Abstract
The development of the vertebrate inner ear depends on the precise expression of fibroblast growth factors. In a mutagenesis screen for zebrafish with abnormalities of inner-ear development and behavior, we isolated a mutant line, ru622, whose phenotypic characteristics resembled those of null mutants for the gene encoding fibroblast growth factor 8 (Fgf8): an inconsistent startle response, circular swimming, fused otoliths, and abnormal semicircular canals. Positional cloning disclosed that the mutant gene encodes the transcriptional corepressor Atrophin2. Both the Fgf8 protein and zebrafish “similar expression to fgf genes” protein (Sef), an antagonist of fibroblast growth factors induced by Fgf8 itself, were found to be overexpressed in ru622 mutants. We therefore hypothesized that an excess of Sef eliminates Fgf8 signals and produces an fgf8 null phenotype in ru622 mutants. In support of this idea, we could rescue larvae whose atrophin2 expression had been diminished with morpholinos by reducing the expression of Sef as well. We propose that Atrophin2 plays a role in the feedback regulation of Fgf8 signaling. When mutation of the atrophin2 gene results in the overexpression of both Fgf8 and Sef, the excessive Sef inhibits Fgf8 signaling. The resultant imbalance of Fgf8 and Sef signals then underlies the abnormal aural development observed in ru622.
Keywords: auditory system, hearing, vestibular system
The complex structure of the inner ear is largely conserved across vertebrate species. In an adult animal, the inner ear contains several distinct sensory epithelia that are variously dedicated to the detection of linear acceleration, angular acceleration, and sound. Each sensory epithelium includes mechanosensory hair cells that are separated by nonsensory supporting cells and innervated by afferent terminals from the eighth cranial nerve (reviewed in ref. 1).
The ear's development involves dramatic morphological changes, first from a simple epithelial sheet to a closed otic vesicle, and subsequently to the more complex topology of the adult labyrinth (2). Classical genetics offers one fruitful avenue to the identification of genes whose products play important roles in this progression. The zebrafish is particularly useful in this context, for the availability of large numbers of progeny facilitates genetic mapping. This species is also valuable in developmental studies owing to its rapid external development and the optical transparency of its larvae. Finally, because the inner ear of the zebrafish resembles those of other vertebrates, mechanisms elucidated in this organism are likely to prove of general applicability.
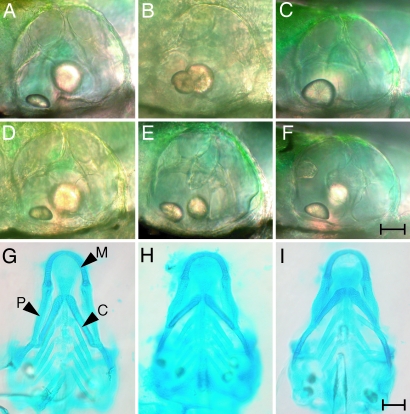
The development of the zebrafish's inner ear commences during somitogenesis with the formation of the otic placode, an area of thickened ectoderm adjacent to the hindbrain that soon forms the hollow otic vesicle (3, 4). Two otoliths originate at the 21-somite stage as tiny, glistening granules of calcium carbonate at the anterior and posterior ends of the otic vesicle's lumen. By 3 days postfertilization (dpf), the anterior otolith has become a prolate ellipsoid and the larger, posterior otolith an oblate ellipsoid (Fig. 1). At the same time, the otic vesicle's structure changes dramatically during the formation of the semicircular canals (5). Three finger-like protrusions first extend from the anterior, posterior, and lateral walls of the otic vesicle. After these protrusions have fused together, another extends from the ventral wall and at 3 dpf fuses with the others at the center of the otocyst. By 5 dpf, sensory cristae that detect angular acceleration have developed in each of the three semicircular canals.
Fig. 1.
Morphological features of WT (A, D, and G), ru622 mutant (B, E, and H), and fgf8 knockdown (C, F, and I) larvae at 5 dpf. (A) The otic vesicle of a WT animal contains two otoliths, a smaller, anterior one lying in the horizontal plane and a larger, posterior one oriented vertically. (B and C) By contrast, an ru622 mutant (B) or a larva treated with a morpholino against fgf8 (C) sometimes has only a single otolith. (D) The semicircular canals of a WT animal begin to be delineated at this stage by three pillars extending from the anterior, posterior, and ventral walls of the otic vesicle and by a dorsolateral septum protruding from the dorsal wall to the center of the otic vesicle. (E and F) In an ru622 mutant (E) or an fgf8 knockdown larva (F), the formation of semicircular canals is abnormal. (G) Alcian blue staining reveals the pattern of cartilage formation in the rostral region of a WT larva. P, palatoquadrate cartilage; C, ceratohyal cartilage; M, Meckel's cartilage. (H and I) An ru622 mutant (H) or an fgf8 knockdown larva (I) is distinguished by shorter palatoquadrate cartilages and the orientation of the ceratohyal cartilages at an increased angle. (Scale bars: 20 μm for A–F; 100 μm for G–I.)
