Abstract
This study is aimed at establishing a sensitive approach to detect disseminated tumor cells in peripheral blood and evaluate its clinical significance. A total of 198 blood samples including 168 from colorectal carcinoma (CRC) patients and 30 from healthy volunteers were examined by quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) to evaluate the expression of carcinoembryonic antigen (CEA), cytokeratin 20 (CK20) and cytokeratin 19 (CK19) mRNA. CEA mRNA was detected in 35.8% of patients and 3.3% of controls, CK20 mRNA in 28.3% of patients and 6.7% of controls, and CK19 mRNA in 41.9% of patients and 3.3% of controls. CEA and CK20 mRNA positive ratio increased with the advancing Dukes stages, but there was no significant difference in positive ratio between any two stages (P>0.05). Also, relatively high positive ratio of CEA, CK20 and CK19 mRNA expression was observed in some CRC patients with earlier Dukes stages. A higher positive ratio was obtained when two or three detection markers were combined compared to a single marker. Our study indicates that quantitative real-time RT-PCR detection for CEA, CK20 and CK19 mRNA in peripheral blood is a valuable tool for monitoring early stage dissemination of CRC cells in blood circulation.
Keywords: Colorectal carcinoma, Real-time RT-PCR, CEA mRNA, CK20 mRNA, CK19 mRNA
INTRODUCTION
Despite advances in novel therapeutic approaches for colorectal cancer (CRC) patients, tumor dissemination via the bloodstream or lymphatic circulation to distant organ is still the major cause of death. Therefore, there is urgent need to establish sensitive methods for early detection of disseminated tumor cells or micrometastasis in peripheral blood or lymphatic circulation of CRC patients.
Detection of disseminated tumor cells in blood circulation of CRC patients has been achieved primarily using immunocytological (Leather et al., 1993) or flow cytometry (FCM) (Chen et al., 1999) based techniques. A major limitation of these assays is their limited sensitivity. To establish alternative approaches with higher sensitivity, polymerase chain reaction (PCR)-based assays that have been used as PCR technique are highly sensitive and clinically useful for detecting cancer markers in circulating tumor cells (Rosenberg et al., 2000; Schuster et al., 2004; Giribaldi et al., 2006). The markers employed for PCR detection of tumor cells are based on specific traits of the tissue from which the tumor originates, such as the cytokeratins (CKs)—CK18, CK19 and CK20 (Funaki et al., 1997; Ikeguchi et al., 2003; Ikeda et al., 2006) generally used for detection of most epithelial cell-type tumors. In gastrointestinal carcinomas, the tumor-associated antigens such as CEA and carbohydrateantigen are usually employed. Among the above mentioned markers, CEA, CK20 and CK19 mRNA are satisfactory for PCR detection for CRC (Funaki et al., 1997; Miura et al., 2003; Kijima et al., 2005; Holdenrieder et al., 2005).
In the present study, we used quantitative real-time RT-PCR (Schuster et al., 2004; Dandachi et al., 2005) method to detect CEA, CK20 and CK19 mRNA expression in peripheral blood of CRC patients. The mRNA expression level was correlated with Dukes stage to investigate the clinicopathological significance of quantitative RT-PCR data, and then we assessed the early-stage diagnostic value of quantitative RT-PCR for the above three markers taken in combination to increase the assay positivity.
MATERIALS AND METHODS
Patients
A total of 168 CRC patients (female to male ratio 69:99; median age 59.8 years, SD: 13.6 years, range: 30~86 years) admitted to the Department of Oncology, at the Second Affiliated Hospital of Zhejiang University from January 2001 to May 2003, were included in this study. All patients were informed about the study and gave written consent for the investigation in accordance with the ethical guidelines at Zhejiang University. Tumor samples were examined microscopically by certified pathologists after surgical resection and were classified according to Dukes staging. Thirty healthy volunteers (female to male ratio 11:19; median age 60.1 years, SD: 9.9 years, range: 34~77 years) were included as controls.
