have evaluated a restricted number of polymorphisms, primarily in genes implicated in the metabolism of tobacco-associated carcinogens and protection of DNA from carcinogen-induced damage. To identify novel lung cancer susceptibility alleles, we extended our search to include genes with relevance to cancer biology, evaluating only nsSNPs that have a higher probability of being directly causal. We acknowledge that the loci considered as candidates will be based on current preconceptions of cancer biology, and it is likely that other genes may influence tumor development. The number of candidate loci will inevitably increase with advances in cancer biology.
The number of nsSNPs that displayed significant association with lung cancer risk was greater than that expected, supporting the tenet that polymorphic variation contributes to lung cancer susceptibility. This assertion is supported by the fact that associations were stronger when the analysis was restricted to those cases with a family history of lung cancer. We cannot exclude the possibility that some of the associations detected are a consequence of LD with causal mutations. It is noteworthy that the SNPs in BAT3, BAT4, and MSH5, which were all associated with lung cancer risk, were in strong LD.
Of the 64 SNPs found to be associated with lung cancer risk, several reside in genes involved in either apoptosis (BARD1 and death associated transcription factor 1 [DATF1]), or the DNA damage-response pathway (BRCA2, MSH4, MSH5, XRCC4), thereby having relevance to the pathobiology of lung cancer a priori.
There is evidence that several of the associated SNPs directly impact on the structure and function of the expressed protein, and are therefore likely to be directly responsible for the observed association. SNPs BRCA2 K3326X and THBS1 N700S are pre-eminent in this respect. The K3326X polymorphism in BRCA2 results in loss of the terminal 91 amino acids of the expressed protein. The C-terminal region of BRCA2 is involved in the nuclear colocalization of Fanconi anemia complementation group D2 (FANCD2) (Wang et al. 2004) and cells lacking the terminal 188 amino acids of BRCA2 are hypersensitive to radiation (Morimatsu et al. 1998). SNP K3326X has been reported to play a role in BRCA2-related Fanconi's anemia (Howlett et al. 2002) and recently reported to increase the risk of pancreatic cancer (Martin et al. 2005). The N700S SNP of THBS1, encoding the anti-angiogenic protein thrombospondin, impacts on calcium binding vital for the normal function of THBS1, and has been established to critically affect the structure and function of the expressed protein (Stenina et al. 2005).
For 37 SNPs correlated with lung cancer risk, evidence that they are deleterious is supported by predictions of functionality based on the PolyPhen and/or SIFT programs. Although in silico predictions about the functional consequences of amino acid changes are in part speculative, such algorithms have been demonstrated in benchmarking studies to successfully categorize 80% of amino acid substitutions (Xi et al. 2004). Two of these 37 putatively deleterious substitutions, A1718T (rs2274750) and P464L (rs4647932), were located in genes Tenascin C (TNC) and fibroblast growth-factor receptor-like 1 (FGFRL1), respectively. Both of these genes have been shown to be differentially expressed in the various lung cancer histologies (Garber et al. 2001) and form part of the extended GH-IGF pathway, with FGFRL1 binding to fibroblast growth factor 2 (FGF2) and TNC interacting with epidermal growth factor receptor (EGFR) and IGFBP5.
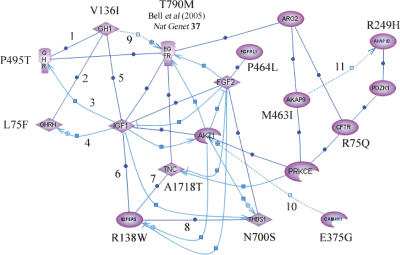
Eleven of the 64 associated SNPs map to genes encoding pivotal components of the GH-IGF1 pathways (Fig. 1). The absence of suitable nsSNPs in AKT1, ARG2, FGF2, IGF1, PZDK1, and PRKCE did not permit us to examine whether variants in these genes also contribute to lung cancer susceptibility. The prior probability of identifying a significant association with lung cancer risk for a series of 11 SNPs mapping to a single defined pathway of genes is intuitively small. The assertion that polymorphic variation and subsequent dysregulation in the GH-IGF axis could be associated with risk of lung cancer is not without precedent. IGF1, which is up-regulated by GH, regulates cellular proliferation and apoptosis and has been shown to increase tumor growth (Khandwala et al. 2000). Elevated levels of circulating IGF1 have been shown to confer an increased risk of various tumors including breast (Toniolo et al. 2000), colorectal (Ma et al. 1999), lung (Yu et al. 1999), and prostate cancers (Chan et al. 1998). Furthermore, polymorphic variation in IGFBP3 has been reported to increase risk of non-small cell lung cancer (NSCLC) (Moon et al. 2006). Recently, Bell et al. (2005) demonstrated that inherited susceptibility to lung cancer may be associated with acquisition of drug resistance mediated by EGFR T790M. While no studies have reported an association between IGFBP5 variants and cancer to date, it is noteworthy that IGFBP5 is required for regulation of cell-specific IGF responses during lung development (Schuller et al. 1995).
