Abstract
Asynchronous DNA replication is an epigenetically determined feature found in all cases of monoallelic expression, including genomic imprinting, X-inactivation, and random monoallelic expression of autosomal genes such as immunoglobulins and olfactory receptor genes. Most genes of the latter class were identified in experiments focused on genes functioning in the chemosensory and immune systems. We performed an unbiased survey of asynchronous replication in the mouse genome, excluding known asynchronously replicated genes. Fully 10% (eight of 80) of the genes tested exhibited asynchronous replication. A common feature of the newly identified asynchronously replicated areas is their proximity to areas of tandem gene duplication. Testing of other clustered areas supported the idea that such regions are enriched with asynchronously replicated genes.
Asynchronous DNA replication is a hallmark of mammalian genes that exhibit monoallelic expression. Such genes can be divided into three broad classes. For imprinted genes, transcriptional activity of an allele depends on whether it was inherited from the father or from the mother (Bartolomei and Tilghman 1997). With X-inactivation in females, cells render one of their two copies of the X chromosome inactive to compensate for the double dosage of X-linked genes. This epigenetic choice is maintained in each cell’s progeny, making the animal’s tissues mosaic for expression of X-linked genes (Lyon 1994). Finally, a number of autosomal genes exhibit random monoallelic expression.
In many respects, random monoallelic expression is an autosomal analog of X-inactivation. With equal probability, cells make a decision whether the paternal or maternal allele will be active, and the choice is epigenetically maintained in each cell’s progeny (Bix and Locksley 1998; Gimelbrant et al. 2005). Asynchronous replication, which is associated with random monoallelic expression, is coordinated in a chromosome-wide manner: Such loci on the same chromosome are replicated in concert early, while on the opposite chromosome, the same loci are replicated later during S-phase (Singh et al. 2003; Ensminger and Chess 2004). As with the establishment of the early- and late-replicated X chromosomes, the early or late replication timing is set for such genes in early embryogenesis and maintained in clonal cell populations.
Initially, random monoallelic expression was described as “allelic exclusion” of immunoglobulins (Pernis et al. 1965): In a given lymphocyte, either the paternal or maternal allele of the Ig locus would be functional, with half the cells making either choice. The discovery of monoallelic expression in olfactory receptor (OR) genes (Chess et al. 1994) demonstrated that this epigenetic phenomenon is not limited to immunoglobulins. With ∼1000 individual members (Young and Trask 2002), the OR family accounts for about 4% of all known mouse genes, making the genes subject to random monoallelic expression a sizable fraction of the genome. However, all the subsequently identified genes of this class also belonged to the chemosensory system [vomeronasal receptors (Rodriguez et al. 1999)] or to the immune system [e.g., natural killer cell receptors Ly49A (curently Kira1), and Ly49C (currently Kira3), (Held et al. 1995), Toll-like receptor Tlr4 (Pereira et al. 2003), and interleukin 4 (Bix and Locksley 1998; Riviere et al. 1998)].
One possibility is that random monoallelic expression is limited to the genes specific to these two systems, and thus the known list of the genes subject to monoallelic expression and asynchronous DNA replication is nearly complete. If that is not the case, a sampling of genes that do not belong to the known list should yield novel examples of the genes of this type.
Unlike monoallelic expression, asynchronous DNA replication is independent of whether a gene is expressed in a given cell type or not. To take one example, OR genes are asynchronously replicated in fibroblasts, embryonic stem (ES) cells, and lymphocytes (Mostoslavsky et al. 2001; Singh et al. 2003), whereas their transcription is restricted to olfactory sensory neurons. Importantly, random asynchronous DNA replication is maintained to the same degree as monoallelic expression: In a given clonal cell line all cells would replicate the maternal copy of an OR locus early, and the paternal copy late, even as the opposite would be the case in the progeny of another cell (Singh et al. 2003).
We have shown before that S-phase fractionation of the cells from unsynchronized clonal ES cell lines can be used to detect random asynchronous replication (Singh et al. 2003; Gimelbrant et al. 2005). Here, we report the use of this approach for a survey of the mouse genome to obtain an estimate of the fraction of asynchronously replicated genes in a mammalian genome.
Results
Survey of asynchronous replication in the mouse genome
For each gene analyzed, to distinguish between synchronous and asynchronous DNA replication, we measured relative maternal and paternal allele content in eight cell-cycle fractions from a clonal 129×CastF1 ES cell line. Since the 129/SvJ (129) and Cast/Ei (Cast) mouse strains are quite divergent, this F1 hybrid is rich in single nucleotide polymorphisms (SNPs) (Lindblad-Toh et al. 2000). For each interrogated SNP, a primer-extension reaction was performed on PCR-amplified DNA from each cell cycle fraction, and the relative content of paternal (Cast) and maternal (129) allele was determined by mass spectrometry.
For a locus with synchronous DNA replication, one expects to observe equal amounts of each allele throughout the S phase. For asynchronously replicated genes, when analyzing a clonal cell line, one or more of the fractions should differ, with the early replicating allele becoming more abundant until the other allele is replicated. This skewing can be as large as 2:1 (66%:33%), reflecting the particular point of the S phase when one allele has been completely replicated and the other allele is completely unreplicated. In practice, the observed skewing is often smaller, probably because of the limited resolution of cell fractionation. Take, for example, the M12 olfactory receptor gene, for which the fractional content of the early replicated allele increases to 55% in the first fraction of the S phase (Fig. 1A). Following on this and other examples, we chose a cutoff where an allele had to reach greater than 55% or less than 45% abundance (in one or more contiguous fractions of the S-phase) in order for that gene to be considered asynchronously replicated (and chosen for corroboration using a FISH assay, see below).