Isolation of mononuclear cells from peripheral blood
Mononuclear cells were isolated from 5 ml heparinized whole blood by density centrifugation using a lymphocyte-separating medium (Huajing Corp., Shanghai, China). Cells were washed three times with phosphate buffered saline (PBS), centrifuged at 2000 r/min for 5 min, and cell pellet was collected for RNA extraction.
RNA extraction and reverse transcription (RT)
Total RNA was extracted using TRIzol reagent (Life Technologies, Gaithersburg, MD) following the manufacturer’s instructions. One to two micrograms of total RNA was reverse-transcribed by moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega Corp., Madison, WI) according to the manufacturer’s protocol using oligo(dT)15 as reverse transcription primer. The RT reaction mixture was incubated for 60 min at 42 °C.
Primers and probes
Real-time PCR primers and fluorogenic probes were included in the CEA/CK20/CK19 real-time detection kit (Jiusheng Corp., Shanghai, China). The fluorogenic probes contained a reporter dye (FAM) covalently attached at the 5′ end and a quencher dye (TAMRA) covalently attached at the 3′ end.
Standards for quantitative real-time PCR
Total RNA was extracted from healthy human volunteers, and CEA, CK20 and CK19 cDNA fragments were generated by reverse transcription. The amplified products were cloned into pGEM Teasy T-vector (Promega Corp., Madison, WI). Ligated fragments were transformed into DH5a competent cells. The exact sequence of the inserted plasmids were analysed by sequencing with M13 universal primers. Serial dilutions from the resulting plasmids were used as standard curves, each containing a known amount of input copy number.
Quantitative real-time PCR (TaqMan)
PCR reactions were performed in the ABI Prism 7700 sequence detector (Perkin Elmer/Applied Biosystems, Foster City, CA). PCR amplifications were performed in a total volume of 50 μl consisting of 5 μl template cDNA and 45 μl PCR master mixture (CEA/CK20/CK19 real-time detection kit, Jiusheng Corp., Shanghai, China) containing Tag DNA polymerase, dNTP mixture, reaction buffer, forward and reverse primers and probes.
Cycling conditions were 10 min at 95 °C initial denaturation, followed by 40 cycles of 30 s 95 °C denaturation, 15 s 60 °C combined annealing and 15 s 72 °C primer extension.
Statistical analysis
The statistical differences of CEA, CK20 and CK19 mRNA positive ratios in peripheral blood between CRC patients and healthy volunteers, between different Dukes stages, and between single marker detection and combined detection were calculated with the chi-square test. A P value <0.05 was considered significant.
RESULTS
Real-time PCR standard curves of CEA, CK20 and CK19 mRNA
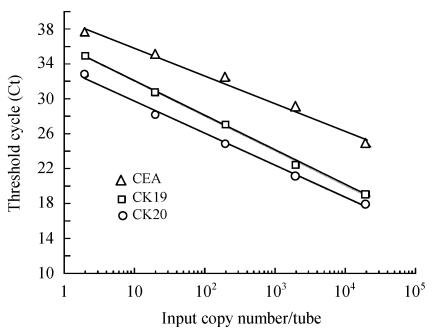
Quantitative real-time PCR monitoring the fluorescent signal of each cycle allowed sensitive and specific detection of CEA, CK20 and CK19 mRNA in the peripheral blood samples of patients. The calculated threshold cycle (Ct) reflects quantity of the starting targets (Fig.1) with lower Ct values reflecting a greater amount of starting target molecules (Oki et al., 2002).
Fig. 1.
Standard curves for CEA, CK20 and CK19 estimation. Each curve was constructed using data from five external standards by plotting the Ct (threshold cycle) value against the input cDNA concentration (serial dilutions of pGEM Teasy T-vector) of samples
Fig.1 presents the Ct value plotted versus the input cDNA concentration (serial dilutions of pGEM Teasy T-vector) of each sample. The Ct value decreased linearly with increasing target quantity from 2 copies/tube to 0.2 million copies/tube. Thus, in this system, target molecules could be detected at a sensitivity of at least 2 copies/tube. The CEA, CK20 and CK19 mRNA values for patient samples were calculated with reference to standard curve (Fig.1).