Figure 1.
Inter-relationship between genes involved in the GH-IGF pathway containing SNPs associated with risk of lung cancer. Interactions were established using Pathway Assist software and are color-coded as follows: blue (expression), gray (regulation), and red (protein binding). Supporting publications are indicated with the corresponding NCBI Entrez PubMed ID in square brackets. (1) Binding [12,888,636]; (2) Binding [11,832,396]; (3) Expression [11,849,991]; (4) Expression [11,606,442]; (5) Binding [11,126,270]; (6) Binding [15,140,223]; (7) Binding [10,982,804]; (8) Binding [11,751,588]; (9) Regulation [14,517,795]; (10) Regulation [11,395,482]; (11) Regulation [15,047,863]. Validated nsSNPs with frequency data from Caucasian populations were not available in dbSNP Build 123 for genes AKT1, ARG2, FGF2, IGF1, PZDK1, and PRKCE. Bell et al. (2005) found association between lung cancer and SNP T790M.
While it is desirable to validate our findings through analysis of additional large data sets, our study provides evidence that inherited predisposition to lung cancer is in part mediated through low-penetrance alleles and specifically identifies variants in genes comprising the GH-IGF pathway as susceptibility alleles.
Methods
Patients and control subjects
Patients with lung cancer were ascertained from the Genetic Lung Cancer Predisposition Study (GELCAPS) based in the United Kingdom (UK). Information on clinico-pathological characteristics and family history was collected using standardized questionnaires (Matakidou et al. 2005). In total, 1529 individuals with lung cancer were included in the study (506 males and 1023 females, median age at diagnosis 63 yr, range 26–92 yr). Case selection was prioritized firstly by family history and secondly, by early age-at-diagnosis. A total of 573 cases (38%) had a parent or sibling affected with lung cancer. Only 97 (6.3%) of 1529 cases were nonsmokers. Histology information was available for 1489 of the lung cancer cases—387 were small-cell cases and 1098 were non-small-cell cases (of which 483 were squamous and 343 adenocarcinomas).
A total of 2707 healthy individuals were recruited through either the Royal Marsden Hospital Trust/Institute of Cancer Research Family History and DNA Registry (1999–2004; http://intra-test.icr.ac.uk/tissueres/patient_blood.html), the National Study of Colorectal Cancer Genetics Trial (2004; http://www.ncrn.org.uk/portfolio/data.asp?ID=1269) or GELCAPS, all established within the UK. The control group contained 836 (31%) males and 1871 (69%) females, median age 59 yr (range 21–92 yr). None of the controls reported a personal history of cancer. All cases and controls were British Caucasians and there were no obvious demographic differences between groups in terms of place of residence within the UK. All study participants provided written informed consent. Ethical approval for the study was obtained from the London Multi-Center Research Ethics Committee (MREC/98/2/67) in accordance with the tenets of the Declaration of Helsinki. DNA was extracted from blood samples using conventional methodologies and quantified using PicoGreen (Invitrogen).