Figure 1.
Survey of the mouse genome for asynchronous DNA replication. (A) Detection of asynchronous and synchronous replication by quantitative genotyping of cell-cycle fractions. An unsynchronized, clonal population of ES cells from 129×CastF1 mouse was FACS-sorted into eight fractions according to DNA content: G0/G1, G2, and six S-phase fractions. DNA from each fraction was analyzed for relative allele content in a SNP within a gene of interest using Sequenom primer-extension approach. For each cell-cycle fraction, the relative content of the 129 allele is shown (mean ± SEM; n = 4). To correct for detection bias and confirm linearity of allele detection, each experiment included known mixes of parental genomic DNA. Synchronously replicated genes, even those with noisy profiles such as Pep4 (currently Pepd), showed less then ±5% difference in any of the S-phase fractions. Known asynchronously replicated loci, e.g., M12 olfactory receptor (OR) had at least one fraction with one allele significantly more abundant than the other. See also Gimelbrant et al. (2005) for additional examples of replication profiles. Note that since we had prior knowledge of the OR replication timing status, these loci were only used as internal controls and were not included in the survey set. Based on the results for the ORs, a threshold of ±5% was chosen to distinguish synchronous and asynchronous replication (gray bar). (B) Replication profiles of novel asynchronously replicated loci identified in the survey. Replication profile of Catns (p120 catenin) obtained in the survey has been published elsewhere (Gimelbrant et al. 2005). (C) Distribution of the surveyed loci in the mouse genome. All the assessed loci (tick marks; the width of the marks is not to scale) are depicted according to their placement on the mouse genome assembly (NCBI build 33). The full list of assessed loci is in Supplemental Table S1. Acrosomes are to the left. Asynchronously replicated genes identified in this survey are denoted by larger tick marks and gene symbols.
For the survey, we screened a set of 80 genes that had been randomly selected from known transcripts polymorphic in 129×CastF1 (Lindblad-Toh et al. 2000); the set has genes from each mouse autosome (Fig. 1C; Supplemental Table 1). The survey set excluded known monoallelically expressed and asynchronously replicated genes, such as odorant receptor genes. We have described (Gimelbrant et al. 2005) asynchronous replication and monoallelic expression in mouse and human cells of one of the genes found in the survey, Catns (note that we have used “p120 catenin” to refer to both mouse Catns and its human ortholog CTNND1).
We find that of the 80 assessed genes, eight exhibited asynchronous DNA replication as indicated by one or more fractions of S-phase displaying fractional allele content differing from 50%:50% by more than the threshold 5% (Fig. 1B). Some of these genes are widely expressed, such as cell adhesion molecule Catns and NFκB inhibitor kinase Chuk; others are expressed in a more restricted manner, such as mast cell protease Mcpt5 (currently Cma1), Fgf3, transcription factor Tcf1, ectonucleotide phosphatase Enpp1, melanocortin receptor Mc3r, and somatostatin receptor Sstr3. For some of the eight genes, the prevalence of the early replicated allele was near the theoretical limit of 2:1 (e.g., Sstr3 and Chuk, Fig. 1B). For other genes, the skewing was more modest, similar to that observed for the M12 OR gene.
Corroboration of asynchronous replication with FISH assay
To check the replication synchrony status with an independent approach, we used a fluorescence in situ hybridization (FISH) replication timing assay (Selig et al. 1992; Kitsberg et al. 1993). In this assay, the numbers of the hybridization signals for a locus of interest are counted in S-phase interphase nuclei labeled with BrdU. A configuration where the locus on one chromosome is visualized as a single hybridization spot and that on the other chromosome is visualized as a doublet is diagnostic of asynchronous replication. We will refer to this configuration as SD for singlet–doublet. For most genes (whose alleles are synchronously replicated), SD nuclei are relatively rare (about 10%–20%), and the predominant patterns of hybridization are either two single spots (SS), representing two unreplicated alleles, or two doublets (DD), each representing a newly replicated allele. Asynchronously replicated genes reveal a higher range (30%–45%) of SD cells. Note that the fraction of cells with a visible doublet signal for a given allele may be influenced by differences in sister chromatid cohesion, especially when different FISH protocols are used (Azuara et al. 2003). These protocols (known as three-dimensional FISH) use substantially different, milder cell-fixation and denaturation conditions to visualize cohesion. By contrast, the FISH conditions we use are designed to minimize the detection of differences in sister chromatid cohesion (see Methods).
Probes for all eight of the candidate genes revealed a high percentage of primary fibroblast nuclei with the SD pattern [Catns 44% SD (Gimelbrant et al. 2005), Mc3r 41%, Tcf1 35%, Fgf3 41%, Enpp1 48%, Mcpt5 44%, Sstr3 45%, Chuk 45%; Supplemental Fig. S1], providing independent corroboration of their asynchronous replication. As is characteristic for asynchronously replicated genes studied previously, the high SD fraction can be observed in a variety of cell types of various genetic backgrounds. In addition to primary fibroblasts, similar results were observed in 129×CastF1 and 129 ES cells, Abl-MLV transformed lymphocytes (129×CastF1 and 129×SpretF1), and 129×CastF1 and C57BL/6 SV40 large T- and E1A-transformed fibroblasts, using both PCR-generated products and bacterial artificial chromosome clones as probes (not shown).
