Abstract
MicroRNAs (miRNAs) are endogenous noncoding RNAs, which negatively regulate gene expression. To determine genomewide miRNA DNA copy number abnormalities in cancer, 283 known human miRNA genes were analyzed by high-resolution array-based comparative genomic hybridization in 227 human ovarian cancer, breast cancer, and melanoma specimens. A high proportion of genomic loci containing miRNA genes exhibited DNA copy number alterations in ovarian cancer (37.1%), breast cancer (72.8%), and melanoma (85.9%), where copy number alterations observed in >15% tumors were considered significant for each miRNA gene. We identified 41 miRNA genes with gene copy number changes that were shared among the three cancer types (26 with gains and 15 with losses) as well as miRNA genes with copy number changes that were unique to each tumor type. Importantly, we show that miRNA copy changes correlate with miRNA expression. Finally, we identified high frequency copy number abnormalities of Dicer1, Argonaute2, and other miRNA-associated genes in breast and ovarian cancer as well as melanoma. These findings support the notion that copy number alterations of miRNAs and their regulatory genes are highly prevalent in cancer and may account partly for the frequent miRNA gene deregulation reported in several tumor types.
Keywords: genome, noncoding RNA, comparative genomic hybridization
MicroRNAs (miRNAs) are endogenous ≈22-nt noncoding small RNAs, which regulate gene expression in a sequence-specific manner (1–8). With >300 already identified, the human genome may contain up to 1,000 miRNAs (8). Vertebrate miRNA targets are thought to be plentiful in number (9–13). Up to one-third of human mRNAs are predicted to be miRNA targets (12). Each miRNA can target ≈200 transcripts directly or indirectly (14, 15), whereas more than one miRNA can converge on a single protein-coding gene target (9–13). Therefore, the potential regulatory circuitry afforded by miRNA is enormous. Increasing evidence indicates that miRNAs, in fact, may be key regulators of various fundamental biological processes (1–8).
The expression of miRNAs is highly specific for tissues and developmental stages (5–7) and has allowed recently for molecular classification of tumors (16, 17). Little is known regarding how miRNA expression is regulated. Primary miRNA transcripts are generated by polymerase II (18). These transcripts are capped, polyadenylated, and usually several thousand bases in length (19). A portion of miRNAs are located within introns of pre-mRNAs and are likely transcribed together with the cognate protein-coding genes (2, 10). Some miRNAs are clustered and transcribed as multicistronic primary transcripts, but the majority of human miRNAs are not clustered and are transcribed independently (5–7).
The biogenesis and function of miRNAs require a common set of proteins. Drosha, an RNase III endonuclease, is responsible for processing primary miRNAs in the nucleus and releasing ≈70-nt precursor miRNAs (20). Drosha associates with the dsRNA-binding protein DGCR8 in human (21) or Pasha in flies (22) to form the microprocessor complex. Precursor miRNAs are transported to the cytoplasm by exportin-5 (23, 24) and cleaved by the RNase III endonuclease Dicer, releasing ≈22-nt mature dsmiRNA (25). One strand of the miRNA duplex is subsequently incorporated into the effector complex RNA-induced silencing complex (RISC) that mediates target gene expression. Argonaute2, a key component of RISC, may function as an endonuclease that cleaves target mRNAs (26, 27).
Increasing evidence shows that expression of miRNA genes is deregulated in human cancer (28–31). Specific over- or underexpression has been shown to correlate with particular tumor types (16, 17, 32–34). miRNA overexpression could result in down-regulation of tumor suppressor genes, whereas their underexpression could lead to oncogene up-regulation (28–31). For example, let-7, down-regulated in lung cancer (35–37), suppresses Ras (36); mir-15 and mir-16, deleted or down-regulated in leukemia (38), suppress BCL2 (39); mir-17-5p and mir-20a control the balance of cell death and proliferation driven by the proto-oncogene c-Myc (40). Clear evidence indicates that miRNA polycistron mir-17-92 serves as an oncogene in lymphoma (33) and lung cancer (41); mir-372 and mir-373 are novel oncogenes in testicular germ cell tumors by numbing p53 pathway (42). Most importantly, miRNA expression signatures can predict outcome (35, 37, 43). These data strongly suggest that miRNAs play an important role in human cancer.