To identify proteins involved in the development and operation of the inner ear, we conducted a mutagenesis screen in zebrafish and isolated 16 mutant lines. In the present study, we report the phenotype of one line, ru622, identify the affected gene, and test hypotheses about the function of the gene's product.
Results
Phenotype of ru622 Mutants.
The zebrafish mutant line ru622 was isolated in a mutagenesis screen on the basis of an inconsistent acoustic startle reflex at 5 dpf. Most WT larvae responded to acoustic stimuli, such as gentle taps on the edge of the dish that contained them, with a stereotyped escape reaction (6). Homozygous ru622 mutant larvae, however, did not react consistently to acoustic stimulation. Furthermore, the mutant animals usually remained on their sides at the bottom of their dish or swam upside-down or in circles, all phenotypes that are associated with abnormalities of vestibular function (7).
Homozygous mutant animals appeared grossly normal and were responsive to tactile stimulation. Perhaps because their abnormal swimming prevented effective feeding, however, they generally died on the seventh day of development. The structure of their inner ears was almost normal save that in 8 of 28 ru622 mutants each ear contained only one, often misshapen otolith (Fig. 1 A and B). This phenotype may have resulted from the fusion of the two otoliths that occur normally; cellular labeling disclosed that the anterior and posterior maculae often came into close contact in mutant animals (data not shown). Moreover, 20 of 65 mutants had abnormal semicircular canals (Fig. 1 D and E). The epithelial protrusions that define the canals were sometimes absent; when present, they failed to elongate or fuse. The two phenotypes occurred independently; some larvae displayed only the otolithic phenotype, some showed only the semicircular-canal phenotype, and some had both.
Close examination of mutant larvae disclosed that their head structure was also abnormal. In particular, the palatoquadrate cartilages were shorter and the ceratohyal cartilages were oriented at a greater angle than in WT animals (Fig. 1 G and H).
Like ru622 mutants, some larvae bearing the acerebellar mutation that impairs zebrafish fibroblast growth factor 8 (Fgf8) signaling have fused otoliths and abnormal epithelial protrusions in the semicircular canals (8). Injection of morpholinos against fgf8 yields a similar phenotype (Fig. 1 C and F). Like acerebellar mutants (9) and ru622 mutants, larvae injected with morpholinos against fgf8 show defects of cartilage formation in the first and second pharyngeal arches (Fig. 1I).
Hair-Cell Morphology and Function.
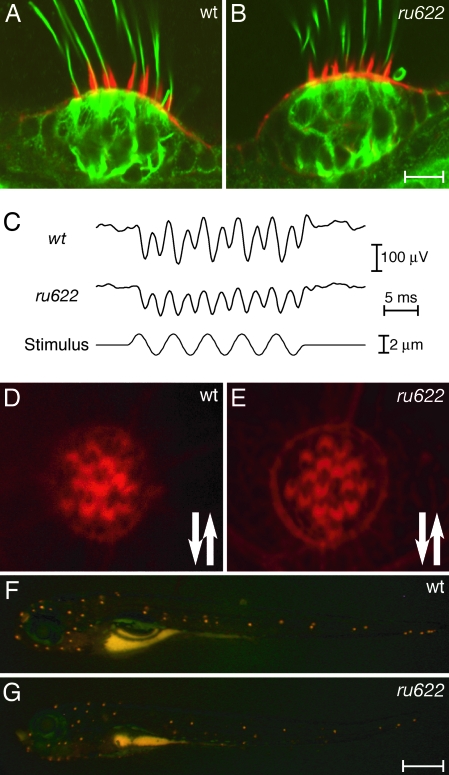
The abnormalities of ru622 mutants suggested that the development or functioning of sensory hair cells was affected. To determine whether the structure of hair bundles was disturbed by the mutation, we labeled the stereocilia of ru622 mutants with phalloidin and their kinocilia with antibodies against acetylated tubulin. At the light-microscopic level, the hair bundles of mutants appeared to be normal (Fig. 2A and B).
Fig. 2.
Phenotypic characteristics of the ru622 mutation. (A) In a confocal image of the crista in the lateral semicircular canal of a larva at 5 dpf, Alexa Fluor 568-phalloidin (red) labels the stereocilia and an antiserum against acetylated tubulin (green) labels the kinocilia and cellular cytoskeletons. (B) The crista of a mutant larva displays similar features. (C) Extracellular microphonic recordings at 5 dpf indicate that the transduction current of a WT otic vesicle is approximately twice that of an ru622 mutant. (D) In a confocal image of the apical surface of a WT neuromast stained with Alexa Fluor 568-phalloidin, the axis of each hair bundle's polarity is denoted by a notch in the actin-rich cuticular plate corresponding to the position of the kinocilium. The arrows indicate the polarities observed. (E) The hair bundles of an ru622 mutant display normal polarization. (D) Fluorescence imaging of a WT larva exposed to 4-(4-(diethylamino)styryl)-N-methylpyridinium iodide reveals a stereotyped pattern of labeled neuromasts. (F) The labeling pattern is similar in an ru622 mutant, but the average number of labeled neuromasts is smaller than in a WT larva. (Scale bars: 10 μm for A, B, D, and E; 500 μm for F and G.)