Corroboration of the asynchronous DNA replication of all eight candidate genes with the FISH assay indicates that the threshold in the fractionation assay used for the initial screen (Fig. 1) resulted in no false positives. As for the possibility of false negatives in the original screen, several genes called as synchronous, but near the threshold (Pep4, Fig. 1A; Oprm, not shown; Ccnf) were confirmed as synchronous in FISH assay (Supplemental Fig. S1; Gimelbrant et al. 2005). In addition, other genes that the survey called as synchronous (Hoxa10 and Ins1) have been previously assessed as synchronous in the FISH assay (Singh et al. 2003; Deltour et al. 2004). Still, it is possible that some assays were false negatives, and thus our finding of 10% asynchronous genes can be taken as a low-end estimate.
Expression of the novel asynchronously replicated genes
Given the association of asynchronous replication and monoallelic expression, we assessed the novel asynchronously replicated genes for imprinted or random monoallelic expression. First, we assessed the possibility that the novel asynchronously replicated loci are imprinted in a parent-specific manner by assessing their allele-specific expression in tissues from 129×CastF1 mice. Some of these genes had been previously assessed in tissues from hybrids of several mouse strains, including Cast/Ei, and did not show allele-specific bias in transcript level (Tcf1, Chuk, Sstr3; Cowles et al. 2002). To test the rest of the novel asynchronously replicated genes for imprinting, we measured their allele-specific transcript levels in brain, liver, spleen, heart, skeletal muscle, and gonads using primer extension approach (not shown). None exhibited a significant allele-specific transcript level bias, indicating that the asynchronously replicated genes identified in this study are not subject to genomic imprinting.
Unlike imprinting, random monoallelic expression cannot be observed in homogenized tissue samples, because of the mosaic nature of choice of the active allele. However, the choice of the transcriptionally active allele is clonally stable, and thus clonal cell populations can be used to assess whether a gene is subject to random monoallelic expression. We assessed allele-specific expression of the novel asynchronously replicated genes in clonal lines we generated from Abl-MLV transformed B lymphocytes and SV-40 transformed fibroblasts. These cells are the only differentiated mouse cell types that can be easily generated and subcloned. In these cells, only two of the genes were expressed at detectable level. The Catns gene was expressed monoallelically in over half the Abelson cell lines analyzed; other clonal cells’ lines expressed Catns biallelically (Gimelbrant et al. 2005). The Chuk gene was expressed from both alleles in all cell lines tested (not shown).
We also tested the allele-specific transcript level bias of several genes we found to be synchronously replicated (Caml, Ccnf, Eif3s10, Emp1, Fancc, Hira, Kcnab1, Nfkb1, Pep4, Siah1a, Sntb2, Stat5a, and Usf2). As expected, their expression was biallelic in Abelson clonal cell lines (not shown), consistent with previous studies indicating that synchronously replicated genes are biallelically expressed.
Asynchronously replicated genes are found in areas of tandem gene duplication
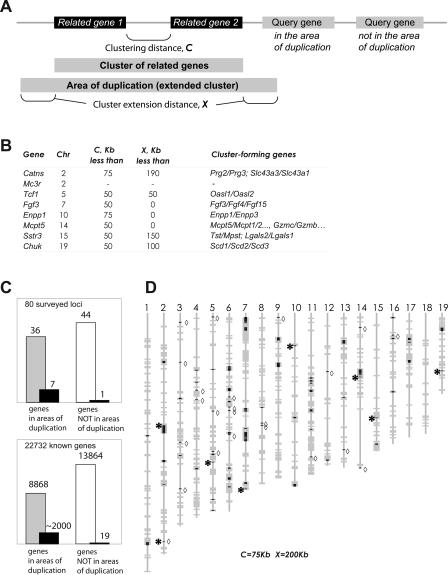
A striking feature of the novel asynchronously replicated genes identified in our survey is that seven out of the eight are situated near a cluster of related genes or are themselves a part of such a cluster. For example, Tcf1 is about 10 Kb from two closely related genes, Oasl1 and Oasl2, that are separated by ∼15 Kb. In another example, the Mcpt5 gene is located in a chromosome 14 array of multiple related protease genes. The definition of cluster that we used and these spatial relationships are summarized in Figure 2A,B.
Figure 2.