The mechanisms underlying miRNA gene deregulation in cancer are not well understood. Although genomic alterations are critical in oncogenesis (44, 45), studies so far have focused mostly on protein-coding genes. Because more than one-half of the miRNAs have been aligned to genomic fragile sites or regions associated with cancers (30), genome copy abnormalities could involve miRNA genes. Over the past several years, array comparative genomic hybridization (aCGH) has proven its value for analyzing DNA copy number variations (46). In the present study, we investigated genomewide DNA copy number abnormalities of genomic regions containing 283 known human miRNA genes in 227 human cancer samples by using a recently described high-resolution aCGH (47). This approach was generated by using BAC clones with: (i) unambiguous mapping data across genome builds based primarily on range-defining sequence anchors; (ii) the capacity for reproducible, sensitive copy number determination; and (iii) even spacing across the genome, so that the collection has no gap >2 Mb and a mean spacing of <1 Mb (47). An aCGH platform with this resolution is a powerful tool to screen cancers for genetic changes and provides a direct link to the human genome sequence for immediate identification of genes in regions exhibiting copy number changes.
Here we provide experimental genomewide documentation of DNA copy alterations involving miRNA genes in epithelial cancers. We demonstrate a high frequency of copy number abnormalities in regions containing miRNA and their associated genes in breast cancer, ovarian cancer, and melanoma. Importantly, we show that miRNA gene copy changes are concordant with miRNA gene transcriptional expression. Given that miRNA genes may target genes critical for oncogenesis, the present data support the notion that genomic alterations of miRNA and associated genes are partly responsible for miRNA deregulation in cancer and may constitute a critical step in cancer development.
Results
High-resolution aCGH (47) was used to determine the DNA copy number abnormalities of genomic regions containing known miRNA genes in 227 cancer specimens, including 109 ovarian cancer specimens (93 primary tumors and 16 cell lines), 73 breast cancer specimens (55 primary tumors and 18 cell lines), and 45 primary cultured melanoma cell lines. Tumor DNA and reference DNA were labeled with Cy3 and Cy5, respectively (Fig. 5A, which is published as supporting information on the PNAS web site). Reverse dye experiments were done to validate results for all samples. The fluorescence intensity ratio of tumor to reference DNA <0.8 was considered copy number loss, whereas >1.2 was a gain. A circular binary segmentation algorithm (48) was applied to raw log2 ratio data. This algorithm recursively identifies breakpoints and splits chromosomes into subsegments based on a maximum t statistic. A permutation-generated reference distribution is used to define the statistical significance of the estimated splits and decide whether to split at each stage (48). This process removed clone-based dependence and allowed accurate estimations of copy number for the entire genome. Fig. 5B depicts the bioinformatic approach, with transitions between diploid DNA content and circular binary segmentation calls of gain or loss regions. The genomic loci of 283 known human miRNA genes located in autosomes were identified in the miRNA registry (49) (Fig. 5 C and D). Copy number alterations observed in >15% tumors were considered significant for each miRNA gene. In summary, 37.1% (105 of 283) of miRNA genes were located in regions that exhibited DNA copy number abnormalities in ovarian cancer (Fig. 1; see also Tables 1–3, which are published as supporting information on the PNAS web site), 72.8% (206 of 283) in breast cancer (Fig. 6 and Tables 4 and 5, which are published as supporting information on the PNAS web site; see also Table 1), and 85.9% (243 of 283) in melanomas (Fig. 7 and Tables 6 and 7, which are published as supporting information on the PNAS web site; see also Table 1).
Fig. 1.
High frequency miRNA gene copy number alterations in ovarian cancer. aCGH frequency plots of ovarian cancer specimens are shown. Green represents gain, and red represents loss. Stars indicate miRNA genes.