The functioning of hair cells in the ears of ru622 mutants was examined by recording the microphonic potentials evoked by mechanical stimulation at 5 dpf (Fig. 2C). Although microphonic signals were present in mutants, they were consistently diminished in comparison with those of WT fish.
The lateral-line system, which underlies a fish's sensitivity to predators, prey, and obstacles, also has hair cells whose hair bundles are precisely oriented with respect to the body axes (10, 11). Labeling of the hair bundles in ru622 mutants disclosed that the vectorial orientation of these cells was normal (Fig. 2 D and E). The functioning of lateral-line hair cells was assessed by observing the uptake of 4-(4-(diethylamino)styryl)-N-methylpyridinium iodide, a fluorophore that enters through normal transduction channels. The number of neuromasts labeled on each side of a mutant, 23 ± 2.0 (mean ± SD, n = 64) was slightly but significantly smaller than the value for a WT animal, 28 ± 1.5 (n = 50; P < 0.001 by Student's one-tailed t test). The intensity of 4-(4-(diethylamino)styryl)-N-methylpyridinium iodide labeling was also diminished in mutant neuromasts, possibly as a result of a reduction in the number of hair cells (Fig. 2 F and G).
Identification of the Mutant Gene.
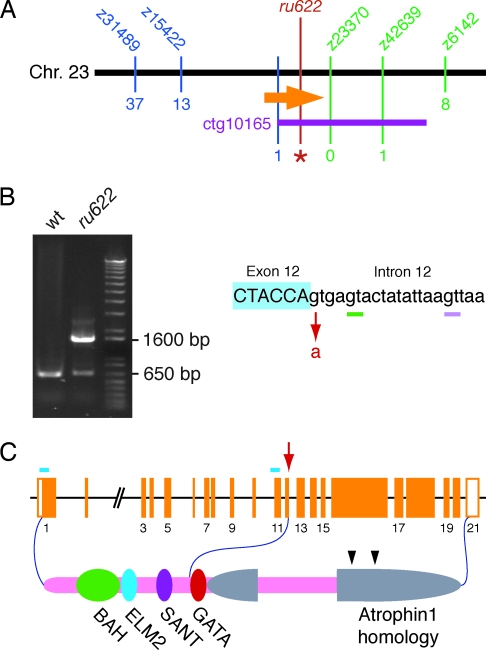
We used positional cloning to identify the mutation responsible for the ru622 phenotype. By low- and intermediate-resolution genetic mapping, we linked the mutation to the simple sequence-length polymorphic markers z15422 and z42693 on chromosome 23 (Fig. 3A). Through the genotyping of 2,247 mutant animals, which yielded 13 recombinants for z15422 and one for z42693, we calculated these markers to be separated by ≈0.3 cM. Although this region includes another marker, z23370, no recombinations occurred among the genotyped larvae between this marker and the mutation. A bacterial artificial chromosome contig from the zebrafish-genome project, ctg10165, was found to include both z23370 and z42693. The 13 recombinants for z15422 were tested for recombination with single-nucleotide polymorphisms identified at the end of this contig nearer z23370. Because one larva displayed a recombination event between these polymorphisms and the mutation, ctg10165 contained the complete candidate region bearing the mutation.
Fig. 3.
Identification of the basis of the ru622 mutation. (A) A map of the genomic region surrounding the ru622 locus on zebrafish chromosome 23 displays the number of recombinations with respect to five simple sequence-length polymorphisms (blue and green) and a single-nucleotide polymorphism near the end of the bacterial artificial chromosome contig ctg10165 (purple). A total of 2,247 larvae were examined for each marker save z6142, for which only 1,164 animals were analyzed. The orange arrow represents the atrophin2 gene and the red line and asterisk denote the ru622 mutation. (B) When WT RNA is used as a template, RT-PCR amplification of a cDNA region extending from exon 8 to exon 13 produces a product ≈650 bp in length (Left). A reaction using cDNA from ru622 mutants yields a product of similar size and another ≈1,600 bp long. The ru622 mutation occurred at the boundary between exon 12 and intron 12, where the splice donor GT was changed to AT (Right). The smaller RT-PCR band from the ru622 reaction includes two products. In one, splicing occurs 4 bp downstream from the mutated splice site (green bar). In the other, splicing instead occurs 16 bp downstream (pink bar). The large RT-PCR product contains the entirety of intron 12. (C) The genomic structure of the zebrafish atrophin2 gene is represented as orange boxes. The arrow indicates the site of the ru622 mutation; the blue bars denote the targets of morpholinos directed against the translation start site and the splicing site between intron 10 and exon 11. Atrophin2 includes a bromo-adjacent homology domain (BAH), an EGL27 and Mta1 homology 2 domain (ELM2), a SWI3, ADA2, N-CoR and TFIIIB domain (SANT), and a zinc finger domain (GATA). The two arrowheads indicate RE repeats.