Asynchronously replicated genes are associated with regions of tandem gene duplication. (A) Defining the areas of gene duplication. Two or more genes are considered clustered if they encode related proteins (relatedness of proteins determined by BLASTP analysis of the complete nonredundant set of mouse protein-coding genes; see details in the text) and are no more than C Kb apart in the genome. To define an area of gene duplication, such a cluster is extended by X Kb in either direction. A gene is counted as belonging to an area of duplication if it overlaps in whole or partially with such an area; in the hypothetical depicted example, three genes fall within an area of gene duplication, and one does not. (B) Determining the values of parameters C and X for the whole-genome analysis. Minimal values of these parameters for each of the novel asynchronously replicated genes are shown. When the clustering parameters are set at C = 75 Kb, X = 200 Kb, then all these loci, with the exception of Mc3r, fall within the gene duplication areas. (C) Summary of the whole-genome clustering analysis. (Top) Of the 80 loci assessed in this study, 36 were in the areas of gene duplication (gray bar), and the rest were outside of such areas (white bar). Of the genes we identified as asynchronously replicated (black bars), seven were in the areas of duplication. (Bottom) 39% of mouse genes (8868 of 22,732 sequences analyzed) are in the areas of gene duplication (gray bar), with clustering parameters set at C = 75 Kb and X = 200 Kb uniformly applied to the locations of genes on the mouse genome assembly (NCBI build 33). The rest of the genes were not in such areas (white bar). Note that of known and presumed genes with asynchronous replication or random monoallelic expression (black bars) a vast majority are in the areas of gene duplication. (D) Distribution of the areas of gene duplication on the mouse genome. Locations of these areas were plotted on the mouse genome assembly (gray bars; acrosomes at top). A complete set of coordinates is in Supplemental Table S2. The duplicated areas largely overlap with the known or presumed loci with asynchronous replication (black tick marks): chemosensory receptor genes and pseudogenes, and known asynchronously replicated, monoallelically expressed genes from the immune system, as well as loci identified in this study as asynchronously replicated (asterisk to the left of the locus). Of ∼2000 known or presumed asynchronously replicated loci in the genome, we found only 19 that are apparent exceptions to this rule (diamond marks to the right of the locus).
To gauge the statistical significance of the observation that seven out of eight genes are in or near clusters of related genes, one must determine the expectation for a mouse gene to be located in such a genomic area. To accomplish this, we first determined the criteria that define the seven observed genes as being in or near the clusters, and then applied these criteria to all genes in the mouse genome. We derived the following set of criteria: Two related genes were considered to form a cluster if they were separated by less than 75 Kb; a gene would be considered within an area of duplication if it was within 200 Kb of the cluster (see Fig. 2A and Methods for details). The relatedness of genes was determined using a standard approach for detecting protein families (Lynch and Conery 2000): Each protein sequence was queried in a BLAST search against the complete set of mouse gene products; if expectation value E was below a threshold (e.g., 10−15), these proteins were considered related.
Applying these criteria to a nonredundant complete set of mouse genes (see Methods for definition), we find that 39% of mouse genes are in or near clusters of related genes (Fig. 3C). The probability that seven or eight observations out of eight belong to a group with the expected frequency of 0.39 is P < 0.01 (binomial distribution). Our findings were robust—they were not sensitive to small perturbations in procedure or in data sets. A different set of genes, defined as “Known” in Ensembl, yielded similar results (32%; P < 0.002). We also applied a different, pairwise procedure to find relatedness (Gu et al. 2002) with similar results.
Figure 3.
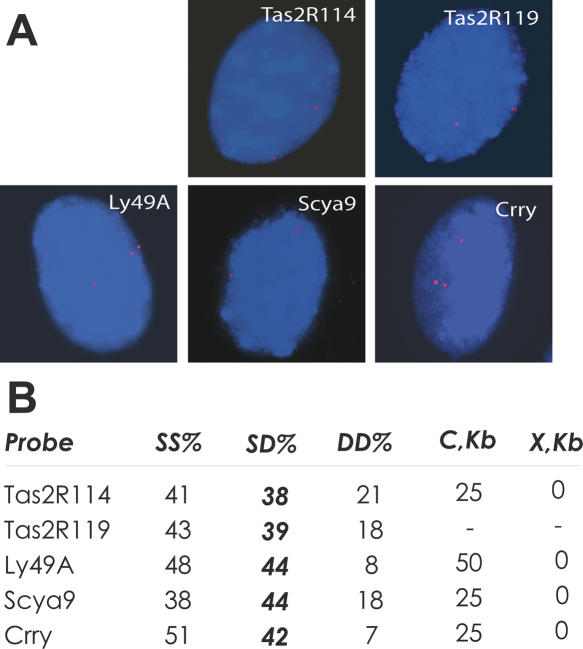
Sampling of genes in areas of tandem gene duplication reveals their asynchronous replication. (A) Mouse primary fibroblast nuclei with singlet–double (SD) hybridization pattern are shown. Red depicts PCR-generated Cy3-labeled probes for the denoted gene; blue depicts DAPI. (Supplemental Fig. S3A shows a high resolution version of this figure.) (B) Summary of the FISH assay for asynchronous replication in primary mouse fibroblasts and summary of spatial relationships (defined by the parameters C and X; see also Supplemental Fig. S3B) of the analyzed genes with clusters of related genes. (SS) Percentage of BrdU-positive nuclei with single–single hybridization signal; (SD) same for SD signal; (DD) same for double–double signal. In each case, 80–110 BrdU-positive nuclei were counted.
Thus, the asynchronous DNA replication is significantly correlated with the genomic areas with arrayed gene duplication. Of course, this conclusion is further strengthened when the previously known genes subject to random monoallelic expression and asynchronous replication are considered (Fig. 2C,D). The areas of gene duplication, as defined here, cover nearly 500 Mb of mouse genome, about 17% (Fig. 2D; Supplemental Table S2). A vast majority of the asynchronously replicated genes, including ORs, vomeronasal receptors, and the variety of immune system genes are located within this 17%. Of about 2000 such genes, only 19 are outside of the areas of tandem gene duplication.