Selection of candidate genes and SNPs
We have previously established a publicly accessible PICS (Predicted Impact of Coding SNPs) database (http://www.icr.ac.uk/cancgen/molgen/MolPopGen_PICS_database.htm) of potentially functional nsSNPs in genes with relevance to cancer biology (Rudd et al. 2005). Briefly, candidate cancer genes were identified by interrogating the Gene Ontology Consortium database (http://www.geneontology.org; Ashburner et al. 2000), Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg; Kanehisa et al. 2004), Stratagene’s Interaction Explorer Pathway Assist Program (http://www.iobion.com/news/hotnews. html?cmd=Retrieve&dopt=Abstract), National Center for Biotechnology Information (NCBI) Entrez Gene database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=gene; Maglott et al. 2005), and the CancerGene database (http://caroll.vjf.cnrs.fr/cancergene/HOME.html). A total of 9537 validated nsSNPs with minor allele frequency (MAF) data were identified within 21,506 LocusLink annotated genes in NCBI dbSNP Build 123 (http://www.ncbi.nlm.nih.gov/SNP/; Sherry et al. 2001). Filtering this list and linking it to 7080 candidate cancer genes yielded 3666 validated nsSNPs with MAF ≥ 0.01 in Caucasian populations. The functional impact of nsSNPs was predicted using the in silico computational tools PolyPhen (http://www.bork.embl-heidelberg.de/PolyPhen/; Ramensky et al. 2002) and SIFT (version 2.1; http://blocks.fhcrc.org/sift/SIFT.html; Ng and Henikoff 2001). Using the PICS database and published work on resequencing of DNA repair genes (Ford et al. 2000; Kuschel et al. 2002; Mohrenweiser et al. 2002; Fearnhead et al. 2004; Savas et al. 2004) we prioritized a set of 1476 nsSNPs for the current study. For those SNPs yet to be documented in the latest release of NCBI dbSNP (Build 125), we have submitted complete genotype information including MAF to NCBI and assigned the resultant dbSNP ‘ss’ designations accordingly. Annotated flanking sequence information for each SNP was derived from the University of California Santa Cruz (UCSC) Human Genome Browser (Assembly hg17; http://genome.ucsc.edu/cgi-bin/hgGateway).
SNP genotyping and data manipulation
Genotyping of samples was performed using customized Illumina Sentrix Bead Arrays according to the manufacturer’s protocols. DNA samples with GenCall scores <0.25 at any locus were considered “no calls.” A DNA sample was deemed to have failed if it generated genotypes at <95% of loci. A SNP was deemed to have failed if <95% of DNA samples generated a genotype at the locus. Conversion of genotype data into formats suitable for processing was performed using in-house Perl scripts (available upon request). Conventional statistical manipulations were undertaken in STATA (version 8; http://www.stata.com), S-Plus (version 7; http://www.insightful.com), or R (version 2.0.0; http://www.r-project.org).
Population stratification
Genotypic frequencies in control subjects for each SNP were tested for departure from Hardy-Weinberg equilibrium (HWE) using a χ2 test or Fisher’s exact test, where an expected cell count was less than five. SNPs that violate HWE in the control population can indicate selection bias or genotyping errors; these were removed from further analysis. To detect and control for possible population stratification, we used the genomic control approach (Devlin and Roeder 1999), using all SNPs to estimate the stratification parameter  and its associated 95% confidence interval (CI).
and its associated 95% confidence interval (CI).
Risk of lung cancer associated with nsSNPs
The most efficient test of association depends on the true mode of allelic inheritance. Since this is not known, we based our analyses on the difference between allelic frequencies in cases and controls using a χ2 test with one degree of freedom, or Fisher’s exact test if the expected numbers in individual cells were less than five. We denote this test statistic TA with corresponding P-value PA. We also investigated two further tests based on 2 × 2 tables combining the heterozygotes with either the common or rare homozygotes to derive the statistics TR and TD with corresponding P-values PR and PD, which are most powerful under recessive or dominant models, respectively. The risks associated with each SNP were estimated by allelic, dominant, and recessive odds ratios (ORs) using unconditional logistic regression. Associated 95% confidence intervals (CI) were calculated in each case. Where it was not possible to calculate ORs by asymptotic methods, an exact approach was implemented using LogXact software (http://www.cytel.com; Cytel Corporation).
To increase the power to detect associations, we further analyzed case and control genotypes adopting a set-association approach, combining the largest TA statistics from individual tests into a single genome-wide statistic to model the joint effects of individual loci on lung cancer risk. Set-association analysis was conducted using the Sumstat program (Hoh et al. 2001), performing 50,000 iterations, and setting the maximum possible number of terms in the sum to be 100. The significance of this statistic was estimated through permutation, adjusting for the number of terms in the set being, a priori, unknown.
