Abstract
A patented kinetic uricase method was evaluated for serum uric acid assay. Initial absorbance of the reaction mixture before uricase action (A 0) was obtained by correcting the absorbance at 293 nm measured before the addition of uricase solution, and background absorbance (A b) was predicted by an integrated method. Uric acid concentration in reaction solution was calculated from ΔA, the difference between A 0 and A b, using the absorptivity preset for uric acid. This kinetic uricase method exhibited CV<4.3% and recovery of 100%. Lipids, bilirubin, hemoglobin, ascorbic acid, reduced glutathione and xanthine <0.32 mmol/L in serum had no significant effects. ΔA linearly responded to 1.2 to 37.5 μmol/L uric acid in reaction solution containing 15 μl serum. The slope of linear response was consistent with the absorptivity preset for uric acid while the intercept was consistent with that for serum alone. Uric acid concentrations in clinic sera by different uricase methods positively correlated to each other. By Bland-Altman analysis, this kinetic uricase method accorded with that by quantifying the total change of UV absorbance on the completion of uricase reaction. These results demonstrated that this kinetic uricase method is reliable for serum uric acid assay with enhanced resistance to both xanthine and other common errors, wider range of linear response and much lower cost.
Keywords: Background absorbance, Bland-Altman analysis, Kinetic uricase method, Reaction curve fitting, Serum uric acid assay
INTRODUCTION
Uricase method is widely used to monitor serum uric acid for laboratory diagnosis of gout, kidney function and hyperuricemia-associated diseases. It is easy to quantify uric acid by its distinctive absorbance at 293 nm or to quantify hydrogen peroxide, one of the uricase products, by peroxidase-coupled analysis. There is a direct uricase method for quantifying the decrease of UV absorbance at 293 nm of uric acid upon uricase action and an indirect uricase method for quantifying the amount of hydrogen peroxide formed by uricase action. Indirect uricase method is sensitive to bilirubin, ascorbate, glutathione, lipids, hemoglobin, and peroxide in serum besides xanthine (Aoki et al., 1992; Bartl et al., 1979; Kroll and Elin, 1994; Sanders et al., 1980; Spain and Wu, 1986). The direct equilibrium method by quantifying the difference between initial UV absorbance of the reaction solution before uricase action (A 0) and background absorbance after completion of uricase reaction (A b) is resistant to most of these interferences except xanthine, but has lower efficiency and precision (Duncan et al., 1982). The kinetic uricase method by fitting the classical Michaelis-Menten rate equation to the uricase reaction curve of UV absorbance is limited by background absorbance and unsatisfactory lower limit (Hamilton and Pardue, 1982).
By predicting A b of the uricase reaction solution with an integrated method (Liao et al., 2001; 2003a; 2003b; 2005a), a new kinetic uricase method was proposed for uric acid assay with A 0 estimated through extrapolation according to the initial rate and preset lag time of steady-state reaction (Liao, 2005; Liao et al., 2005b). This kinetic uricase method showed much lower cost and resistance to most common errors. However, it was unclear whether this kinetic uricase method accorded with other established uricase methods for uric acid assay in clinic sera. Hyperuricemic serum is rich in xanthine (Hande et al., 1979), but this direct kinetic uricase method is sensitive to xanthine when uric acid concentration in reaction solution is comparable to uricase K m (Michelis-Menten constant) (Liao, 2005; Liao et al., 2005b). Moreover, the detection limit is much higher than the lower limit of linear response, and the use of lower uricase activity to reduce the detection limit results in lower efficiency (Liao et al., 2005b). Herein, by correcting the absorbance measured before the addition of uricase solution to determine the desired A 0 and using uricase of higher K m, this kinetic uricase method was evaluated for uric acid assay in clinic sera.
MATERIALS AND METHODS
Chemicals and materials
Bacillus fastidious uricase (EC 1.7.3.3) with K m of 65 μmol/L was Fluka 94310 (Zhao et al., 2005), uric acid was from ICN. Other chemicals were domestic analytical reagents.