Assessment of replication timing in additional areas of gene duplication
We went on to assess several areas of gene duplication for replication timing using the FISH assay. Consistent with the above analysis of the correlation between gene duplication and asynchronous replication, we found that a taste receptor gene Tas2R114, an array of natural killer receptor genes (containing Ly49A) previously shown to be monoallelically expressed (Held et al. 1995; Held and Raulet 1997), complement receptor Crry, and a cluster of cytokine precursor genes including Scya9 (currently Ccl9) were indeed all asynchronously replicated (Fig. 3; see Supplemental Fig. S2 for clustering data). These observations support the conclusion that clusters of tandemly duplicated genes are enriched in genes that exhibit asynchronous replication. Our preliminary data indicate that Scya9 is monoallelically expressed in Abelson clonal cell lines (not shown).
Asynchronous replication has been observed for standalone members of large, duplicated families of asynchronously replicated genes, such as ORs (Ensminger and Chess 2004). We tested a member of taste receptor family, Tas2R119, which is not near any cluster of duplicated genes, and found its replication to be asynchronous as well (Fig. 3).
Discussion
Over the last decade, the emergence of a class of autosomal genes subject to random monoallelic expression and asynchronous replication has raised the question of how prevalent this type of gene regulation is. To address this issue we surveyed 80 autosomal genes for asynchronous DNA replication, using a direct method. We measured relative allele content in several fractions of S phase using quantitative genotyping platform (Sequenom) (Xiong et al. 1998; Singh et al. 2003; Gimelbrant et al. 2005). We found that eight of 80 assessed loci were asynchronously replicated. This allows us to estimate (α = 0.95; binomial distribution) that 4.4%–18.7% of (non-OR) mouse genes are in the regions of asynchronous DNA replication. We had excluded from our surveyed gene set known asynchronously replicated genes, such as odorant receptors. Thus, the estimate from our survey is in addition to ∼4% of all genes taken up by known asynchronously replicated genes, suggesting that the fraction of genes in the mouse genome that are subject to asynchronous replication could range from 8% to as high as 23%. We should emphasize that we selected SNPs within known genes; thus, our frequency estimate is for the genes, as opposed to the genome as a whole.
There is no apparent common gene function or structural motif that distinguishes the newly identified asynchronously replicated loci, nor can they be grouped with previously identified loci. The 80 genes analyzed here represent a sampling of the genes in the genome; when the entire genome is assessed, we expect to find hundreds more genes subject to asynchronous replication. Based on our findings, these genes are likely to have a wide variety of functions and various patterns of expression.
Our estimate of the prevalence of asynchronously replicated genes is conservative. The initial S-phase fractionation screen (see Fig. 1) did not generate false positives; each was corroborated by the independent, FISH-based assay for replication timing (Supplemental Fig. S1). However, it is possible that some of the synchronous replication calls were false negatives. Our preliminary analyses indicated that clonal ES cell lines behaved similarly to clonal somatic cell lines in that the decision of which allele is early replicated and which is late appears to be stable (Singh et al. 2003; N. Singh, unpubl.). More recently, instability of asynchronous replication in several loci has been observed in some ES cell clones (Gribnau et al. 2005). Nevertheless, the 129×CastF1 ES cell clone ES-23 used for the survey displayed stability of direction of asynchronous replication in multiple loci that allowed us to conduct the survey. By design, the cell-cycle fractionation assay cannot detect replication asynchrony that is not consistently in the same direction throughout the monoclonal cell population used for cell-cycle fractionation. Thus, we cannot rule out the possibility that instability within the clonal cell population led to some false negatives. In addition, it is possible that the resolution of six S-phase fractions was insufficient to detect replication asynchrony for some asynchronously replicated loci.
The SNP-based assay directly measures asynchronous replication in a very small locus: the PCR-amplified area flanking the SNP, about 100–200 bp. However, this small sample represents a fairly large replication timing domain (e.g., Simon and Cedar 1999). Consistent with this, in FISH-based assays at candidate loci, both PCR-amplified probes (about 10 Kb; Fig. 2) and bacterial artificial chromosome-based probes (100–150 Kb; not shown) yielded SD scores characteristic for asynchronous replication.
The eight genes identified in this survey as asynchronously replicated were assessed for imprinted monoallelic expression: We and others (Cowles et al. 2002) found no evidence of parent-specific imprinting in several tested tissues. As for the random monoallelic expression, six of the eight novel asynchronously replicated genes were not expressed in available clonal cell lines. We were, however, able to assess two of these genes in such lines. Chuk showed biallelic expression in lymphocytes and fibroblasts; Catns showed random monoallelic expression in lymphocytes (but, interestingly, not in fibroblasts) (Gimelbrant et al. 2005).
From the survey data, we also observe that asynchronously replicated genes are enriched in areas of the genome containing tandem gene duplications. Of course, not every gene in such areas is asynchronously replicated, but the enrichment is about 10-fold, compared with that of the nonclustered areas (see Fig. 2C). This observation is further strengthened by the previously known clustering of the vast majority of the rest of the asynchronously replicated genes (odorant receptor genes and others, Fig. 2D).