Of the five expressed sequence tags localized to the ≈280-kb candidate region, one was identified by homology with the corresponding genes of other species as the atrophin2 gene. Reverse transcription and PCRs were performed on WT and mutant larvae to examine this gene's coding region. In contrast to the single band amplified from WT animals, mutants yielded two bands (Fig. 3B); this result raised the possibility of a mutation at an exon–intron boundary. Sequencing disclosed that one product, ≈1,600 bp in length, included the entirety of the ≈1-kb intron 12. In the other, smaller products amplified from mutants, splicing was found to have occurred at two GT dinucleotides downstream from the ordinary boundary between exons 12 and 13. As a result, the shorter transcripts from ru622 mutants included either a 4-bp or a 16-bp insertion between exons 12 and 13. Sequence analysis of the exon–intron boundaries of the mutant atrophin2 gene demonstrated a single-base change (G to A), the putative mutation, at the 5′ splicing site of intron 12.
Known as RERE in humans, Atrophin2 was originally identified as a homolog of Atrophin1, the protein affected in the neurodegenerative disease dentatorubral-pallidoluysian atrophy (12). The coding region of the zebrafish's atrophin2 gene comprises 21 exons spanning ≈110 kb of the genome (Fig. 3C). The human RERE gene includes an additional exon following the first; the transcript lacking this exon represents a minor splicing variant. The zebrafish genome appears to lack the corresponding exon, however, for it could be found neither by our cDNA cloning procedure nor by searching genomic databases.
The predicted Atrophin2 protein of zebrafish is 1,521 aa in length. Its amino-terminal portion includes four domains that resemble those of metastasis-associated protein 2 (Mta2): the bromo-adjacent homology (BAH) domain; the EGL27 and Mta1 homology 2 (ELM2) domain; the SWI3, ADA2, N-CoR, and TFIIIB (SANT) domain; and the zinc-finger (GATA) domain (Fig. 3C). The carboxyl-terminal half of Atophin2 resembles Atrophin1 and shares arginine-glutamic acid (RE) dipeptide repeats.
The mutant transcripts all include stop codons shortly after exon 12 and should therefore produce truncated proteins (Fig. 3B). To test the supposition that deficiency of Atrophin2 underlies the ru622 mutant phenotype, we injected WT eggs with a morpholino designed to block splicing at the boundary between intron 10 and exon 11. As anticipated, morpholino-treated larvae showed the same behavioral and morphological phenotypes as ru622 mutants (data not shown). Moreover, morpholino injection reduced the number of hair cells in the anterior macula at 5 dpf from 57 ± 3 (n = 4) to 47 ± 7 hair cells (n = 9; P < 0.01 by Student's one-tailed t test).
Expression Pattern of Zebrafish atrophin2.
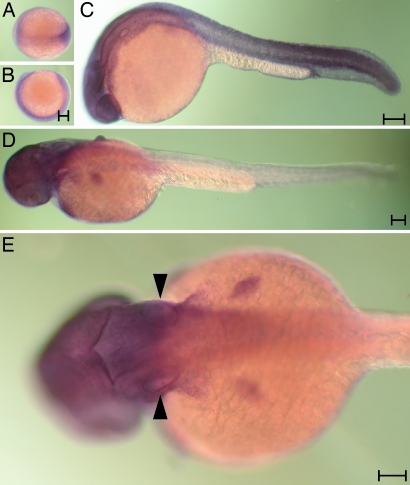
We used in situ hybridization to examine the expression pattern of mRNA encoding Atrophin2 in zebrafish. At all of the developmental stages tested, including the late gastrula and 24 or 36 h postfertilization (hpf), essentially every cell expressed atrophin2, although the expression in the trunk was weak at the last stage (Fig. 4). The epithelia of the otic vesicle were labeled strongly. The labeling pattern was identical when an antisense probe to the 5′ portion of the mRNA was used. The corresponding sense probes, however, produced no signal at any stage (data not shown).
Fig. 4.
The expression pattern of the atrophin2 gene. Whole-mount in situ hybridization was conducted with a 3′ probe at various developmental stages. (A and B) Labeling of the germ band is apparent in a dorsal (A) or a lateral view (B) of the late gastrula. (C) At 24 hpf, essentially all tissue except the yolk is labeled. (D) Labeling diminishes from the caudal extreme by 36 hpf. (E) Arrowheads highlight the labeling of the otic vesicles in a dorsal view of a larva at 36 hpf. (Scale bar: 100 μm.)
In Situ Hybridization with Probes for fgf8 and sef.