Specimens
Clinic sera were collected in the clinic laboratory of the First Affiliated Hospital of Chongqing University of Medical Sciences, and stored below −10 °C, thawed and centrifuged to remove denatured proteins before analysis. By this kinetic uricase method, uric acid from 0.68 to 0.99 mmol/L in six clinic sera showed a decrease of (5.4±3.0)% after 6-day storage at 4 °C (P<0.05 by paired t-test). Therefore, 189 clinic sera with the widest distribution of uric acid concentrations (screened by the indirect equilibrium method) in the pool of over 600 sera were analyzed by the following three methods within 12 h to examine their agreements with each other.
Indirect equilibrium method
The two-reagent kit containing ascorbate oxidase for serum uric acid assay by peroxidase-coupled assay of hydrogen peroxide was from Biosino Biotechnology and Science INC (Changping, Beijing 102200, China). It was used on HITACHI 7170 Autoanalyzer with 8 μl serum in 398 μl reaction solution for analysis. For the comparison of recoveries and the ranges of linear response with all methods, the reaction solution was amplified to 1.2 ml with the same sample ratio to measure absorbance using common spectrophotometer.
Direct equilibrium method
URCA uric acid Flex® cartridge from Dade Behring (Newark, DE 19714, USA) was used on Dade Behring Dimension® RXL Clinical Chemistry System to measure the total change of absorbance at 293 nm upon uricase action (Duncan et al., 1982). Operations followed the stated protocol with 17 μl serum in 406 μl reaction solution for analysis.
Kinetic uricase method by predicting background absorbance
1. Monitoring uricase reaction curve and estimating A 0
Absorptivity of uric acid at 293 nm was fixed at 11.5 (L·cm)/mmol throughout all analysis. The buffer and assay of uricase activity were the same as before (Liao et al., 2005a; Zhao et al., 2005). Bacillus fastidious uricase at final 0.03 U/ml had no detectable absorbance at 293 nm. Final reaction mixture contained 1.18 ml buffer, 15 μl clinic serum, and 5 μl uricase solution to give final 0.020 U/ml uricase unless stated otherwise. Reaction solution without uricase was mixed in quartz cell before the absorbance at 293 nm was taken, and the subtraction of 0.3% from this absorbance to correct the effect of volume change upon the addition of uricase solution yielded the desired A 0. After the addition of uricase solution, the reaction was initiated by mixing solutions in quartz cell. The reaction curve was monitored 30 s after the initiation of reaction at 10 to 30 s interval within 5 min at (25±0.5) °C unless stated otherwise. The highest absorbance was limited to 1.270. For direct assay of A b, the absorbance after 40 min reaction at uricase activity of no less than 0.020 U/ml was measured again and directly taken as A b.
2. Estimation of A b by the integrated method
K m for Bacillus fastidious uricase was fixed at 65 μmol/L unless stated otherwise (Zhao et al., 2005). By taking that more than 45 s after the initiation of reaction as the initial datum for analysis, background absorbance (A b) was estimated by fitting integrated Michaelis-Menten rate equation with the predictor variable of reaction time to uricase reaction curve that contained more than five data with absorbance drop above 0.001 (Liao, 2005; Liao et al., 2005b). If there were less than five absorbance data with change above 0.001 at uricase >0.02 U/ml for reaction curve fitting, the absorbance after 5 min reaction minus 0.001 was directly taken as A b. Uric acid concentration was calculated from ΔA, the difference between A 0 and A b, using absorptivity preset for uric acid at 293 nm.
3. Programming
The program was written in Visual Basic 6.0 with plotting of data to examine outliers. Both K m and the range of data for fitting with the integrated rate equation were adjustable. Results were output to a specified text file for further analysis.
Statistical analysis
The results were mean±SD. The lower limit was thrice the standard error of estimate, the upper limit was that with CV<5% and deviation from the linear plot below twice the standard error of estimate, respectively. The agreements between any two methods were examined according to Bland-Altman using SAS 8.0 (Bland and Altman, 1995).