The observed correlation of replication asynchrony and gene duplication led us to test the idea that genes in areas of gene duplication are promising candidates for asynchronous replication and monoallelic expression. Indeed, sampling several areas of gene duplication, we detected asynchronous replication in several such loci (Fig. 3). In addition, assessment of asynchronous replication with multiple short and long probes across nearly a 3 Mb clustered region on chromosome 6 showed that this whole extended region was asynchronously replicated (A. Gimelbrant, unpubl.). This indicates that clustering of related genes is a valuable predictor of the epigenetic status of such a genomic area.
If duplication is associated with asynchronous replication, we might expect that many portions of the genome that contain nongenic repetitive sequences would also replicate asynchronously. Indeed, there are some indications that is the case: Genomic areas of monoallelically expressed genes are also enriched with LINE element repeats (Allen et al. 2003), and the highly repetitive centromeric structures had been shown to be asynchronously replicated (Haaf and Ward 1994).
Regarding the rare (19/2000; Fig. 2C,D) asynchronously replicated genes that are outside of the areas of gene duplication, note that they belong to large gene families, other members of which are clustered together and asynchronously replicated. We have shown such “family association” for taste receptor loci (Fig. 3) and for members of the OR gene family (Ensminger and Chess 2004), and it is likely true for such genes as Mc3r (other family members Mc2r and Mc5r are about 40 kb apart from each other) and Tlr4 [which we previously showed to be asynchronously replicated (Gimelbrant et al. 2005); related genes Tlr1 and Tlr6 are within 20 kb; see Supplemental Fig. S2].
The correlation between asynchronous DNA replication and tandem gene duplication raises intriguing possibilities regarding the links between the two processes. Cross-species comparisons and detailed analysis of the evolutionary relationships between tandemly duplicated and nonduplicated members of the same families of asynchronously replicated gene families should provide clues of the presence of a cause–effect relationship.
Gene duplication events are “evolutionary factories,” producing raw material for increasing organismal complexity. Many highly clustered, asynchronously replicated, monoallelically expressed gene families are examples of rapid evolutionary change. Olfactory receptor genes display different evolutionary histories in primates and rodents (Young et al. 2002); a cluster of mast cell proteases (including Mcpt5) has apparently undergone an expansion in mouse but not in human (Supplemental Fig. S3). If asynchronous replication affects the rate of further tandem duplication (e.g., distinct chromatin states of the differentially replicated regions could lower recombination rate in these regions), it would play an important role in the balance between allowing expansion of gene families and maintaining genomic stability.
Methods
Animals and cells
To obtain F1 animals, 129/SvJ females were crossed with CAST/Ei males (Jackson Laboratories). Immortalized B-cell lines were generated from bone marrow of 6-wk-old mice or from embryonic day 14 liver by infecting primary culture with Abl-MLV virus (Rosenberg et al. 1975). Once established, cells were subcloned using single-cell FACS sorting. Clonal 129×CastF1 ES cells used for the replication timing analysis were provided by A. Wutz (IMP, Vienna). Fibroblasts were derived from 129 or 129×CastF1 embryos; some lines were transformed with SV40 large-T antigen.
Replication timing–FISH
Replication timing–FISH was performed essentially as described (Selig et al. 1992; Ensminger and Chess 2004). Note that recent work (Azuara et al. 2003) has demonstrated that sister chromatid cohesion can also be observed through a FISH approach known as three-dimensional FISH; however, the conditions under which such detection is done should not be confused with the methods used here and by others in the field to measure replication timing. For three-dimensional FISH, cells are subjected to fixation with paraformaldehyde designed to preserve the nuclear architecture. The preservation of nuclear structure, while often a desirable aim, may interfere with the measurement of replication timing. Such an interpretation is supported by a recent observation (Azuara et al. 2003) that the FISH-based assay of replication timing not only gives different results than three-dimensional FISH, but it also more closely reflects results they obtained from direct measurements of replication timing. Thus, the best measurements of DNA replication using FISH are likely to be made under conditions that disrupt the structures that might interfere with the separation of replicated sister chromatids.
Briefly, cells were given a 45-min pulse of BrdU before fixation with cold methanol–acetic acid mix (3:1). The DNA probe (BAC, cosmid, or PCR product as noted) was labeled by nick-translation (Amersham kit, Piscataway, NJ) with Cy3-dCTP (Molecular Probes, Carlsbad, CA). After hybridization, the nuclei were stained with anti-BrdU antibodies (BD) and counterstained with DAPI. For replication timing analysis, only BrdU-positive cells were counted. The list of probes and primers is available upon request.
Replication timing–primer extension
Replication timing–primer extension was based on the assay of Xiong at al. (1998), as modified by us previously (Singh et al. 2003; Gimelbrant et al. 2005). Unsynchronized clonal ES cells from 129×CastF1 mouse were fixed in 75% EtOH (Latt 1973), stained with DAPI in the presence of RNAse A and sorted using FACS into eight fractions, each containing at least 5 × 105 cells. From each cell-cycle fraction, DNA was prepared, its quantity normalized using SybrGreen reagent (Molecular Probes), and regions containing the SNP PCR amplified. The relative amount of the two parental alleles was determined by primer extension using PCR products as template. Detection was by MALDI-TOF mass spectrometry on Sequenom platform (Tang et al. 1999; Cowles et al. 2002). In each individual reaction, the ratio of peak heights corresponding to the two alleles was calibrated with a series of mixes with known composition of purified genomic DNA from parental strains (Jackson Labs): 60:40, 50:50, and 40:60 mix. DNA from each point in cell cycle was amplified and measured in quadruplicate.