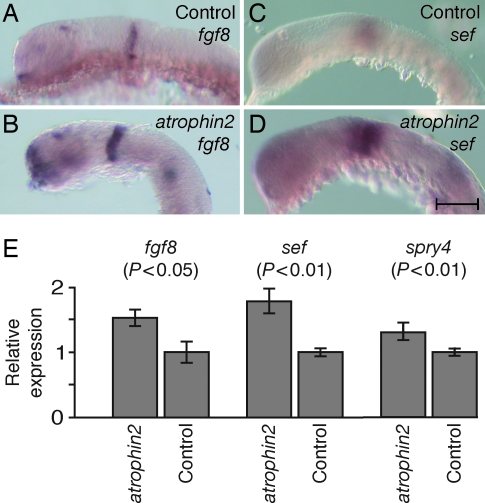
Because murine atrophin2 mutants display misregulation of fgf8 expression (13), we examined the expression pattern of zebrafish fgf8 by in situ hybridization. Consistent with the demonstrated function of Atrophin2 as a transcriptional corepressor (13–15), the expression of fgf8 mRNA was broader in fish injected with the morpholino directed against atrophin2 splicing than in control animals (Fig. 5A and B). As estimated by quantitative PCR, animals exposed to the atrophin2 morpholino expressed an increased concentration of fgf8 transcripts in comparison with control fish (Fig. 5E). Because the expression pattern of fgf8 mRNA in the ru622 mutant was opposite that expected from the phenotype, and Fgf8 is known to activate antagonists of fibroblast growth factor signaling (16–18), we next examined the possibility that the overexpression of fgf8 in the ru622 mutant induced overproduction of an antagonist. As we expected, the zebrafish “similar expression to fgf genes” protein (Sef), a known antagonist of fibroblast growth factor, was expressed with a similar pattern but more robustly in fish injected with the morpholino against atrophin2 splicing than in control animals (Fig. 5 C and D). In situ hybridization indicated that the expression of the transcription factor pax2.1, which is expressed at the midbrain-hindbrain boundary and in the otic vesicle at the same developmental stage, was not affected by overexpression of fgf8 (data not shown). Quantitative PCR confirmed that exposure to the atrophin2 morpholino significantly increased the expression of transcripts for fgf8, sef, and sprouty4, another antagonist of Fgf signaling (Fig. 5E).
Fig. 5.
The expression of fgf8 and sef genes in WT and atrophin2 knockdown embryos. (A) In a control embryo at 24 hpf, fgf8 transcripts are detected in the telencephalon, optic stalk, isthmus, otic vesicle, developing somites, and tail bud. (B) fgf8 is expressed more broadly in an atrophin2 knockdown embryo. (C) A 26-somite control embryo treated with sef probe displays prominent labeling of the optic cup and otic vesicle. (D) An atrophin2 knockdown embryo shows a similar pattern of labeling but more extensive expression than in control fish. (E) Quantitative PCR analysis confirms the increased expression of messages for fgf8, sef, and sprouty4 in atrophin2 knockdown animals; significance values were obtained with Student's one-tailed t test. Control animals were injected with water. The error bars indicate standard deviations. (Scale bar: 100 μm.)
Rescue of the ru622 Phenotype.
If overproduction of Sef eliminates Fgf8 signals, resulting in a phenocopy of the fgf8 null mutation, then it should be possible to rescue affected larvae by reducing the amount of Sef protein. A morpholino against sef was therefore coinjected with that directed against atrophin2 splicing. At 5 dpf, seven of eight (88%) larvae injected with the atrophin2 morpholino alone showed the ru622 phenotype of abnormal equilibrium. Coinjection reduced the fraction of abnormal animals to 23 of 53 (43%) for 1 ng of the sef morpholino and to 10 of 34 (29%) for 5 ng. In a control experiment, the use of 5 ng of a sef morpholino with five mismatches resulted in 25 of 34 (74%) abnormal animals. These results suggest that overproduced Sef protein eliminates Fgf8 signals, causing ru622 fish to manifest a phenotype similar to that of fgf8 null mutants.
Discussion
Originally identified in a mutagenesis screen on the basis of defective startle responses, zebrafish of the ru622 line have been shown to be affected by mutation of the atrophin2 gene. Conserved throughout the metazoans, atrophins act as transcriptional corepressors whose principal effects, especially in the mouse, involve signaling by fibroblast growth factors (13–15). One indication that the ru622 mutation acts through this pathway in zebrafish is the similarity of its phenotype to that associated with the acerebellar mutation, which inactivates the fgf8 gene (8, 9). Both ru622 mutants and fgf8 morphants display similar defects in the formation of otoliths, semicircular canals, and head cartilage (Fig. 1). Unlike ru622 mutants, however, acerebellar larvae lose their midbrain-hindbrain boundary. It is possible that maternally supplied Atrophin2 prevents such defects in early development.