Multiple testing
Standard approaches to adjust for multiple testing such as the Bonferroni correction are known to be conservative due to their reliance on the assumption of independence between tests, which can lead to type I errors. To control error rate, we adopted an empirical Monte Carlo simulation approach (Churchill and Doerge 1994) based on 10,000 permutations, which takes into account the fact that tests may be correlated due to the presence of LD throughout the genome. At each iteration, case and control labels are permuted at random and maximum test statistics TAmax, TDmax, and TRmax are determined. Significance levels of the observed statistics from the original data are then estimated by the proportion of permutation samples with Tmax larger than that in the observed data. Although this approach adjusts for multiple testing for each of the three statistics separately, the consequent increase in false-positive rate is expected to be small due to the strong dependence between tests.
Assessment of linkage disequilibrium between SNPs
To identify SNPs in high LD, we calculated the pairwise LD measure D′ between consecutive pairs of markers throughout the genome using the expectation–maximization algorithm to estimate two-locus haplotype frequencies. We computed D′ for SNPs with MAF >0.1%, as the distribution of LD estimates for SNPs with smaller MAF was found to be unstable. For the purposes of this study, a pair of SNPs was defined as being in high LD if they had pairwise LD measure D′ > 0.5. This information was used to investigate the relationship between haplotypes and disease status. Specifically, haplotypes were reconstructed using a Markov chain Monte Carlo method, and their frequencies in cases and controls compared by permutation testing using the program PHASE (Stephens et al. 2001; Stephens and Donnelly 2003).
Covariates and interactions
Information on a number of covariates was available for the cases, including family history of lung cancer, histology, age at diagnosis, smoking history, and asbestos exposure. Analyses were only undertaken for subgroups with a sample size >300. The test statistics TA, TR, and TD were computed for all subgroups, together with ORs and their associated 95% CIs. The set association approach was also implemented for each subgroup.
Under certain conditions, a two-stage process incorporating estimates of pairwise interaction between significant SNPs can yield greater power to detect association (Marchini et al. 2005). To investigate epistatic interactions, each pair of SNPs that showed significant allelic association at the 5% level were tested fitting a saturated logistic regression model, and the log likelihood ratio statistic for comparison with the main effects model computed. This was compared against a χ2 distribution with 1 d.f. Statistics were then adjusted for multiple testing using a Bonferroni correction.
Acknowledgments
Funding for this work was undertaken with support from Cancer Research UK, the Arbib Foundation, HEAL, the National Cancer Research Network, the European Union Network of Excellence, and the Institute of Cancer Research. A.M. was the recipient of a clinical research fellowship from the Allan J. Lerner Fund. We gratefully acknowledge the participation of patient and control individuals. The authors are indebted to Richard Coleman, Christina Fleischmann, Olivia Fletcher, Nick Hearle, Nichola Johnson, Rosalind Mutch, Claire Palle, Julian Peto, Mobshra Qureshi, Elaine Ryder-Mills, Hayley Spendlove, and Remben Talaban for sample ascertainment. We thank Jurg Ott for access to a recompiled version of his Sumstat program.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5120106