Expression of CEA, CK20 and CK19 mRNA in peripheral blood of CRC patients and healthy volunteers
We detected CEA mRNA in 95 CRC patients, CK20 mRNA in 46 patients, and CK19 mRNA in 148 patients. These three markers were detected simultaneously in 30 healthy volunteers. The positive ratio of CEA, CK20 and CK19 mRNA in CRC patients are significantly higher than that in healthy volunteers (Table 1). However, the positive ratio of the three markers did not differ significantly from each other.
Table 1.
Expression of CEA, CK20 and CK19 mRNA in peripheral blood of healthy volunteers and CRC patients
| Healthy volunteers |
CRC patients |
P value | |||
| n | Positive ratio (%) | n | Positive ratio (%) | ||
| CEA | 30 | 1 (3.3%) | 95 | 34 (35.8%) | 0.001 |
| CK20 | 30 | 2 (6.7%) | 46 | 13 (28.3%) | 0.021 |
| CK19 | 30 | 1 (3.3%) | 148 | 62 (41.9%) | 0.000 |
Expression of CEA, CK20 and CK19 mRNA in peripheral blood of CRC patients at different Dukes stages
Among the three mRNA markers we detected, CEA and CK20 mRNA positive ratio increased with the advancing Dukes stages (Table 2), but there was no significant difference between any two stages (P>0.05). CK19 mRNA positive ratio showed no obvious correlation with the advancing Dukes stages.
Table 2.
Positive ratio of CEA, CK20 and CK19 mRNA expression in peripheral blood of CRC patients at different Dukes stages
| Expression positive ratio (%) (n) |
|||||
| n | Dukes A | Dukes B | Dukes C | Dukes D | |
| CEA | 95 | 16.7 (1/6) | 29.2 (7/24) | 38.5 (15/39) | 42.3 (11/26) |
| CK20 | 46 | 00.0 (0/4) | 16.7 (2/12) | 33.3 (5/15) | 40.0 (6/15) |
| CK19 | 148 | 33.3 (2/6) | 42.2 (19/45) | 40.7 (22/54) | 44.2 (19/43) |
Markers combined analysis helps to increase the positive ratio of our detection
Quantitative RT-PCR based assay has greater sensitivity and specificity than the conventional semi-quantitative RT-PCR methods. We analyzed the positive ratio of the combined markers among 42 CRC patients where the three mRNA were detected simultaneously. As indicated in Table 3, positive ratio of two combined markers showed a significant increase compared to positive ratio of CEA, CK20 or CK19 single marker detection (35.7%, 28.6% and 40.5% of the 42 CRC patients, respectively) (P<0.05, Table 3). However, no significant difference of ratio positivity was seen between any group of two or three markers combined detection (P>0.05).
Table 3.
Positive ratio of markers combined detection
| n | PR* of markers combined detection | |
| CEA+CK19 | 42 | 22 (52.4%) a |
| CEA+CK20 | 42 | 21 (50.0%) b |
| CK19+CK20 | 42 | 23 (54.8%) b |
| CEA+CK19+CK20 | 42 | 25 (59.5%) a, b, c |
PR: Positive ratio
P<0.05, vs PR of CEA
P<0.05, vs PR of CK20
P>0.05, vs PR of two markers combined detection
DISCUSSION
Real-time RT-PCR is a quantitative nucleotide detection technique, which combines the high-efficiency of PCR, the specificity of DNA probe, and the high sensitivity and accurate quantification of spectral analysis. Using a standard curve for the target of interest, relative copy number value can be determined for any unknown sample. Amplification and subsequent data analysis without post-amplification procedures such as gel electrophoresis can be achieved using a sequence detector (ABI Prism). This will theoretically reduce the possibility of laboratory contamination and false positivity.
In the present study, we applied real-time PCR to determine the absolute CEA, CK20 and CK19 mRNA molecules in peripheral blood of CRC patients. The assay sensitivity demonstrated that target molecules can be detected at a sensitivity of at least 2 copies/tube. This sensitivity is sufficiently high to determine very low levels of CEA, CK20 and CK19 mRNA in peripheral blood.