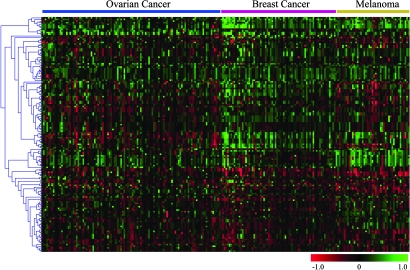
Overall, the genomic alterations involving miRNA genes were distinct among tumor types as evidenced by a heat map condition tree developed by using cluster analysis (Fig. 2). Despite different profiles, some novel miRNA genomic alterations were shared by the three cancer types (Fig. 3). Ovarian and breast cancer shared 41 miRNA genes located in regions with copy number gains and 19 with losses. Furthermore, 26 miRNA genes located in regions with copy number gains and 15 with losses were shared by all three types of cancer. Bioinformatic target prediction for the 41 miRNAs with genomic alterations shared by the three cancer types suggests an important role in oncogenesis (Tables 8 and 9, which are published as supporting information on the PNAS web site). For example, mir-9-1, whose mature miRNA was reported recently to be overexpressed in breast cancer (50), is located in loci amplified in all three tumor types. Its target, hairy and enhancer of split 1 (HES1), predicted by miranda, targetscans, and pictar, is a potential tumor suppressor gene that inhibits breast cancer cell proliferation (51). Furthermore, mir-320 is located in regions with DNA copy number loss in all of the three cancer types. A notable mir-320 target predicted by two independent programs is methyl CpG binding protein 2 (MECP2), which is overexpressed in breast cancer and serves as an oncogene promoting cell proliferation (52).
Fig. 2.
Genetic aberrations of miRNAs in human cancer. Heat map condition tree developed by using gene cluster 2.0 shows aCGH data of all genomic loci containing miRNAs in ovarian cancer, breast cancer, and melanoma specimens. Green and red indicate gain and loss in DNA copy number, respectively.
Fig. 3.
Venn diagrams of miRNA genes with copy number gain and loss shared by two or three types of epithelial cancer.
This study revealed novel genomic alterations involving miRNA genes in epithelial cancers, which are distinct from previous studies in hematologic malignancies. For example, the mir-17-92 polycistron is frequently amplified in B cell lymphoma (33). In contrast, we found that the region containing this polycistron rather was deleted in 16.5% of ovarian cancers, 21.9% of breast cancers, and 20.0% of melanomas. This result suggests that specific miRNA gene alterations underlie specific malignancies. On the other hand, select miRNA alterations can be shared among epithelial and hematologic malignancies. For example, mir-15a and mir-16-1, located within one cluster at 13q14, are deleted or down-regulated in >50% chronic B cell lymphocytic leukemias (38). Our data showed a copy number loss of the regions containing mir-15a and mir-16-1 in 23.9% of ovarian and 24.7% of breast cancers. These miRNAs recently have been shown to negatively regulate BCL2 protein at a posttranscriptional level, and their overexpression induces apoptosis in leukemic cells (39).
Among 283 miRNA genes analyzed, at least 47 miRNAs are located within introns of protein-coding genes, which are likely transcribed together with the cognate protein-coding genes (2, 10). Of these genes, 21 were in regions that exhibited significantly altered copy number in ovarian cancers, 23 in breast cancers, and 38 in melanomas (Table 10, which is published as supporting information on the PNAS web site). This result suggests that copy number changes may occur simultaneously in miRNA and the hosting protein-coding genes in these cancers (5–7, 10). For example, mir-218-1 is located within the tumor suppressor gene SLIT2 (human homologue of Drosophila Slit2), which is frequently inactivated in breast, lung, and colorectal cancer because of allelic loss (53). We found copy number losses of the region containing mir-218-1 and SLIT2 in 15.5% of ovarian cancers, 35.6% of breast cancers, and 33.3% of melanoma lines. Thus, miRNA genes may gain or lose copy numbers simultaneously with respective oncogenes or tumor suppressor genes in cancer.