RESULTS
Estimation of A b by the integrated method
In the pool of over 600 clinic sera, there were only two sera needing 1:1 dilution to give A 0<1.270. After the desired dilution, A 0 varied from 0.300 to 1.130 while A b varied from 0.220 to 1.030. The percentage of residual substrate to initial substrate under analysis was the primary factor to determine the reliability of A b estimated by the integrated method while the lag time to monitor reaction curve showed no effects. With initial uric acid <30 μmol/L, residual substrate <1/5 of the initial one usually gave A b with deviation of <2% from that by direct assay after 40 min reaction. A b was resistant to deviation of 15 μmol/L in preset K m if residual uric acid was below one-fifth of initial uric acid that was <15 μmol/L, and the lower concentration of initial uric acid for analysis gave stronger resistance. The estimation of A b was sensitive to outliers in the reaction curve, and lower content of initial uric acid for analysis led to higher sensitivity to outliers.
For all tested clinic sera to give A 0 below 1.270 after necessary dilution, uric acid in reaction solution at the time 45 s after the addition of uricase was <25 μmol/L, and residual uric acid after 5 min reaction at 0.020 U/ml uricase was low enough for reliable estimation of A b. With uric acid from 0.23 mmol/L to 0.97 mmol/L in clinic sera and reaction curve monitored within 5 min, the increase of uricase up to 0.030 U/ml resulted in no differences in A b from those at 0.020 U/ml uricase (n=3, P>0.1 by paired t-test), respectively, but reaction duration for reliable estimation of A b with these sera was reduced to within 3.5 min at 0.030 U/ml uricase. The shift of reaction temperature to 21 °C at 0.020 U/ml uricase to monitor reaction within 5 min still led to no changes of A b in the presence of 15 μmol/L uric acid in reaction solution.
Comparisons of this kinetic uricase method to other uricase methods
1. Reproducibility
For serum uric acid from 0.35 to 1.0 mmol/L, the within-run variation and between-run CV were <4.3% (n=5), and comparable to automated analysis with other uricase methods.
2. Recoveries and the effects of common serum interferences
By this kinetic uricase method, the recoveries for 0.83 mmol/L serum uric acid were consistent to 100% (P>0.3 by t-test). There were no interferences from common serum substances including lipids, bilirubin, glutathione, ascorbate besides xanthine <0.32 mmol/L (Table 1). Except for slight negative error by lipids and xanthine, the direct equilibrium method showed similar pattern of resistance to these interferences, and the recoveries were consistent to 100% (P>0.3 by t-test). However, there were usually negative deviations with the indirect equilibrium method except that lipids led to slight positive deviation, and the recovery for 0.83 mmol/L serum uric acid was ((95.3±2.7)%, n=4), smaller than 100% (P<0.05 by t-test).
Table 1.