A significant finding in this study is the relatively high positive ratio of CEA, CK20 and CK19 mRNA expression in the peripheral blood of CRC patients at earlier Dukes stages (Table 2). CEA, CK20 and CK19 mRNA were expressed specifically in the normal epithelial cells and malignant tumor cells of epithelial origin and normally are absent in peripheral blood (Funaki et al., 1997; Schuster et al., 2004). Therefore, as for the CRC patients at Dukes A or B stage, the presence of CEA, CK20 or CK19 mRNA indicates the existence of heterotypic malignant tumor cells in the peripheral blood. Theoretically, these patients are at a greater risk of developing metastasis and should be monitored closely. This finding supports the hypothesis that early tumor-cell dissemination occurs in CRC as a systemic disease. However, the presence of protein products of the three markers cannot reveal the occurrence of tumor cell dissemination in peripheral blood because they are expressed at very low levels, including in normal peripheral blood. Our studies demonstrate that CEA, CK20 and CK19 mRNA are superior molecular markers for detection of disseminated CRC cells compared to their protein products. This accords with a 5-year follow-up study reporting that high preoperative CEA, CK20 and CK19 mRNA levels in CRC patients were associated with poor 5-years survival, while low preoperative levels were associated with good survival (Rosenberg et al., 2000; Ishida et al., 2004; Iinuma et al., 2006).
Another interesting finding is that positive ratio of CEA and CK20 mRNA expression in peripheral blood, which increased with advancing Dukes stages. The lack of a significant difference between any of the two stages may be explained by the low number of patients included in this study. The above results indicate a correlation between CEA, CK20 mRNA expression levels and Dukes stages. Our results support that CEA and CK20 mRNA are promising complementary markers for CRC staging and prediction of cancer progression and metastasis. Our ongoing research is investigating this correlation further.
In a previous study, Zippelius et al.(1997) reported that inappropriate transcription of epithelial-specific genes in hematopoietic cells or the presence of pseudogenes could be limiting factors for the specificity of RT-PCR assays. In the present study we have used the quantitative RT-PCR based assay with the detection specificity of CEA, CK20 and CK19 mRNA increasing significantly compared with the specificity of conventional RT-PCR (Table 4). This result accords with previous reports of real-time RT-PCR detection for these same markers (Table 4). However with increase of real-time PCR specificity, the positive ratio of detection decreased when compared with conventional RT-PCR (Table 4). Thus, combined markers detection we report here can possibly improve the problem of decreased positive ratio. The highest positive ratio was obtained using combined detection of two or three markers compared to single marker (Table 3).
Table 4.
Comparison of positive ratio and specificity of three markers in peripheral blood detected by real-time and conventional RT-PCR respectively
| CEA | CK19 | CK20 | ||
| Present study | P (%) | 35.7 | 40.5 | 28.6 |
| S (%) | 96.7 | 96.7 | 93.3 | |
| Previous real-time RT-PCR detection | P (%) | 24.8 (Schuster et al., 2004); 52.9 (Öberg et al., 2004) | 20.2 (Hardingham et al., 2000) | 22.2 (Giribaldi et al., 2006) |
| S (%) | 100 (Schuster et al., 2004) | 97.8 (Stathopoulou et al., 2003) | 100 (Giribaldi et al., 2006) | |
| Conventional RT-PCR detection | P (%) | 69 (Fiorella et al., 2001) | 64 (Wong et al., 2001); 75 (Gradilone et al., 2003) | 30 (Vlems et al., 2002); 44.8 (Zhang et al., 2003) |
| S (%) | 96.7 (Fiorella et al., 2001); 94 (Piva et al., 2000) | 81 (Wong et al., 2001); 71 (Ko et al., 2000) | 78.7 (Vlems et al., 2002);76 (Jung et al., 1999) |
Note: P: Positive ratio (%); S: Specificity (%)
From a clinical point of view, more attention should be given to the significance of quantitative detection of CEA, CK20 and CK19 mRNA: (1) The detection helps monitoring the occurrence of metastasis, recurrence, and therapeutic outcome. The change in CEA, CK20 and CK19 mRNA level could reflect the presence of metastasis or recurrence (Molnar et al., 2003; Iinuma et al., 2006). Real-time RT-PCR based detection facilitates quantifying therapy response and choosing the best treatment option. (2) Early indicator of high-risk patients. A prognosis study on CRC patients reported that among the Dukes A or B patients, about 30%~40% suffered from cancer recurrence or metastasis (Deans et al., 1992). One possible explanation for this observation is the failure of identifying early disseminated tumor cells in blood or lymph circulation by traditional staging methods (e.g. histopathologic and cellular immunological methods). Thus among patients with Dukes A or B stage, increased CEA, CK20 or CK19 mRNA expression in peripheral blood should be considered as a high-risk factor and, hence, adequate treatment and intensive monitoring should be applied to benefit these patients.