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Wilson J.E., Amos C.I., Pinney S.M., Petersen G.M., de Andrade M., Wiest J.S., Fain P., Schwartz A.G., You M., Franklin W., Amos C.I., Pinney S.M., Petersen G.M., de Andrade M., Wiest J.S., Fain P., Schwartz A.G., You M., Franklin W., Pinney S.M., Petersen G.M., de Andrade M., Wiest J.S., Fain P., Schwartz A.G., You M., Franklin W., Petersen G.M., de Andrade M., Wiest J.S., Fain P., Schwartz A.G., You M., Franklin W., de Andrade M., Wiest J.S., Fain P., Schwartz A.G., You M., Franklin W., Wiest J.S., Fain P., Schwartz A.G., You M., Franklin W., Fain P., Schwartz A.G., You M., Franklin W., Schwartz A.G., You M., Franklin W., You M., Franklin W., Franklin W., et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am. J. Hum. Genet. 2004;75:460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D.W., Gore I., Okimoto R.A., Godin-Heymann N., Sordella R., Mulloy R., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J., Gore I., Okimoto R.A., Godin-Heymann N., Sordella R., Mulloy R., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J., Okimoto R.A., Godin-Heymann N., Sordella R., Mulloy R., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J., Godin-Heymann N., Sordella R., Mulloy R., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J., Sordella R., Mulloy R., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J., Mulloy R., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J., Brannigan B.W., Mohapatra G., Settleman J., Mohapatra G., Settleman J., Settleman J., et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat. Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- Chan J.M., Stampfer M.J., Giovannucci E., Gann P.H., Ma J., Wilkinson P., Hennekens C.H., Pollak M., Stampfer M.J., Giovannucci E., Gann P.H., Ma J., Wilkinson P., Hennekens C.H., Pollak M., Giovannucci E., Gann P.H., Ma J., Wilkinson P., Hennekens C.H., Pollak M., Gann P.H., Ma J., Wilkinson P., Hennekens C.H., Pollak M., Ma J., Wilkinson P., Hennekens C.H., Pollak M., Wilkinson P., Hennekens C.H., Pollak M., Hennekens C.H., Pollak M., Pollak M. Plasma insulin-like growth factor-I and prostate cancer risk: A prospective study. Science. 1998;279:563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- Churchill G.A., Doerge R.W., Doerge R.W. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Doll R., Peto R., Peto R. The causes of cancer: Quantitative estimates of avoidable risks of cancer in the United States today. J. Natl. Cancer Inst. 1981;66:1191–1308. [PubMed] [Google Scholar]
- Fearnhead N.S., Wilding J.L., Winney B., Tonks S., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F., Wilding J.L., Winney B., Tonks S., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F., Winney B., Tonks S., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F., Tonks S., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F., Tomlinson I.P., Mortensen N.J., Bodmer W.F., Mortensen N.J., Bodmer W.F., Bodmer W.F. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc. Natl. Acad. Sci. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford B.N., Ruttan C.C., Kyle V.L., Brackley M.E., Glickman B.W., Ruttan C.C., Kyle V.L., Brackley M.E., Glickman B.W., Kyle V.L., Brackley M.E., Glickman B.W., Brackley M.E., Glickman B.W., Glickman B.W. Identification of single nucleotide polymorphisms in human DNA repair genes. Carcinogenesis. 2000;21:1977–1981. doi: 10.1093/carcin/21.11.1977. [DOI] [PubMed] [Google Scholar]