Knocking out the atrophin2 gene disrupts fibroblast growth factor signaling in the mouse. In the neural plate of affected animals, the expression of fgf8 is not appropriately limited to the anterior neural ridge (13). Mutant animals additionally show diminished expression of sonic hedgehog along the anterior midline. Although sonic hedgehog plays a role in inner-ear development (19–22), it is unlikely for two reasons that reduction of this signal accounts for the phenotype of ru622 mutants. First, the otic vesicle's formation is initiated by signals from the hindbrain, where the expression of sonic hedgehog is normal in atrophin2 knockout mice. Second, although zebrafish slow muscle omitted mutants are deficient in responsiveness to all hedgehog ligands (21, 23), ru622 larvae clearly differ phenotypically from these animals. Unlike the inner ears of ru622 and fgf8 null mutants, those of slow muscle omitted animals are symmetrical along the anteroposterior axis; the two otoliths, for example, both resemble the normal anterior otolith (21). Because Atrophin2 is ubiquitously expressed, the localized defects caused by its deficiency may reflect the specificity of interaction between Atrophin2 and nuclear receptors. Atrophin2 binds to several nuclear receptors of subfamily 2, but not to those of subfamily 1 (24). The effects of knocking out atrophin2 gene function on the later development of the mouse's ear unfortunately cannot be investigated, for mutants succumb to heart failure shortly after the appearance of the otic vesicles.
Why might the phenotype of larvae overexpressing fgf8 resemble that of fgf8 null mutants? We hypothesize that Atrophin2 acts in the zebrafish as a corepressor in the regulation of fgf8 and sef expression. Fgf8 evidently down-regulates its own expression by negative feedback: in acerebellar mutants, transcription of the fgf8 gene is elevated (8). If Atrophin2 is involved in this pathway, then the expression of Fgf8 may not be down-regulated in atrophin2 mutants owing to the absence of appropriate feedback. In addition, the binding of Fgf8 to its receptor is known to activate the expression of antagonists to signaling by fibroblast growth factors (16–18). It is therefore plausible that excessive Fgf8 protein in atrophin2 mutants induces additional sef expression as well (Fig. 5B). Although Sef activity should normally silence the Fgf pathway and inhibit Sef expression, an absence of Atrophin2 may disable this negative feedback and permit continuing expression of Sef. The activity of Sef may then dominate that of Fgf8, leaving larvae with an fgf8 null phenotype. This schema is supported by three results. First, our results from quantitative PCR and in situ hybridization indicate that injection of a morpholino against atrophin2 enhances the expression of both fgf8 and sef (Fig. 5). Next, the injection of sef mRNA into embryos produces a phenotype similar to that of fgf8 deficiency, including a smaller otic vesicle (17). Finally, reducing the expression of sef rescues larvae whose atrophin2 expression has been lowered with morpholinos.
The ru622 mutation provides an interesting demonstration of the developmental consequences of imbalanced signaling between a growth factor and its antagonist. A more detailed analysis of the inner ear's sensory organs, including the patterns of hair-cell polarization, afferent and efferent innervation, and neural projection, may reveal additional consequences of the mutation. More importantly, further studies are needed to ascertain how perturbation of fibroblast growth factor signaling culminates in the observed structural and functional phenotypes.
Materials and Methods
Fish Husbandry and Mutagenesis.
Zebrafish were maintained and bred at 28°C by standard procedures (25). The ru622 line was isolated in an F3 screen after mutagenesis with ethylnitrosourea (26, 27).
Alcian Blue Staining.
Larvae were fixed for 12 h at 4°C in 4% (wt/vol) paraformaldehyde in PBS, rinsed with PBS containing 0.1% (vol/vol) Tween, and transferred for 12 h into 0.1% (wt/vol) alcian blue in 80% (vol/vol) ethanol and 20% (vol/vol) glacial acetic acid. The specimens were rinsed in ethanol and rehydrated gradually in an ethanol series. After treatment for several hours with 50 mg/ml trypsin in saturated aqueous sodium tetraborate, larvae were bleached in 3% (vol/vol) H2O2 and 1% (wt/vol) KOH, then placed in 70% (wt/vol) glycerol with 1% (wt/vol) KOH for microscopic observation.
Immunolabeling and Phalloidin Labeling.
Larvae were fixed, permeabilized, and labeled in blocking solution as described (26). Primary mouse monoclonal antibodies against acetylated tubulin (6-11B-1; Sigma) were used at a dilution of 1/1,000; the Alexa Fluor 488-conjugated goat anti-mouse IgG secondary antiserum (Molecular Probes) was used at 1/100. We labeled filamentous actin with 2 units per ml of Alexa Fluor 568-conjugated phalloidin (Molecular Probes).
Microphonic-Potential Recording and Neuromast Labeling.
Microphonic potentials produced by whole maculae in response to vibrational stimulation were measured as described (27) from WT and ru622 mutant larvae at 5 dpf. To label neuromasts (26), we immersed larvae for 5 min in 200 μM 4-(4-(diethylamino)styryl)-N-methylpyridinium iodide (Molecular Probes), then rinsed them several times with water.