In conclusion, quantitative RT-PCR detection for CEA, CK20 and CK19 mRNA in peripheral blood of CRC patients can have a clinical significance in monitoring early stage hematogenous spreading that may further develop into metastasis or recurrence. CEA, CK20 and CK19 mRNA are superior to their protein products as molecular detection markers because their presence appears to be an indicator for disseminated tumor cells in blood circulation.
Footnotes
Project (No. 021103004) supported by the Science and Technology Development Program of Zhejiang Province, China
Note: The first two authors contributed equally to this work
References
- 1.Chen YH, Gao W, Zhou T, Zhao W, Zhao H, Liu D, Felber E. Detection of bone marrow micrometastasis. Hybridoma. 1999;18(5):465–466. doi: 10.1089/hyb.1999.18.465. [DOI] [PubMed] [Google Scholar]
- 2.Dandachi N, Balic M, Stanzer S, Halm M, Resel M, Hinterleitner TA, Samonigg H, Bauernhofer T. Critical evaluation of real-time reverse transcriptase-polymerase chain reaction for the quantitative detection of cytokeratin 20 mRNA in colorectal cancer patients. Mol Diagn. 2005;7(5):631–637. doi: 10.1016/S1525-1578(10)60597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deans GT, Parks TG, Rowlands BJ, Spence RA. Prognostic factors in colorectal cancer. Br J Surg. 1992;79(7):608–613. doi: 10.1002/bjs.1800790706. [DOI] [PubMed] [Google Scholar]
- 4.Fiorella G, Judith K, Simona A, Maria DC, Antonella S, Roberta DA, Maria RA, Maurizio C, Franco G, Fabio C, et al. Detection of blood-borne cells in colorectal cancer patients by nested reverse transcription-polymerase chain reaction for carcinoembryonic antigen messenger RNA: longitudinal analyses and demonstration of its potential importance as an adjunct to multiple serum markers. Cancer Res. 2001;61(6):2523–2532. [PubMed] [Google Scholar]
- 5.Funaki NO, Tanaka J, Itami A, Kasamatsu T, Ohshio G, Onodera H, Monden K, Okino T, Imamura M. Detection of colorectal carcinoma cells in circulating peripheral blood by reverse transcription-polymerase chain reaction targeting cytokeratin-20 mRNA. Life Sci. 1997;60(9):643–652. doi: 10.1016/S0024-3205(96)00700-X. [DOI] [PubMed] [Google Scholar]
- 6.Giribaldi G, Procida S, Ulliers D, Mannu F, Volpatto R, Mandili G, Fanchini L, Bertetto O, Fronda G, Simula L, et al. Specific detection of cytokeratin 20-positive cells in blood of colorectal and breast cancer patients by a high sensitivity real-time reverse transcriptase-polymerase chain reaction method. Mol Diagn. 2006;8(1):105–112. doi: 10.2353/jmoldx.2006.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gradilone A, Gazzaniga P, Silvestri I, Gandini O, Trasatti L, Lauro S, Frati L, Agliano AM. Detection of CK19, CK20 and EGFR mRNAs in peripheral blood of carcinoma patients: correlation with clinical stage of disease. Oncol Rep. 2003;10(1):217–222. [PubMed] [Google Scholar]
- 8.Hardingham JE, Hewett PJ, Sage RE, Finch JL, Nuttall JD, Kotasek D, Dobrovic A. Molecular detection of blood-borne epithelial cells in colorectal cancer patients and in patients with benign bowel disease. Int J Cancer. 2000;89(1):8–13. doi: 10.1002/(SICI)1097-0215(20000120)89:1<8::AID-IJC2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 9.Holdenrieder S, Holubec LJr, Topolcan O, Finek J, Stieber P. Circulating nucleosomes and cytokeratin 19-fragments in patients with colorectal cancer during chemotherapy. Anticancer Res. 2005;25(3A):1795–1801. [PubMed] [Google Scholar]