Effects of common substances on serum uric acid assay by different uricase methods
| Candidate interferences | Cde (mmol/L) | Cdk (mmol/L) | Cie (mmol/L) |
| Serum A+watera | 0.978±0.012 | 0.976±0.011 | 0.934±0.005 |
| Serum A+6.0 mmol/L glutathionea | 0.974±0.011 | 0.966±0.010 | 0.897±0.006* |
| Serum A+3.0 mmol/L ascorbatea | 0.985±0.018 | 0.980±0.012 | 0.827±0.013* |
| Serum A+45 g/L beef serum albumina | 0.982±0.015 | 0.980±0.016 | 0.930±0.008 |
| Serum A+0.32 mmol/L xanthinea | 0.896±0.020* | 0.974±0.016 | 0.916±0.008** |
| Serum A+0.85 mmol/L EDTAa | 0.980±0.012 | 0.978±0.009 | 0.940±0.010 |
| Serum B+waterb | 0.490±0.006 | 0.490±0.007 | 0.486±0.004 |
| Serum C (Hb 1.35 g/L)+waterb | 0.074±0.002 | 0.064±0.004 | 0.066±0.002 |
| Serum B+Serum Cc | 0.563±0.009 | 0.555±0.008 | 0.510±0.006* |
| Serum D (TB 0.17 mmol/L)+waterb | 0.080±0.002 | 0.068±0.009 | 0.064±0.002 |
| Serum B+Serum Dc | 0.571±0.010 | 0.558±0.012 | 0.538±0.007** |
| Serum E (TG 12.0 mmol/L)+waterb | 0.097±0.005 | 0.085±0.008 | 0.080±0.002 |
| Serum B+Serum Ec | 0.561±0.010** | 0.570±0.018 | 0.576±0.004** |
C de: Serum uric acid concentration by the direct equilibrium method; C ie: Serum uric acid concentration by the indirect equilibrium method; C dk: Serum uric acid concentration by this kinetic uricase method. C de, C dk and C ie (in mmol/L) were from assays in triplicate. Serum A and Serum B were artificial hyperuricemic sera made by adding different amounts of solid uric acid into 20 ml serum from healthy individual. Serum C, Serum D, Serum E were clinic sera rich in endogenous interferences, of total bilirubin (TB), total triglycerides (TG), hemoglobin (Hb), respectively.
Indicated P<0.01 vs that without indicated interference
Indicated P<0.05 vs that without indicated interference
Serum A was mixed with water alone or aqueous solution of specified candidate interference at 10:1 ratio to give indicated final concentrations in serum
Serum B or serum rich in endogenous interference (Serum C, Serum D or Serum E, respectively) was mixed with water at 1:1 ratio to determine the amount of endogenous uric acid
Serum B was mixed with serum rich in candidate endogenous interference (Serum C, Serum D or Serum E, respectively) at 1:1 ratio to test the effect of interference
3. Ranges of linear response
By this kinetic uricase method with data monitored within 5 min at 0.020 U/ml uricase, ΔA linearly responded to the concentration of extra uric acid (C u, μmol/L) added into reaction solution containing 15 μl serum (ΔA=0.029+0.0116×C u, r>0.9996, s<0.005). The slope was consistent with the absorptivity preset for uric acid. ΔA for 15 μl serum alone was 0.030±0.001 (n=3). The correction by 0.3% of the mixture absorbance before the addition of uricase solution to determine A 0 yielded the smallest difference between the intercept and ΔA for serum alone. After the contribution from serum alone was corrected, uric acid at 0.6 μmol/L in reaction solution yielded ΔA of 0.007±0.001 (n=3). This signal of ΔA was below the lower limit of linear response. The upper limit for uric acid in reaction solution was >37.5 μmol/L, which suggested that with sample ratio of 1.25%, serum uric acid from 0.10 to 3.0 mmol/L was suitable for analysis without dilution given measurable A 0. When 0.030 U/ml uricase was used, the upper limit was increased to >50 μmol/L in reaction solution while the lower limit was the same.
With the indirect equilibrium method, there was negative deviation at uric acid concentration >25 μmol/L in reaction solution, and the range of linear response was for 0.06 to 1.2 mmol/L serum uric acid. The direct equilibrium method gave linear response to uric acid in reaction solution from 2.3 to 50 μmol/L (ΔA 293=0.033+0.0116×C u, r>0.9996, s<0.009), enabling serum uric acid from 0.06 to 1.2 mmol/L to be suitable for analysis without dilution. However, it exhibited the highest standard error of estimate for linear response.