- Garber M.E., Troyanskaya O.G., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M., de van Rijn M., Rosen G.D., Perou C.M., Whyte R.I., Troyanskaya O.G., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M., de van Rijn M., Rosen G.D., Perou C.M., Whyte R.I., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M., de van Rijn M., Rosen G.D., Perou C.M., Whyte R.I., Petersen S., Thaesler Z., Pacyna-Gengelbach M., de van Rijn M., Rosen G.D., Perou C.M., Whyte R.I., Thaesler Z., Pacyna-Gengelbach M., de van Rijn M., Rosen G.D., Perou C.M., Whyte R.I., Pacyna-Gengelbach M., de van Rijn M., Rosen G.D., Perou C.M., Whyte R.I., de van Rijn M., Rosen G.D., Perou C.M., Whyte R.I., Rosen G.D., Perou C.M., Whyte R.I., Perou C.M., Whyte R.I., Whyte R.I., et al. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh J., Wille A., Ott J., Wille A., Ott J., Ott J. Trimming, weighting, and grouping SNPs in human case-control association studies. Genome Res. 2001;11:2115–2119. doi: 10.1101/gr.204001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett N.G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., Persky N., Grompe M., Joenje H., Pals G., Grompe M., Joenje H., Pals G., Joenje H., Pals G., Pals G., et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Hwang S.J., Cheng L.S., Lozano G., Amos C.I., Gu X., Strong L.C., Cheng L.S., Lozano G., Amos C.I., Gu X., Strong L.C., Lozano G., Amos C.I., Gu X., Strong L.C., Amos C.I., Gu X., Strong L.C., Gu X., Strong L.C., Strong L.C. Lung cancer risk in germline p53 mutation carriers: Association between an inherited cancer predisposition, cigarette smoking, and cancer risk. Hum. Genet. 2003;113:238–243. doi: 10.1007/s00439-003-0968-7. [DOI] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer). IARC Monographs on the evaluation of the carcinogenic risk of chemicals to humans: Tobacco smoking. IARC; Lyon, France: 1986. [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M., Goto S., Kawashima S., Okuno Y., Hattori M., Kawashima S., Okuno Y., Hattori M., Okuno Y., Hattori M., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandwala H.M., McCutcheon I.E., Flyvbjerg A., Friend K.E., McCutcheon I.E., Flyvbjerg A., Friend K.E., Flyvbjerg A., Friend K.E., Friend K.E. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endocr. Rev. 2000;21:215–244. doi: 10.1210/edrv.21.3.0399. [DOI] [PubMed] [Google Scholar]
- Kuschel B., Auranen A., McBride S., Novik K.L., Antoniou A., Lipscombe J.M., Day N.E., Easton D.F., Ponder B.A., Pharoah P.D., Auranen A., McBride S., Novik K.L., Antoniou A., Lipscombe J.M., Day N.E., Easton D.F., Ponder B.A., Pharoah P.D., McBride S., Novik K.L., Antoniou A., Lipscombe J.M., Day N.E., Easton D.F., Ponder B.A., Pharoah P.D., Novik K.L., Antoniou A., Lipscombe J.M., Day N.E., Easton D.F., Ponder B.A., Pharoah P.D., Antoniou A., Lipscombe J.M., Day N.E., Easton D.F., Ponder B.A., Pharoah P.D., Lipscombe J.M., Day N.E., Easton D.F., Ponder B.A., Pharoah P.D., Day N.E., Easton D.F., Ponder B.A., Pharoah P.D., Easton D.F., Ponder B.A., Pharoah P.D., Ponder B.A., Pharoah P.D., Pharoah P.D., et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum. Mol. Genet. 2002;11:1399–1407. doi: 10.1093/hmg/11.12.1399. [DOI] [PubMed] [Google Scholar]
- Ma J., Pollak M.N., Giovannucci E., Chan J.M., Tao Y., Hennekens C.H., Stampfer M.J., Pollak M.N., Giovannucci E., Chan J.M., Tao Y., Hennekens C.H., Stampfer M.J., Giovannucci E., Chan J.M., Tao Y., Hennekens C.H., Stampfer M.J., Chan J.M., Tao Y., Hennekens C.H., Stampfer M.J., Tao Y., Hennekens C.H., Stampfer M.J., Hennekens C.H., Stampfer M.J., Stampfer M.J. Prospective study of colorectal cancer risk in men and plasma levels of insulin-like growth factor (IGF)-I and IGF-binding protein-3. J. Natl. Cancer Inst. 1999;91:620–625. doi: 10.1093/jnci/91.7.620. [DOI] [PubMed] [Google Scholar]