- 10.Iinuma H, Okinaga K, Egami H, Mimori K, Hayashi N, Nishida K, Adachi M, Mori M, Sasako M. Usefulness and clinical significance of quantitative real-time RT-PCR to detect isolated tumor cells in the peripheral blood and tumor drainage blood of patients with colorectal cancer. Int J Oncol. 2006;28(2):297–306. [PubMed] [Google Scholar]
- 11.Ikeda S, Fujimori M, Shibata S, Okajima M, Ishizaki Y, Kurihara T, Miyata Y, Iseki M, Shimizu Y, Tokumoto N, et al. Combined immunohistochemistry of beta-catenin, cytokeratin 7, and cytokeratin 20 is useful in discriminating primary lung adenocarcinomas from metastatic colorectal cancer. BMC Cancer. 2006;6(1):31. doi: 10.1186/1471-2407-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeguchi M, Ohro S, Maeda Y, Fukuda K, Yamaguchi K, Shirai H, Kondo A, Tsujitani S, Kaibara N. Detection of cancer cells in the peripheral blood of gastric cancer patients. Int J Mol Med. 2003;11(2):217–221. [PubMed] [Google Scholar]
- 13.Ishida H, Miwa H, Tatsuta M, Masutani S, Imamura H, Shimizu J, Ezumi K, Kato H, Kawasaki T, Furukawa H, et al. Ki-67 and CEA expression as prognostic markers in Dukes’ C colorectal cancer. Cancer Lett. 2004;207(1):109–115. doi: 10.1016/j.canlet.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 14.Jung R, Petersen K, Kruger W, Wagener C, Zander A, Neumaier M. Detection of micrometastasis by cytokeratin 20 RT-PCR is limited due to stable background transcription in granulocytes. Br J Cancer. 1999;81(5):870–873. doi: 10.1038/sj.bjc.6690778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kijima M, Togo S, Ichikawa Y, Miura M, Yamagishi S, Matsuo K, Tanaka K, Masui H, Ishikawa T, Ike H, et al. Clinical significance of serum CEA protein and CEA mRNA after resection of colorectal liver metastases. Anticancer Res. 2005;25(2B):1327–1332. [PubMed] [Google Scholar]
- 16.Ko Y, Grunewald E, Totzke G, Klinz M, Fronhoffs S, Gouni-Berthold I, Sachinidis A, Vetter H. High percentage of false-positive results of cytokeratin 19 RT-PCR in blood: a model for the analysis of illegitimate gene expression. Oncology. 2000;59(1):81–88. doi: 10.1159/000012126. [DOI] [PubMed] [Google Scholar]
- 17.Leather AJ, Gallegos NC, Kocjan G, Savage F, Smales CS, Hu W, Boulos PB, Northover JM, Phillips RK. Detection and enumeration of circulating tumour cells in colorectal cancer. Br J Surg. 1993;80(6):777–780. doi: 10.1002/bjs.1800800643. [DOI] [PubMed] [Google Scholar]
- 18.Miura M, Ichikawa Y, Tanaka K, Kamiyama M, Hamaguchi Y, Ishikawa T, Yamaguchi S, Togo S, Ike H, Ooki S, et al. Real-time PCR (TaqMan PCR) quantification of carcinoembryonic antigen (CEA) mRNA in the peripheral blood of colorectal cancer patients. Anticancer Res. 2003;23(2B):1271–1276. [PubMed] [Google Scholar]
- 19.Molnar B, Sipos F, Galamb O, Tulassay Z. Molecular detection of circulating cancer cells. Role in diagnosis, prognosis and follow-up of colon cancer patients. Dig Dis. 2003;21(4):320–325. doi: 10.1159/000075355. [DOI] [PubMed] [Google Scholar]