We also examined whether miRNA genes involved by genomic alterations are, in fact, expressed in cancer and whether their transcript expression correlates with DNA copy number. First, we examined expression of mature miRNA transcripts in ovarian cancer samples for which aCGH data were available. Because host cells infiltrating ovarian cancer could contribute to miRNA gene expression changes without contributing to genomic alterations, we first chose to limit our analysis to 16 cell lines. We used TaqMan miRNA arrays, which comprised 140 of the miRNA genes previously analyzed by aCGH. The majority (n = 121) of these genes could be detected at the miRNA level, and only 19 miRNA genes were undetectable in all ovarian cancer cell lines. Similar data were obtained with five ovarian cancer cell lines analyzed with miRNA microarrays, comprising 167 of the miRNA genes analyzed by aCGH.
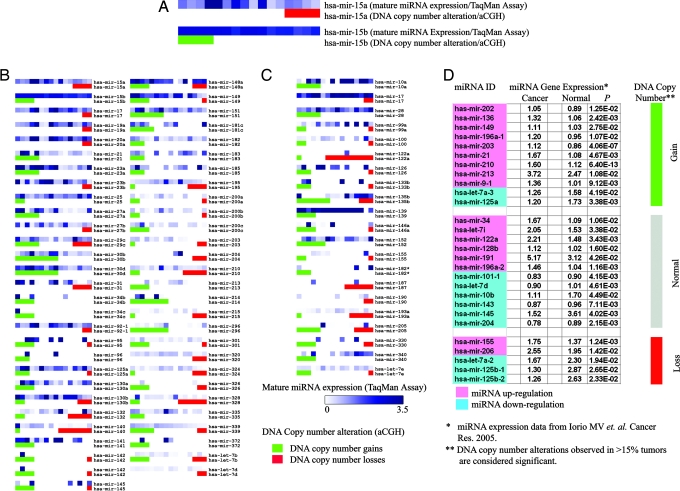
To investigate whether abnormalities in DNA copy number result in aberrant miRNA expression, we examined the level of concordance between miRNA transcript levels and miRNA gene copy number. From the 140 genes analyzed simultaneously by TaqMan miRNA arrays and aCGH, we selected 78 genes for this concordance analysis, eliminating genes that were either: (i) never found to be involved in genomic alterations (thus their transcriptional expression could not be attributed to genomic changes; n = 21); (ii) never detected as mature miRNA in any ovarian cancer line (thus genomic alterations could not be interpreted at a transcriptional level; n = 19); or (iii) multiple copies existed in different genomic loci (thus transcriptional changes could not be linked to genomic alterations of one specific locus; n = 22). For each miRNA, we compared the mature miRNA cell expression level in lines with normal or abnormal DNA copy number. We found that 73.1% (57 of 78) miRNA genes showed concordance between mature miRNA levels and DNA copy number; i.e., mean mature miRNA levels were higher in lines with DNA copy number gain relative to lines without copy number gain for those miRNAs. Similarly, for miRNA that were mostly deleted, mean mature miRNA levels were lower in lines with copy number loss relative to lines without copy number loss (Fig. 4B).
Fig. 4.
Correlation analysis between DNA copy number alteration and miRNA expression. (A–C) Expression of mature miRNA transcripts (by TaqMan miRNA assay) and DNA copy number of loci containing the specific miRNA (by aCGH) in 16 ovarian cancer cell lines. (A) Higher magnification of mir-15a and mir-15b maps. Each spot represents a cell line. Expression levels of mature miRNA transcripts are presented in the upper lane as a heat map in 16 cell lines. DNA status is presented in the lower lane (red, DNA copy number loss; green, DNA copy number gain). Cell lines are ranked based on DNA status (leftmost, amplified; middle, normal; rightmost, deleted gene copy). (B) miRNA genes (n = 57) showing concordance between DNA copy number alterations and miRNA transcript expression. (C) miRNA genes (n = 21) showing no concordance. (D) Comparison of miRNA aCGH data from the present tumor set with expression data of 28 miRNAs overexpressed in a different breast cancer set, as reported by Iorio et al. (50). miRNA genes with normal DNA copy number are marked in gray; gains are marked in green, and losses in red. miRNA transcript overexpression (relative to normal breast) is marked in pink; underexpression (relative to normal breast) is in blue (50).