- Maglott D., Ostell J., Pruitt K.D., Tatusova T., Ostell J., Pruitt K.D., Tatusova T., Pruitt K.D., Tatusova T., Tatusova T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Donnelly P., Cardon L.R., Donnelly P., Cardon L.R., Cardon L.R. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Martin S.T., Matsubayashi H., Rogers C.D., Philips J., Couch F.J., Brune K., Yeo C.J., Kern S.E., Hruban R.H., Goggins M., Matsubayashi H., Rogers C.D., Philips J., Couch F.J., Brune K., Yeo C.J., Kern S.E., Hruban R.H., Goggins M., Rogers C.D., Philips J., Couch F.J., Brune K., Yeo C.J., Kern S.E., Hruban R.H., Goggins M., Philips J., Couch F.J., Brune K., Yeo C.J., Kern S.E., Hruban R.H., Goggins M., Couch F.J., Brune K., Yeo C.J., Kern S.E., Hruban R.H., Goggins M., Brune K., Yeo C.J., Kern S.E., Hruban R.H., Goggins M., Yeo C.J., Kern S.E., Hruban R.H., Goggins M., Kern S.E., Hruban R.H., Goggins M., Hruban R.H., Goggins M., Goggins M. Increased prevalence of the BRCA2 polymorphic stop codon K3326X among individuals with familial pancreatic cancer. Oncogene. 2005;24:3652–3656. doi: 10.1038/sj.onc.1208411. [DOI] [PubMed] [Google Scholar]
- Matakidou A., Eisen T., Houlston R.S., Eisen T., Houlston R.S., Houlston R.S. Systematic review of the relationship between family history and lung cancer risk. Br. J. Cancer. 2005;93:825–833. doi: 10.1038/sj.bjc.6602769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrenweiser H.W., Xi T., Vazquez-Matias J., Jones I.M., Xi T., Vazquez-Matias J., Jones I.M., Vazquez-Matias J., Jones I.M., Jones I.M. Identification of 127 amino acid substitution variants in screening 37 DNA repair genes in humans. Cancer Epidemiol. Biomarkers Prev. 2002;11:1054–1064. [PubMed] [Google Scholar]
- Moon J.W., Chang Y.S., Ahn C.W., Yoo K.N., Shin J.H., Kong J.H., Kim Y.S., Chang J., Kim S.K., Kim H.J., Chang Y.S., Ahn C.W., Yoo K.N., Shin J.H., Kong J.H., Kim Y.S., Chang J., Kim S.K., Kim H.J., Ahn C.W., Yoo K.N., Shin J.H., Kong J.H., Kim Y.S., Chang J., Kim S.K., Kim H.J., Yoo K.N., Shin J.H., Kong J.H., Kim Y.S., Chang J., Kim S.K., Kim H.J., Shin J.H., Kong J.H., Kim Y.S., Chang J., Kim S.K., Kim H.J., Kong J.H., Kim Y.S., Chang J., Kim S.K., Kim H.J., Kim Y.S., Chang J., Kim S.K., Kim H.J., Chang J., Kim S.K., Kim H.J., Kim S.K., Kim H.J., Kim H.J., et al. Promoter-202 A/C polymorphism of insulin-like growth factor binding protein-3 gene and non-small cell lung cancer risk. Int. J. Cancer. 2006;118:353–356. doi: 10.1002/ijc.21339. [DOI] [PubMed] [Google Scholar]
- Morimatsu M., Donoho G., Hasty P., Donoho G., Hasty P., Hasty P. Cells deleted for Brca2 COOH terminus exhibit hypersensitivity to γ-radiation and premature senescence. Cancer Res. 1998;58:3441–3447. [PubMed] [Google Scholar]
- Ng P.C., Henikoff S., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin D.M., Bray F., Ferlay J., Pisani P., Bray F., Ferlay J., Pisani P., Ferlay J., Pisani P., Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Ramensky V., Bork P., Sunyaev S., Bork P., Sunyaev S., Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd M.F., Williams R.D., Webb E.L., Schmidt S., Sellick G.S., Houlston R.S., Williams R.D., Webb E.L., Schmidt S., Sellick G.S., Houlston R.S., Webb E.L., Schmidt S., Sellick G.S., Houlston R.S., Schmidt S., Sellick G.S., Houlston R.S., Sellick G.S., Houlston R.S., Houlston R.S. The predicted impact of coding single nucleotide polymorphisms database. Cancer Epidemiol. Biomarkers Prev. 2005;14:2598–2604. doi: 10.1158/1055-9965.EPI-05-0469. [DOI] [PubMed] [Google Scholar]
- Sanders B.M., Jay M., Draper G.J., Roberts E.M., Jay M., Draper G.J., Roberts E.M., Draper G.J., Roberts E.M., Roberts E.M. Non-ocular cancer in relatives of retinoblastoma patients. Br. J. Cancer. 1989;60:358–365. doi: 10.1038/bjc.1989.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas S., Kim D.Y., Ahmad M.F., Shariff M., Ozcelik H., Kim D.Y., Ahmad M.F., Shariff M., Ozcelik H., Ahmad M.F., Shariff M., Ozcelik H., Shariff M., Ozcelik H., Ozcelik H. Identifying functional genetic variants in DNA repair pathway using protein conservation analysis. Cancer Epidemiol. Biomarkers Prev. 2004;13:801–807. [PubMed] [Google Scholar]