Analysis of gene clustering
Analysis of gene clustering was based on sequences and coordinates from the Ensembl annotation of mouse genome assembly (NCBI build 33). To prevent comparisons of different splice variants of the same gene with each other, only the longest from each set of overlapping transcripts was selected for further analysis, resulting in a nonredundant set of protein sequences of autosomal genes. For comparison, a more restricted “Known” protein set from Ensembl was used without modification. Translations of open reading frames from repeat elements were included in all analyses to include more duplication events generated by any mechanism and thus to ensure that our estimates are conservative. The proteins’ relatedness was determined by WU-BLAST (http://blast.wustl.edu) comparison using the E-value cutoff of 10−15 (the probability that such similarity could be expected by chance in the sequence library of the given size; the complete set of proteins was used as the library). We also applied a different, pairwise procedure to find relatedness (Gu et al. 2002) with similar results. Two (or more) related genes were considered to form a cluster if at least a part of one gene was within 75 Kb from any part of the other gene. A gene was considered to be near a cluster if it was within 200 Kb from any part of a gene forming a cluster.
Acknowledgments
We acknowledge NIH grant support to A.C. We thank H. Skaletsky and G. Bell for advice on bioinformatics analysis, B. Blumenstiel and M. DeFelice for assistance with Sequenom, P. Qi and J. Zucker for technical assistance, and A. Bortvin, C. Cowles, A. Ensminger, I. Kramnik, and members of the Chess laboratory for useful discussions.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5023706
References
- Allen E., Horvath S., Tong F., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Horvath S., Tong F., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Tong F., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Spiteri E., Riggs A.D., Marahrens Y., Riggs A.D., Marahrens Y., Marahrens Y. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl. Acad. Sci. 2003;100:9940–9945. doi: 10.1073/pnas.1737401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V., Brown K.E., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Brown K.E., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Buckle V., Merkenschlager M., Fisher A.G., Merkenschlager M., Fisher A.G., Fisher A.G. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 2003;5:668–674. doi: 10.1038/ncb1006. [DOI] [PubMed] [Google Scholar]
- Bartolomei M.S., Tilghman S.M., Tilghman S.M. Genomic imprinting in mammals. Annu. Rev. Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- Bix M., Locksley R.M., Locksley R.M. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 1998;281:1352–1354. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- Chess A., Simon I., Cedar H., Axel R., Simon I., Cedar H., Axel R., Cedar H., Axel R., Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Cowles C.R., Hirschhorn J.N., Altshuler D., Lander E.S., Hirschhorn J.N., Altshuler D., Lander E.S., Altshuler D., Lander E.S., Lander E.S. Detection of regulatory variation in mouse genes. Nat. Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- Deltour L., Vandamme J., Jouvenot Y., Duvillie B., Kelemen K., Schaerly P., Jami J., Paldi A., Vandamme J., Jouvenot Y., Duvillie B., Kelemen K., Schaerly P., Jami J., Paldi A., Jouvenot Y., Duvillie B., Kelemen K., Schaerly P., Jami J., Paldi A., Duvillie B., Kelemen K., Schaerly P., Jami J., Paldi A., Kelemen K., Schaerly P., Jami J., Paldi A., Schaerly P., Jami J., Paldi A., Jami J., Paldi A., Paldi A. Differential expression and imprinting status of Ins1 and Ins2 genes in extraembryonic tissues of laboratory mice. Gene Expr. Patterns. 2004;5:297–300. doi: 10.1016/j.modgep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Ensminger A.W., Chess A., Chess A. Autosome-pair nonequivalence in human resembles X-chromosome inactivation. Hum. Mol. Genet. 2004;13:651–658. [Google Scholar]
- Gimelbrant A.A., Ensminger A.W., Qi P., Zucker J., Chess A., Ensminger A.W., Qi P., Zucker J., Chess A., Qi P., Zucker J., Chess A., Zucker J., Chess A., Chess A. Monoallelic expression and asynchronous replication of p120 catenin in mouse and human cells. J. Biol. Chem. 2005;280:1354–1359. doi: 10.1074/jbc.M411283200. [DOI] [PubMed] [Google Scholar]
- Gribnau J., Luikenhuis S., Hochedlinger K., Monkhorst K., Jaenisch R., Luikenhuis S., Hochedlinger K., Monkhorst K., Jaenisch R., Hochedlinger K., Monkhorst K., Jaenisch R., Monkhorst K., Jaenisch R., Jaenisch R. X chromosome choice occurs independently of asynchronous replication timing. J. Cell Biol. 2005;168:365–373. doi: 10.1083/jcb.200405117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Cavalcanti A., Chen F.C., Bouman P., Li W.H., Cavalcanti A., Chen F.C., Bouman P., Li W.H., Chen F.C., Bouman P., Li W.H., Bouman P., Li W.H., Li W.H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- Haaf T., Ward D.C., Ward D.C. Structural analysis of α-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet. 1994;3:697–709. doi: 10.1093/hmg/3.5.697. [DOI] [PubMed] [Google Scholar]