Positional Cloning.
In a mapping outcross, ru622 heterozygotes of the AB strain were mated with WT WIK animals. A pair of carriers of the ru622 mutation were identified among the offspring and inbred; their progeny were screened for the behavioral phenotypes and fixed in methanol.
Genomic DNA isolation and PCRs were performed as described (28). Bulked-segregant analysis was conducted with primers (Research Genetics, Huntsville, AL) flanking each of 215 simple sequence-length polymorphisms. After the chromosomal linkage had been established, additional markers were found in databases (Tübingen Map of Zebrafish Genome, http://wwwmap.tuebingen.mpg.de; Massachusetts General Hospital/Cardiovascular Research Center Zebrafish Server, http://zebrafish.mgh.harvard.edu; The Zebrafish Information Network, http://zfin.org; and The Children's Hospital Zebrafish Genome Project Initiative, http://134.174.23.167/zonrhmapper/maps.htm). To identify single-nucleotide polymorphisms and ultimately the mutation, genomic DNA was amplified by the PCR with primers designed with information from the Zebrafish Genome-Sequencing Project (www.ensembl.org/danio_rerio/index.html) and sequenced. This source also provided mapping information on expressed sequence tags and predicted introns and exons with the genescan program.
Morpholino Injection.
Morpholinos (Gene Tools, Philomath, OR) were dissolved in Danieau solution [58 mM NaCl, 0.7 mM KCl, 0.6 mM Ca(NO3)2, 0.4 mM MgCl2, and 5 mM Hepes at pH 7.2] and injected in 1- to 5-ng doses into one- or two-cell embryos with 0.05% (wt/vol) phenol red as a marker. Presented in a 5′-3′ orientation, the morpholino sequences were: atrophin2, TCCTTGGAGGCTGTAAACACAAATT; atrophin2 mismatch control, TCgTTGcAGcCTcTAAACAgAAATT; fgf8, GAGTCTCATGTTTATAGCCTCAGTA; sef, CGCAAGTCTCCGTGACCCAGCCATT; and sef mismatch control, CGgAAcTCTCgGTGAgCCAcCCATT.
In Situ Hybridization.
Whole-mount in situ hybridization was performed on homozygous albino larvae (29). Digoxigenin-labeled RNA probes were synthesized according to the instructions of the manufacturer (Ambion, Austin, TX). The atrophin2 probe included ≈1,000 bp of 3′ coding sequence, whereas the fgf8 probe included the entire coding sequence. The cDNA fragment amplified with the primers TCCTGAAAAAGTAGATTCAGGATTG and ATGTTAAAGGACTCTCACATTCAGG served as a sef probe. Hybridization and washing were performed at 65°C for all probes.
Quantitative PCR.
After total RNA had been prepared from pools of 20 embryos (RNeasy; Qiagen, Valencia, CA), cDNA was synthesized from 1 μg of total RNA (SuperScript First Strand Synthesis; Invitrogen). Real-time quantitative PCR was conducted (ABI Prism 7700; Applied Biosystems) with a melting temperature of 60°C. Reactions were conducted in a total volume of 15 μl with 7.5 μl of iTaq SYBR Green Supermix with ROX (Bio-Rad), 0.5–50 ng of cDNA, and 100 nM each of the forward and reverse primers. Each reaction was run in triplicate; dilution series of cDNA (1:0, 1:10, and 1:100) were used to generate calibration curves for each primer set. The forward and reverse primer sequences were: β-actin, AAGATCATTGCCCCACCTGAG and CCGTTTAGAAGCATTTGCGGT; fgf8, AATCGCAGAGCACAGACCCTT and GGCTTTCGGTCCTCTCCTTTT; sef, GGCGCTCGTTATATGTTGCAA and TTTATTAGGCAGCGGCGGA. The triplicate Ct values for each sample were averaged, and the results were normalized by the amount of product from an endogenous control message (β-actin). The data presented represent a comparison of results from embryos injected with the morpholino against atrophin2 with those from a control group injected with water. Each experiment was conducted three times.