Agreements among different uricase methods
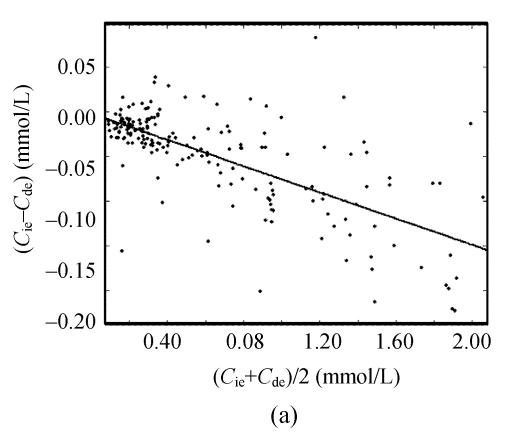
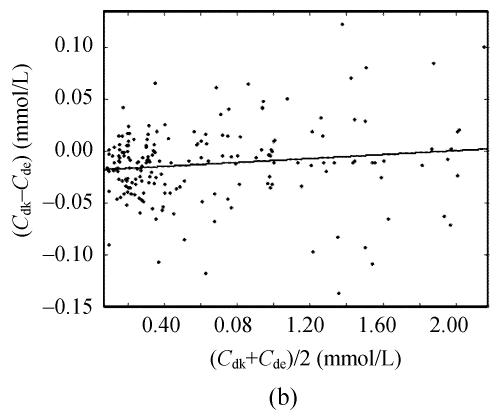
Taking 189 clinic sera into account, there were close positive correlations among serum uric acid concentrations by the direct equilibrium method (C de), by the indirect equilibrium method (C ie) and by this kinetic uricase method (C dk). Deviations between any two methods were <0.25 mmol/L. The slopes for these correlation equations were comparable to their differences in recoveries for 0.83 mmol/L uric acid. However, Bland-Altman analysis demonstrated that there was close correlations of the differences between C ie and C de to their averages (Fig.1a) while there was no significant correlation of the differences between C dk and C de to their averages (Fig.1b).
Fig. 1.
Bland-Altman analysis of the agreement between different uricase methods for the assay of uric acid in 189 clinic sera. (a) The indirect equilibrium method (C ie) vs the direct equilibrium method (C de). Bland-Altman analysis gave C ie−C de=−0.002−0.073×((C ie−C de)/2) (r>0.698, s=0.039) while the correlation equation between them was C ie=−0.001+0.927×C de (r>0.997, s=0.037); (b) This direct kinetic uricase method (C dk) vs the direct equilibrium method (C de). Bland-Altman analysis gave C dk−C de=−0.019+0.010×((C de+C dk)/2) (r<0.137, s=0.037) while the correlation equation between them was C dk=−0.018+1.007×C de (r>0.998, s=0.037)
DISCUSSION
With uricase of higher K m, there was stronger resistance of this kinetic uricase method to the deviation in K m preset for fitting to the reaction curve and the variation of residual substrate for analysis. Xanthine showed comparable inhibition potency on uricase from either Candida utilis or Bacillus fastidious (Liao et al., 2005a; Zhao et al., 2005). By this kinetic uricase method using Bacillus fastidious uricase of higher K m, xanthine in reaction solution at 4.0 μmol/L showed no effects while there was negative deviation when Candida utilis uricase with K m of 12.5 μmol/L was used (Liao et al., 2005b). Moreover, the detection limit for uric acid was below the lower limit of linear response and was independent of uricase activity when A 0 was determined by correcting the absorbance measured before the addition of small amount of uricase solution. The standard error of estimate for this kinetic uricase method was below one-third of that by fitting classical Michaelis-Menten rate equation to uricase reaction curve (Hamilton and Pardue, 1982), and it was comparable to the lowest one of the common uricase method (Sanders et al., 1980). The upper limit for uric acid in reaction solution at 0.030 U/ml uricase was comparable to the highest one of other practical uricase methods (Duncan et al., 1982; Sanders et al., 1980). The upper limit was 30 times of the lower limit at 0.02 U/ml uricase, 40 times of the lower limit at 0.03 U/ml uricase, respectively, suggesting much wider range of linear response than the indirect equilibrium method. Therefore, the use of uricase of higher K m and alternative way to determine A 0 improved this kinetic uricase method.