- Held W., Raulet D.H., Raulet D.H. Expression of the Ly49A gene in murine natural killer cell clones is predominantly but not exclusively mono-allelic. Eur. J. Immunol. 1997;27:2876–2884. doi: 10.1002/eji.1830271120. [DOI] [PubMed] [Google Scholar]
- Held W., Roland J., Raulet D.H., Roland J., Raulet D.H., Raulet D.H. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- Kitsberg D., Selig S., Brandeis M., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Selig S., Brandeis M., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Brandeis M., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Driscoll D.J., Nicholls R.D., Cedar H., Nicholls R.D., Cedar H., Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Latt S.A. Microfluorometric detection of deoxyribonucleic acid replication in human metaphase chromosomes. Proc. Natl. Acad. Sci. 1973;70:3395–3399. doi: 10.1073/pnas.70.12.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K., Winchester E., Daly M.J., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Winchester E., Daly M.J., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Daly M.J., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Ardlie K., Reich D.E., Robinson E., Sklar P., Reich D.E., Robinson E., Sklar P., Robinson E., Sklar P., Sklar P., et al. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J.S., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. The X inactivation centre and X chromosome imprinting. Eur. J. Hum. Genet. 1994;2:255–261. doi: 10.1159/000472369. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R., Singh N., Tenzen T., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Singh N., Tenzen T., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Tenzen T., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Qi P., Reubinoff B.E., Chess A., Cedar H., Reubinoff B.E., Chess A., Cedar H., Chess A., Cedar H., Cedar H., et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- Pereira J.P., Girard R., Chaby R., Cumano A., Vieira P., Girard R., Chaby R., Cumano A., Vieira P., Chaby R., Cumano A., Vieira P., Cumano A., Vieira P., Vieira P. Monoallelic expression of the murine gene encoding Toll-like receptor 4. Nat. Immunol. 2003;4:464–470. doi: 10.1038/ni917. [DOI] [PubMed] [Google Scholar]
- Pernis B., Chiappino G., Kelus A.S., Gell P.G., Chiappino G., Kelus A.S., Gell P.G., Kelus A.S., Gell P.G., Gell P.G. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J. Exp. Med. 1965;122:853–876. doi: 10.1084/jem.122.5.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere I., Sunshine M.J., Littman D.R., Sunshine M.J., Littman D.R., Littman D.R. Regulation of IL-4 expression by activation of individual alleles. Immunity. 1998;9:217–228. doi: 10.1016/s1074-7613(00)80604-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez I., Feinstein P., Mombaerts P., Feinstein P., Mombaerts P., Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 1999;97:199–208. doi: 10.1016/s0092-8674(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Rosenberg N., Baltimore D., Scher C.D., Baltimore D., Scher C.D., Scher C.D. In vitro transformation of lymphoid cells by Abelson murine leukemia virus. Proc. Natl. Acad. Sci. 1975;72:1932–1936. doi: 10.1073/pnas.72.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig S., Okumura K., Ward D.C., Cedar H., Okumura K., Ward D.C., Cedar H., Ward D.C., Cedar H., Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Cedar H., Cedar H.1999. Temporal order of DNA replication. In Concepts in eukaryotic DNA replication (ed. M.L. DePamphilis), pp. 387–408. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY [Google Scholar]
- Singh N., Ebrahimi F.A., Gimelbrant A.A., Ensminger A.W., Tackett M.R., Qi P., Gribnau J., Chess A., Ebrahimi F.A., Gimelbrant A.A., Ensminger A.W., Tackett M.R., Qi P., Gribnau J., Chess A., Gimelbrant A.A., Ensminger A.W., Tackett M.R., Qi P., Gribnau J., Chess A., Ensminger A.W., Tackett M.R., Qi P., Gribnau J., Chess A., Tackett M.R., Qi P., Gribnau J., Chess A., Qi P., Gribnau J., Chess A., Gribnau J., Chess A., Chess A. Coordination of the random asynchronous replication of autosomal loci. Nat. Genet. 2003;33:339–341. doi: 10.1038/ng1102. [DOI] [PubMed] [Google Scholar]
- Tang K., Fu D.J., Julien D., Braun A., Cantor C.R., Koster H., Fu D.J., Julien D., Braun A., Cantor C.R., Koster H., Julien D., Braun A., Cantor C.R., Koster H., Braun A., Cantor C.R., Koster H., Cantor C.R., Koster H., Koster H. Chip-based genotyping by mass spectrometry. Proc. Natl. Acad. Sci. 1999;96:10016–10020. doi: 10.1073/pnas.96.18.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Tsark W., Singer-Sam J., Riggs A.D., Tsark W., Singer-Sam J., Riggs A.D., Singer-Sam J., Riggs A.D., Riggs A.D. Differential replication timing of X-linked genes measured by a novel method using single-nucleotide primer extension. Nucleic Acids Res. 1998;26:684–686. doi: 10.1093/nar/26.2.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.M., Trask B.J., Trask B.J. The sense of smell: Genomics of vertebrate odorant receptors. Hum. Mol. Genet. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]
- Young J.M., Friedman C., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Friedman C., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Ross J.A., Tonnes-Priddy L., Trask B.J., Tonnes-Priddy L., Trask B.J., Trask B.J. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]