- 20.Öberg AN, Lindmark GE, Israelsson AC, Hammarstrom SG, Hammarstrom ML. Detection of occult tumour cells in lymph nodes of colorectal cancer patients using real-time quantitative RT-PCR for CEA and CK20 mRNAS. Int J Cancer. 2004;111(1):101–110. doi: 10.1002/ijc.20231. [DOI] [PubMed] [Google Scholar]
- 21.Oki E, Maehara Y, Tokunaga E, Shibahara K, Hasuda S, Kakeji Y, Sugimachi K. Detection of disseminated cancer cells in bone marrow of gastric cancer using real time quantitative reverse transcriptase polymerase chain reaction. Cancer Lett. 2002;188(1-2):191–198. doi: 10.1016/S0304-3835(02)00057-5. [DOI] [PubMed] [Google Scholar]
- 22.Piva MG, Navaglia F, Basso D, Fogar P, Roveroni G, Gallo N, Zambon CF, Pedrazzoli S, Plebani M. CEA mRNA identification in peripheral blood is feasible for colorectal, but not for gastric or pancreatic cancer staging. Oncology. 2000;59(4):323–328. doi: 10.1159/000012190. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg R, Hoos A, Mueller J, Nekarda H. Impact of cytokeratin-20 and carcinoembryonic antigen mRNA detection by RT-PCR in regional lymph nodes of patients with colorectal cancer. Br J Cancer. 2000;83(10):1323–1329. doi: 10.1054/bjoc.2000.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuster R, Max N, Mann B, Heufelder K, Thilo F, Grone J, Rokos F, Buhr HJ, Thiel E, Keilholz U. Quantitative real-time RT-PCR for detection of disseminated tumor cells in peripheral blood of patients with colorectal cancer using different mRNA markers. Int J Cancer. 2004;108(2):219–227. doi: 10.1002/ijc.11547. [DOI] [PubMed] [Google Scholar]
- 25.Stathopoulou A, Gizi A, Perraki M, Apostolaki S, Malamos N, Mavroudis D, Georgoulias V, Lianidou ES. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin Cancer Res. 2003;9(14):5145–5151. [PubMed] [Google Scholar]
- 26.Vlems FA, Diepstra JHS, Cornelissen IMHA, Ruers TJMM, Ligtenberg JL, Punt CJA, Krieken JHJM, Wobbes T, Muijen GNP. Limitations of cytokeratin 20 RT-PCR to detect disseminated tumour cells in blood and bone marrow of patients with colorectal cancer: expression in controls and downregulation in tumour tissue. Molecular Pathology. 2002;55(3):156–163. doi: 10.1136/mp.55.3.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong IH, Yeo W, Chan AT, Johnson PJ. Quantitative relationship of the circulating tumor burden assessed by reverse transcription-polymerase chain reaction for cytokeratin 19 mRNA in peripheral blood of colorectal cancer patients with Dukes’ stage, serum carcinoembryonic antigen level and tumor progression. Cancer Lett. 2001;162(1):65–73. doi: 10.1016/S0304-3835(00)00630-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhang XW, Fan P, Yang HY, Yang L, Chen GY. Significance of detecting disseminated tumor cells in peripheral blood of gastric and colorectal cancer patients. Zhonghua Zhong Liu Za Zhi. 2003;25(1):66–69. (in Chinese) [PubMed] [Google Scholar]
- 29.Zippelius A, Kufer P, Honold G, Kollermann MW, Oberneder R, Schlimok G, Riethmuller G, Pantel K. Limitations of reverse-transcriptase polymerase chain reaction analyses for detection of micrometastatic epithelial cancer cells in bone marrow. J Clin Oncol. 1997;15(7):2701–2708. doi: 10.1200/JCO.1997.15.7.2701. [DOI] [PubMed] [Google Scholar]