Next, we examined whether miRNA genomic alterations were associated with transcriptional expression in primary tumors. As proof of principle, we used real-time quantitative PCR to analyze let-7a3, let-7f2, mir-9-1 and mir-213 precursor miRNA expression in 57 ovarian cancers for which aCGH data were available. We found that expression of three of four precursor miRNAs (let-7a3, mir-9-1 and mir-213) were concordant to corresponding genomic alterations (Fig. 8, which is published as supporting information on the PNAS web site). Finally, we compared our breast cancer aCGH data with an unrelated tumor set that has been analyzed previously for miRNA expression by miRNA microarrays (50). As recently reported by Iorio et al. (50), 28 miRNAs were differentially expressed between breast cancer and normal breast tissue (Fig. 4D). Among these 28 miRNA genes, 11 exhibited copy number gains in our study; 9 of these 11 (81.8%) miRNAs had been demonstrated by Iorio et al. (50) to be significantly up-regulated for transcript abundance. Similarly, five of their significant miRNAs exhibited copy number losses in our study; three of these five (60.0%) miRNAs had been demonstrated to be significantly down-regulated for transcript abundance by Iorio et al. (50). Collectively, these data suggest that DNA copy number alterations may be a critical factor affecting expression of miRNAs in cancer.
For some miRNAs, additional factors may affect miRNA expression in the absence of genomic alterations. To test how frequently protein-coding genes involved in miRNA biogenesis and function are involved by genomic alterations in cancer, we analyzed DNA copy number alterations in loci harboring miRNA-associated genes. We identified significantly frequent genomic alterations involving the regions containing these genes (Fig. 9, which is published as supporting information on the PNAS web site). For example, Dicer1 and Argonaute 2 exhibited gains in DNA copy number by 24.8% and 51.5%, respectively, in ovarian tumors. These data suggest that DNA copy number alterations of miRNA-associated genes also might serve as an alternative mechanism to affect miRNA expression in human cancers.
Discussion
In summary, our data suggest that high-frequency copy number abnormalities occur in miRNA-containing regions throughout the genome in a range of human epithelial cancers. We identified shared abnormalities in miRNA-containing genomic loci among ovarian cancer, breast cancer, and melanoma. We also identified distinct differences in genomic alterations in miRNA-containing genomic loci between epithelial cancers and previously described hematologic malignancies. Furthermore, we found DNA copy alterations of loci containing miRNA genes that were unique to specific epithelial cancers. Importantly, for many miRNAs, DNA copy changes correlated with miRNA transcript expression. These findings support the notion that copy number alterations of miRNAs may account partly for the frequent miRNA gene deregulation reported in several tumor types. Clearly, additional factors are involved in miRNA deregulation in cancer, as we could not identify a concordance between copy number and transcript levels for many miRNAs. Our data show that protein-coding genes involved in miRNA biogenesis, including Dicer1 and Argonaute2, also may participate in complex interactions that regulate miRNA expression in tumors, together with additional mechanisms that may regulate miRNA at the epigenetic, transcriptional, posttranscriptional, and translational levels. We performed bioinformatic analyses by using available tools to predict targets for miRNAs that emerged as significant in this study, such as the miRNAs that were shared among all tumor types. We found predicted targets that are known protein-coding genes implicated in oncogenesis. This notion is in agreement with previous reports investigating miRNAs in cancer, where significant miRNAs were predicted to target protein-coding genes involved in malignant transformation. At this point, the mechanisms underlying the high frequency alteration of miRNA genes observed in cancer genome remain unclear. In light of a previous report that miRNAs are frequently located in fragile sites and genomic regions involved in cancers (30), one potential explanation is that genomic aberrations preferentially involve regions containing miRNA genes at a high density. Alternatively, clones with miRNA amplifications or deletions are selected because of the biological advantage that is afforded by these miRNA expression changes.