- Schuller A.G., van Neck J.W., Beukenholdt R.W., Zwarthoff E.C., Drop S.L., van Neck J.W., Beukenholdt R.W., Zwarthoff E.C., Drop S.L., Beukenholdt R.W., Zwarthoff E.C., Drop S.L., Zwarthoff E.C., Drop S.L., Drop S.L. IGF, type I IGF receptor and IGF-binding protein mRNA expression in the developing mouse lung. J. Mol. Endocrinol. 1995;14:349–355. doi: 10.1677/jme.0.0140349. [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker L., Phan L., Smigielski E.M., Sirokin K., Ward M.H., Kholodov M., Baker L., Phan L., Smigielski E.M., Sirokin K., Kholodov M., Baker L., Phan L., Smigielski E.M., Sirokin K., Baker L., Phan L., Smigielski E.M., Sirokin K., Phan L., Smigielski E.M., Sirokin K., Smigielski E.M., Sirokin K., Sirokin K. dSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenina O.I., Ustinov V., Krukovets I., Marinic T., Topol E.J., Plow E.F., Ustinov V., Krukovets I., Marinic T., Topol E.J., Plow E.F., Krukovets I., Marinic T., Topol E.J., Plow E.F., Marinic T., Topol E.J., Plow E.F., Topol E.J., Plow E.F., Plow E.F. Polymorphisms A387P in thrombospondin-4 and N700S in thrombospondin-1 perturb calcium binding sites. FASEB J. 2005;19:1893–1895. doi: 10.1096/fj.05-3712fje. [DOI] [PubMed] [Google Scholar]
- Stephens M., Donnelly P., Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Smith N.J., Donnelly P., Smith N.J., Donnelly P., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya M., Shiraishi S., Teramoto T., Miki Y., Shiraishi S., Teramoto T., Miki Y., Teramoto T., Miki Y., Miki Y. Bloom’s syndrome with porokeratosis of Mibelli and multiple cancers of the skin, lung and colon. Clin. Genet. 1987;31:35–44. doi: 10.1111/j.1399-0004.1987.tb02764.x. [DOI] [PubMed] [Google Scholar]
- Toniolo P., Bruning P.F., Akhmedkhanov A., Bonfrer J.M., Koenig K.L., Lukanova A., Shore R.E., Zeleniuch-Jacquotte A., Bruning P.F., Akhmedkhanov A., Bonfrer J.M., Koenig K.L., Lukanova A., Shore R.E., Zeleniuch-Jacquotte A., Akhmedkhanov A., Bonfrer J.M., Koenig K.L., Lukanova A., Shore R.E., Zeleniuch-Jacquotte A., Bonfrer J.M., Koenig K.L., Lukanova A., Shore R.E., Zeleniuch-Jacquotte A., Koenig K.L., Lukanova A., Shore R.E., Zeleniuch-Jacquotte A., Lukanova A., Shore R.E., Zeleniuch-Jacquotte A., Shore R.E., Zeleniuch-Jacquotte A., Zeleniuch-Jacquotte A. Serum insulin-like growth factor-I and breast cancer. Int. J. Cancer. 2000;88:828–832. doi: 10.1002/1097-0215(20001201)88:5<828::aid-ijc22>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Wang X., Andreassen P.R., D’Andrea A.D., Andreassen P.R., D’Andrea A.D., D’Andrea A.D. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi T., Jones I.M., Mohrenweiser H.W., Jones I.M., Mohrenweiser H.W., Mohrenweiser H.W. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83:970–979. doi: 10.1016/j.ygeno.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Yamanaka A., Hirai T., Ohtake Y., Kitagawa M., Hirai T., Ohtake Y., Kitagawa M., Ohtake Y., Kitagawa M., Kitagawa M. Lung cancer associated with Werner’s syndrome: A case report and review of the literature. Jpn. J. Clin. Oncol. 1997;27:415–418. doi: 10.1093/jjco/27.6.415. [DOI] [PubMed] [Google Scholar]
- Yu H., Spitz M.R., Mistry J., Gu J., Hong W.K., Wu X., Spitz M.R., Mistry J., Gu J., Hong W.K., Wu X., Mistry J., Gu J., Hong W.K., Wu X., Gu J., Hong W.K., Wu X., Hong W.K., Wu X., Wu X. Plasma levels of insulin-like growth factor-I and lung cancer risk: A case-control analysis. J. Natl. Cancer Inst. 1999;91:151–156. doi: 10.1093/jnci/91.2.151. [DOI] [PubMed] [Google Scholar]