Each reducing agent in serum is quite low, but the total equivalents of reducing agents in serum are not negligible. There was negative deviation with the indirect uricase method due to reducing agents like glutathione and ascorbate. The recovery for higher concentration of serum uric acid by the indirect equilibrium method was lower than those for direct uricase methods. Moreover, hyperuricemic serum is usually rich in xanthine (Hande et al., 1979). The equilibrium method is preferred to uricase method of lower K m, which showed higher sensitivity to xanthine (Liao et al., 2005a; Zhao et al., 2005). Thus the indirect equilibrium method may be prone to negative deviation by the actions of common serum interferences. The direct equilibrium method showed stronger resistance to common errors than other available uricase methods, and was once used as a candidate reference method for uric acid assay (Duncan et al., 1982). Bland-Altman analysis suggested that this kinetic uricase method agreed with the direct equilibrium method aside from its even stronger resistance to serum xanthine while the indirect equilibrium method disagreed with the direct equilibrium method (Bland and Altman, 1995). Therefore, this kinetic uricase method may exhibit higher resistance to common errors than other uricase methods.
This kinetic uricase method can reliably predict A b with clinic sera, and is resistant to the variation of uricase activity or reaction temperature. The standard error of estimate by this kinetic uricase method is smaller than that by the direct equilibrium method (Duncan et al., 1982) or by fitting classical Michaelis-Menten rate equation to uricase reaction curve (Hamilton and Pardue, 1982). The estimation of both A 0 and A b for the same reaction solution led to close positive covariance between A 0 and A b, which may give lower random error in ΔA (Liao et al., 2005b). This property and the resistance to common errors may account for its satisfactory reproducibility. Within 5 min reaction, uricase activity above 0.5 U/ml was required for the upper limit of 50 μmol/L uric acid in reaction solution by the direct equilibrium method (Duncan et al., 1982). This kinetic uricase method needed uricase <0.030 U/ml within 3.5 min reaction to give comparable upper limit. There is no need for other auxiliary enzyme or toxic chemical. Therefore, this kinetic uricase method showed much lower cost for even higher efficiency.
Autoanalyzer like Dimension® series from Dade Behring was supplied with UV light at 293 nm, which may enable automated analysis with this kinetic uricase method. This kinetic uricase method preferred for analyzing data with substrate below one-fifth of uricase K m. In the pool of over 600 clinic sera screened by the indirect equilibrium method, the lowest uric acid concentration was ~0.07 mmol/L while the highest uric acid concentration was ~2.16 mmol/L. This dynamic range suggested that sample ratio of 2% was desirable and that uric acid in reaction solution should be <50 μmol/L for most cases. At the initial datum for analysis by this kinetic uricase method, 40 s after the addition of uricase, uric acid concentration should be lower that the initial one. Thus uricase with K m slightly above 0.20 mmol/L and resistance to xanthine as stronger as possible was desired. With such an uricase, this kinetic uricase method may be reliable for serum uric acid assay in routine practice with enhanced resistance to both xanthine and other common errors, wider range of linear response, improved reproducibility and much lower cost for even higher efficiency. Such uricases were already available, and they conferred on the resistance of this kinetic uricase method to xanthine as high as 30 μmol/L in reaction solution (Zhao et al., 2006). Therefore, this kinetic uricase method may be developed into a practical method for routine assay of serum uric acid after optimization and standardization.