In summary, the present work shows that (i) genomic alterations involving miRNA are highly frequent in epithelial cancers; (ii) they appear to be shared, in part, among epithelial cancers and, in part, among tumor-specific cancers; and (iii) they result in changes in mature miRNA expression. Because some of our predicted targets for miRNA genes involved in DNA copy number changes are protein-coding genes with known involvement in oncogenesis, our work suggests that genomic alteration of miRNA genes may constitute a critical step in cancer development. Genetic alterations of miRNAs thus may promote and/or enhance alteration of protein-encoding gene expression in cancer, accelerating malignant transformation and/or tumor growth (28–31). Rescued expression of down-regulated or functionally deficient miRNAs and/or inhibition of overexpressed miRNAs may contribute to rebalanced expression of large gene clusters implicated in oncogenesis and tumor progression. Therefore, our results may contribute to a better understanding of the pathogenesis and the identification of biomarkers and targets for human cancer. Targeting of miRNAs may provide an important therapeutic stratagem for human cancer.
Materials and Methods
Patients and Specimens.
We evaluated 93 stage-III and -IV epithelial ovarian cancer specimens from untreated patients undergoing debulking surgery. Sixty-two specimens were provided by D. Katsaros (University of Turin). Additional ovarian cancer (n = 31) and breast ductal carcinoma (n = 55) specimens were provided by B.L.W. All ovarian and breast tumors were from primary sites. Specimens were immediately snap-frozen and stored at −80°C. Primary melanoma cell lines (n = 45) were obtained from M. Herlyn (Wistar Institute, Philadelphia). Ovarian (n = 16) and breast (n = 18) cancer cell lines were provided by G.C. and B.L.W.
BAC Array Platforms.
BAC clones included in the “1 Mb–array” platform were described in ref. 47. Detailed information is provided in Supporting Methods, which is published as supporting information on the PNAS web site.
aCGH and Circular Binary Segmentation.
Genomic DNA was isolated from frozen tumors or cultured cells by overnight digestion, phenol-chloroform extraction, and ethanol precipitation. One microgram of tumor and reference DNA were labeled with Cy3 or Cy5, respectively (Amersham Pharmacia, Piscataway, NJ), by using the BioPrime random-primed labeling kit (Invitrogen). In parallel experiments, tumor DNA and reference DNA were labeled with the opposite dye to account for the difference in dye incorporation and provide additional data for analysis. Labeled tumor and reference DNA were combined and precipitated with human Cot-1 DNA to reduce nonspecific binding. DNA was resuspended and hybridized to the array for 72 h at 37°C on a rotating platform. Images were scanned with an Axon 4500 microarray scanner (Axon Instruments, Union City, CA) and analyzed with genepix (Axon Instruments). Tumor/reference DNA fluorescent intensity ratios <0.8 or >1.2 were considered as alteration. For each sample, copy number estimates were made by the circular binary segmentation method with the DNAcopy package in r programming language (48). Additional analyses and visualization of aCGH data were done by using the cghanalyzer suite described in ref. 54.
miRNA Database.
The genomic loci of human miRNA genes were identified in the miRNA registry (The microRNA Registry, release 7.1, October 2005) (49).
Low Molecular Weight RNA Isolation and miRNA Microarray.
Detailed information is provided in Supporting Methods.
Total RNA Isolation and Quantitative Real-Time RT-PCR.
Detailed information is provided in Supporting Methods.
TaqMan miRNA Assay.
Total RNA was isolated from 1 × 106 cultured cells with TRIzol reagent. Expression of 155 mature miRNAs in 16 human ovarian cancer cell lines were analyzed by TaqMan miRNA Assay (Applied Biosystems) under conditions defined by the supplier. Detailed information is provided in Supporting Methods.
Bioinformatic Analysis.
mRNA targets were predicted for 41 miRNAs of interest by using four well known miRNA target prediction programs: diana-microt, targetscans, miranda, and pictar. Detailed information is provided in Supporting Methods.
Protein Isolation and Western Blot.
Detailed information is provided in Supporting Methods.