Acknowledgments
We thank Ms. P. Espitia and Mr. A. Afolalu for outstanding fish husbandry, Dr. M. Vologodskaia and Ms. Y. Castellanos for technical assistance, Dr. H. López-Schier for help with immunolabeling, and Dr. S. Koshida for providing the in situ hybridization probe for zebrafish fgf8. Dr. B. B. Riley, Dr. H. Takeda, and the members of our research group provided valuable comments on the manuscript. This investigation was supported by National Institutes of Health Grant DC00241. Y.A. is an Associate and A.J.H. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- dpf
days postfertilization
- hpf
hours postfertilization
- Fgf8
zebrafish fibroblast growth factor 8
- Sef
zebrafish “similar expression to fgf genes” protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Hudspeth A. J. Nature. 1998;341:397–404. doi: 10.1038/341397a0. [DOI] [PubMed] [Google Scholar]
- 2.Barald K. F., Kelley M. W. Development (Cambridge, U.K.) 2004;131:4119–4130. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- 3.Haddon C., Lewis J. J. Comp. Neurol. 1996;365:113–128. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Whitfield T. T., Riley B. B., Chiang M.-Y., Phillips B. Dev. Dyn. 2002;223:427–458. doi: 10.1002/dvdy.10073. [DOI] [PubMed] [Google Scholar]
- 5.Haddon C., Lewis J. Development (Cambridge, U.K.) 1991;112:541–550. doi: 10.1242/dev.112.2.541. [DOI] [PubMed] [Google Scholar]
- 6.Kimmel C. B., Patterson J., Kimmel R. O. Dev. Physiol. 1974;7:47–60. doi: 10.1002/dev.420070109. [DOI] [PubMed] [Google Scholar]
- 7.Nicolson T., Rüsch A., Friedrich R. W., Granato M., Ruppersberg J. P., Nüsslein-Volhard C. Neuron. 1998;20:271–283. doi: 10.1016/s0896-6273(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 8.Leger S., Brand M. Mech. Dev. 2002;119:91–108. doi: 10.1016/s0925-4773(02)00343-x. [DOI] [PubMed] [Google Scholar]
- 9.Walshe J., Mason I. Dev. Biol. 2003;264:522–536. doi: 10.1016/j.ydbio.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Ghysen A., Dambly-Chaudiere C. Curr. Opin. Neurobiol. 2004;14:67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 11.López-Schier H., Starr C. J., Kappler J. A., Kollmar R., Hudspeth A. J. Dev. Cell. 2004;7:401–412. doi: 10.1016/j.devcel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Yanagisawa H., Bundo M., Miyashima T., Okamura-Ohno Y., Todokoro K., Tokunaga K., Yamada M. Hum. Mol. Genet. 2000;9:1433–1442. doi: 10.1093/hmg/9.9.1433. [DOI] [PubMed] [Google Scholar]
- 13.Zoltewicz J. S., Stewart N. J., Leung R., Peterson A. S. Development (Cambridge, U.K.) 2004;131:3–14. doi: 10.1242/dev.00908. [DOI] [PubMed] [Google Scholar]
- 14.Erkner A., Roure A., Charroux B., Delaage M., Holway N., Core N., Vola C., Angelats C., Pages F., Fasano L., Kerridge S. Development (Cambridge, U.K.) 2002;129:1119–1129. doi: 10.1242/dev.129.5.1119. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S., Xu L., Lee J., Xu T. Cell. 2002;108:45–56. doi: 10.1016/s0092-8674(01)00630-4. [DOI] [PubMed] [Google Scholar]
- 16.Furthauer M., Reifers F., Brand M., Thisse B., Thisse C. Development (Cambridge, U.K.) 2001;128:2175–2186. doi: 10.1242/dev.128.12.2175. [DOI] [PubMed] [Google Scholar]
- 17.Furthauer M., Lin W., Ang S. L., Thisse B., Thisse C. Nat. Cell Biol. 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- 18.Tsang M., Friesel R., Kudoh T., Dawid I. B. Nat. Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- 19.Riccomagno M. M., Martinu L., Mulheisen M., Wu D. K., Epstein D. J. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W., Li G., Chien J. S., Raft S., Zhang H., Chiang C., Frenz D. A. Dev. Biol. 2002;248:240–250. doi: 10.1006/dbio.2002.0733. [DOI] [PubMed] [Google Scholar]
- 21.Hammond K. L., Loynes H. E., Folarin A. A., Smith J., Whitfield T. T. Development (Cambridge, U.K.) 2003;130:1403–1417. doi: 10.1242/dev.00360. [DOI] [PubMed] [Google Scholar]
- 22.Riccomagno M. M., Takada S., Epstein D. J. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barresi M. J., Stickney H. L., Devoto S. H. Development (Cambridge, U.K.) 2000;127:2189–2199. doi: 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Rajan H., Pitman J. L., McKeown M., Tsai C.-C. Genes Dev. 2006;20:525–530. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerfield M. The Zebrafish Book. Eugene: University of Oregon Press; 1995. [Google Scholar]
- 26.Kappler J. A., Starr C. J., Chan D. K., Kollmar R., Hudspeth A. J. Proc. Natl. Acad. Sci. USA. 2004;101:13056–13061. doi: 10.1073/pnas.0405224101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr C. J., Kappler J. A., Chan D. K., Kollmar R., Hudspeth A. J. Proc. Natl. Acad. Sci. USA. 2004;101:2572–2577. doi: 10.1073/pnas.0308474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J., Talbot W. S., Schier A. F. Cell. 1998;92:214–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- 29.Prince V. E., Moens C. B., Kimmel C. B., Ho R. K. Development (Cambridge, U.K.) 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]