Footnotes
Project (No. 30200266) supported by the National Natural Science Foundation of China
References
- 1.Aoki Y, Ihara H, Nakamura H, Aoki T, Yoshlda M. Effects of serum bilirubin on determination of uric acid by the uricase-peroxidase coupled reaction. Clin Chem. 1992;38(7):1350–1352. [PubMed] [Google Scholar]
- 2.Bartl K, Brandhuber M, Ziegenhorn J. Improved automated kinetic determination of uric acid in serum by use of uricase/catalase/aldehyde dehydrogenase. Clin Chem. 1979;25(4):619–621. [PubMed] [Google Scholar]
- 3.Bland JM, Altman DG. Comparing methods of measurement: why plotting difference against standard method is misleading. The Lancet. 1995;346(8982):1085–1087. doi: 10.1016/S0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- 4.Duncan PH, Gochman N, Cooper T, Smith E, Bayse D. A candidate reference method for uric acid in serum. I. Optimization and evaluation. Clin Chem. 1982;28(2):284–290. [PubMed] [Google Scholar]
- 5.Hamilton SD, Pardue HL. Kinetic method having a linear range for substrate concentration that exceeds Michaelis-Menten constant. Clin Chem. 1982;28(12):2359–2365. [PubMed] [Google Scholar]
- 6.Hande KR, Perini F, Putterman G, Eln R. Hyperxanthinemia interferes with serum uric acid determinations by the uricase method. Clin Chem. 1979;25(8):1492–1494. [PubMed] [Google Scholar]
- 7.Kroll MH, Elin RJ. Interference with clinical laboratory analysis. Clin Chem. 1994;40(11):1996–2005. [Erratum appeared in Clin. Chem., 1995, 41(5):770] [PubMed] [Google Scholar]
- 8.Liao F. The Method for Enzymatic Analysis of Uric Acid in Body Fluids by Predicting the Background Absorbance. ZL 03135649.4.2005-08-31. China Patent. 2005
- 9.Liao F, Liu WL, Zhou QX, Zeng ZC, Zuo YP. Assay of serum arylesterase activity by fitting reaction curve to an integrated rate equation. Clin Chim Acta. 2001;314(1-2):67–76. doi: 10.1016/S0009-8981(01)00631-3. [DOI] [PubMed] [Google Scholar]
- 10.Liao F, Tian KC, Yang X, Zhou QX, Zeng ZC, Zuo YP. Kinetic substrate quantification by fitting the enzyme reaction curve to the integrated Michaelis-Menten equation. Anal Bioanal Chem. 2003;375(6):756–762. doi: 10.1007/s00216-003-1829-x. [DOI] [PubMed] [Google Scholar]
- 11.Liao F, Li JC, Kang GF, Zeng ZC, Zuo YP. Measurement of mouse liver glutathione S-transferase activity by the integrated method. J Med Coll PLA. 2003;18(5):295–300. [Google Scholar]
- 12.Liao F, Zhu XY, Wang YM, Zuo YP. The comparison of the estimation of enzyme kinetic parameters by fitting reaction curve to the integrated Michaelis-Menten rate equations of different predictor variables. J Biochem Biophys Methods. 2005;62(1):13–24. doi: 10.1016/j.jbbm.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Liao F, Zhao YS, Zhao LN, Tao J, Zhu XY, Wang YM, Zuo YP. Kinetic method for enzymatic analysis by predicting background using uricase as model. J Med Coll PLA. 2005;20(6):338–344. [Google Scholar]
- 14.Sanders GT, Pasman AJ, Hoek FJ. Determination of uric acid with uricase and peroxidase. Clin Chim Acta. 1980;101(2-3):299–303. doi: 10.1016/0009-8981(80)90257-0. [DOI] [PubMed] [Google Scholar]
- 15.Spain MA, Wu AH. Bilirubin interference with determination of uric acid, cholesterol, and triglycerides in commercial peroxidase-coupled assays, and the effect of ferrocyanide. Clin Chem. 1986;32(3):518–521. [PubMed] [Google Scholar]
- 16.Zhao LN, Liao F, Zhao YS. Characterization of the inhibition of xanthine on uricase by a linear kinetic method. J Fourth Mil Med Univ. 2005;26(15):1363–1365. (in Chinese) [Google Scholar]
- 17.Zhao YS, Zhao LN, Yang GQ, Tao J, Bu YQ, Liao F. The characterization of the extracellular uricase from Bacillus fastidious and its application to direct kinetic assay of serum uric acid. J Huazhong Univ Sci Tech Med Ed. 2006;35 (in press)(in Chinese) [Google Scholar]