Supplementary Material
Acknowledgments
We thank Dr. M. Herlyn for melanoma cell lines. This work was supported by the Ovarian Cancer Research Fund (OCRF), the Abramson Family Cancer Research Institute, the Pennsylvania Department of Health, the Breast Cancer Research Foundation, and Specialized Program of Research Excellence (SPORE) on Skin Cancer Grant P50-CA093372. L.Z. was supported by SPORE P50-CA083638 and the OCRF. D.K. was supported by Associazione Italiana per la Ricerca sul Cancro. A.H. and M.S.M. were supported by National Science Foundation Grant DBI-0238295.
Abbreviations
- aCGH
array comparative genomic hybridization
- miRNA
microRNA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lee R. C., Feinbaum R. L., Ambros V. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 4.Lee R. C., Ambros V. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.He L., Hannon G. J. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Zamore P. D., Haley B. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 9.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 10.John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiriakidou M., Nelson P. T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis B. P., Burge C. B., Bartel D. P. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 14.Bartel D. P., Chen C. Z. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 15.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Volinia S., Calin G. A., Liu C.-G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. Proc. Natl. Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., Kim V. N. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim V. N., Nam J. W. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V. N. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 21.Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 22.Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 23.Yi R., Qin Y., Macara I. G., Cullen B. R. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund E., Guttinger S., Calado A., Dahlberg J. E., Kutay U. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 26.Tabara H., Sarkissian M., Kelly W. G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C. C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 27.Hammond S. M., Boettcher S., Caudy A. A., Kobayashi R., Hannon G. J. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 28.Croce C. M., Calin G. A. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 29.Gregory R. I., Shiekhattar R. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 30.Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McManus M. T. Semin. Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 32.Calin G. A., Liu C.-G., Sevignani C., Ferracin M., Felli N., Dumitru C. D., Shimizu M., Cimmino A., Zupo S., Dono M., et al. Proc. Natl. Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cummins J. M., He Y., Leary R. J., Pagliarini R., Diaz L. A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A. E., Labourier E., et al. Proc. Natl. Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., et al. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 36.Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., et al. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 38.Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., et al. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 41.Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 42.Voorhoeve P. M., le Sage C., Schrier M., Gillis A. J., Stoop H., Nagel R., Liu Y. P., van Duijse J., Drost J., Griekspoor A., et al. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 43.Calin G. A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S. E., Iorio M. V., Visone R., Sever N. I., Fabbri M., et al. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 44.Albertson D. G., Collins C., McCormick F., Gray J. W. Nat. Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 45.Bentley D. R. Nature. 2004;429:440–445. doi: 10.1038/nature02622. [DOI] [PubMed] [Google Scholar]
- 46.Pinkel D., Albertson D. G. Nat. Genet. 2005;37(Suppl. 1):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 47.Greshock J., Naylor T. L., Margolin A., Diskin S., Cleaver S. H., Futreal P. A., deJong P. J., Zhao S., Liebman M., Weber B. L. Genome Res. 2004;14:179–187. doi: 10.1101/gr.1847304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olshen A. B., Venkatraman E. S., Lucito R., Wigler M. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 49.Griffiths-Jones S. Nucleic Acids Res. 2004;32(Database issue):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., et al. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 51.Hartman J., Muller P., Foster J. S., Wimalasena J., Gustafsson J. A., Strom A. Oncogene. 2004;23:8826–8833. doi: 10.1038/sj.onc.1208139. [DOI] [PubMed] [Google Scholar]
- 52.Muller H. M., Fiegl H., Goebel G., Hubalek M. M., Widschwendter A., Muller-Holzner E., Marth C., Widschwendter M. Br. J. Cancer. 2003;89:1934–1939. doi: 10.1038/sj.bjc.6601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dallol A., Da Silva N. F., Viacava P., Minna J. D., Bieche I., Maher E. R., Latif F. Cancer Res. 2002;62:5874–5880. [PubMed] [Google Scholar]
- 54.Margolin A. A., Greshock J., Naylor T. L., Mosse Y., Maris J. M., Bignell G., Saeed A. I., Quackenbush J., Weber B. L. Bioinformatics. 2005;21:3308–3311. doi: 10.1093/bioinformatics/bti500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.