Abstract
This study investigates age-related macular degeneration (AMD) genetic risk factors through identification of a functional single-nucleotide polymorphism (SNP) and its disease association. We chose ERCC6 because of its roles in the aging process, DNA repair, and ocular degeneration from the gene disruption. Bioinformatics indicated a putative binding-element alteration on the sequence containing C−6530>G SNP in the 5′ flanking region of ERCC6 from Sp1 on the C allele to SP1, GATA-1, and OCT-1 on the G allele. Electrophoretic mobility shift assays displayed distinctive C and G allele-binding patterns to nuclear proteins. Luciferase expression was higher in the vector construct containing the G allele than that containing the C allele. A cohort of 460 advanced AMD cases and 269 age-matched controls was examined along with pathologically diagnosed 57 AMD and 18 age-matched non-AMD archived cases. ERCC6 C−6530>G was associated with AMD susceptibility, both independently and through interaction with an SNP (rs380390) in the complement factor H (CFH) intron reported to be highly associated with AMD. A disease odds ratio of 23 was conferred by homozygozity for risk alleles at both ERCC6 and CFH compared with homozygozity for nonrisk alleles. Enhanced ERCC6 expression was observed in lymphocytes from healthy donors bearing ERCC6 C−6530>G alleles. Intense immunostaining of ERCC6 was also found in AMD eyes from ERCC6 C−6530>G carriers. The strong AMD predisposition conferred by the ERCC6 and CFH SNPs may result from biological epistasis, because ERCC6 functions in universal transcription as a component of RNA pol I transcription complex.
Keywords: Cockayne syndrome, single nucleotide polymorphism, gene regulation, interaction, DNA repair
Age-related macular degeneration (AMD) is a leading cause of visual impairment and blindness in Western countries among the elderly (1). AMD is a significant health problem in the United States, with a current estimate of ≈1.75 million persons with advanced AMD in the general population and ≈7.3 million people with early stages of AMD defined by large retinal drusen. By the year 2020, it is estimated that ≈2.95 million people will suffer advanced AMD, and an additional 6.4 million white individuals will have the early stages of AMD in at least one eye (2).
The etiology of AMD remains elusive. To date, age and cigarette smoking have been identified as AMD risk factors (3–8). Various studies have indicated a significant genetic contribution to AMD risk (9). For example, it has been reported that advanced AMD is more prevalent in Caucasian populations as compared with other races that possess higher levels of uveal pigmentation and skin melanin (10). Furthermore, the population-attributed AMD risk related to genetic factors was 23% (11, 12). First-degree relatives of patients with late AMD developed the disease at an increased rate at a relatively young age (12, 13). The higher occurrence of AMD among monozygotic twins and first-degree relatives of AMD patients as compared with spouses and unrelated individuals also indicates a significant genetic component to AMD risk (14, 15). Studies have successfully demonstrated associations between AMD and various single nucleotide polymorphisms (SNPs) (9, 16–21).
The effects of oxidative stress have been implicated in AMD pathological processes (22–25). The retina is particularly susceptible to oxidative stress because of its high consumption of oxygen, high proportion of polyunsaturated fatty acids, and exposure to irradiation (22). Oxidative stress induces various types of DNA damage that contribute significantly to aging and age-related disorders (26). The ERCC6 gene (alternate name CSB) plays roles in the aging process (27) and is known to function extensively in repair of damaged DNA (28–30). The disruption of ERCC6 is causal to a subtype of Cockayne syndrome, an autosomal, recessive human disorder marked by striking somatic and neurological impairment (31). Clinical symptoms of Cockayne syndrome include hypersensitivity to sunlight, severe postnatal growth failure of the soma and brain, and progressive multiorgan and retinal degeneration (32–34).
Activation of the complement cascade leads to the production of a number of proteins that contribute to inflammation. Complement regulatory proteins are present to restrict complement activation at multiple points in the cascades of the three pathways. Complement factor H (CFH) inhibits the formation and accelerates the decay of alternative pathway C3 convertases and serves as a cofactor for the factor I-mediated cleavage and inactivation of C3b (35). The CFH gene is located on chromosome 1q31, which is within the AMD locus identified by the genetic linkage approach (36, 37). Recently, several studies have shown a strong association of CFH SNPs with AMD (17–20, 38, 39). Among them, a CFH intron G/C SNP (rs380390) and CFH-tyr402his are in complete linkage disequilibrium (LD), and both SNPs have been identified to be significantly associated with AMD by using a whole genome scanning approach (20).
In this study, we chose ERCC6 as a candidate gene for AMD pathogenesis because of its roles in the aging process, DNA repair, and ocular manifestations, which resulted from the disruption of this gene. Bioinformatics analysis on common variations in the 5′ flanking region of the gene was used to identify functional SNPs in potential regulatory elements. We performed functional studies on ERCC6 C−6530>G (rs3793784) and confirmed that the SNP conferred a distinct change in regulation of gene expression in vitro and in vivo. Intense immunoreactions against ERCC6 were also detected in AMD eyes from ERCC6 C−6530>G carriers. A case-control study has determined an association between ERCC6 C−6530>G and sporadic advanced AMD cases by using age- and gender-matched control. An AMD-associated SNP in CFH confirmed by multiple studies was also tested in our cohort, and its interaction with ERCC6 C−6530>G was analyzed.
Results
In silico analysis with alibaba2.1 indicated that the C to G alteration in ERCC6−6530 introduced a change of putative transcriptional factor binding patterns around the sequences flanking the SNP (Table 4, which is published as supporting information on the PNAS web site). The C allele corresponded to a possible Sp1 binding element, whereas the G allele corresponded to a possible binding element for Sp1, as well as Oct-1 and GATA-1.
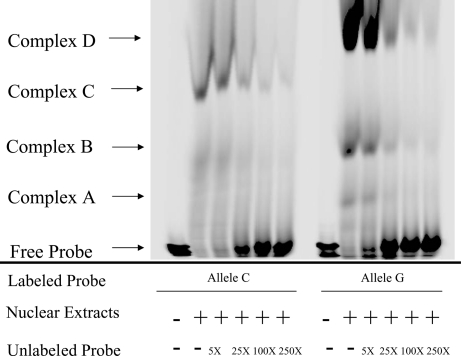
To determine whether this SNP really affects transcription factor binding behavior, we conducted electrophoretic mobility-shift assay (EMSA) to analyze binding of oligo probes containing ERCC6−6530C or ERCC6–6530G to nuclear proteins extracted from a human retinal pigment epithelial (RPE) cell line (ARPE-19). When the infrared dye-labeled probe ERCC6−6530C was incubated with ARPE-19 extracts, a DNA-protein complex was formed (Fig. 1, allele C, complex C). In contrast, when the labeled probe ERCC6−6530G was applied, a distinctive binding pattern was observed (Fig. 1, allele G, complexes A, B, and D), suggesting that the SNP and its adjacent DNA sequences may bind to different regulatory factors within the ERCC6 5′ flanking region. Increasing amounts of the unlabeled probe competed with the binding of the labeled probe, indicating a specific interaction between the DNA and the protein (Fig. 1).
Fig. 1.
EMSA using nuclear extracts from human pigmented epithelium cells (ARPE-19). The oligos containing ERCC6−6530C or ERCC6−6530G were labeled with infrared dye (IRD) and annealed to their complement oligo to generate double-strand DNA. The nuclear extract was incubated with the labeled probes and titrated with different amounts of unlabeled probes. The reaction mixture was subjected to gel electrophoresis, and IRD signal was collected with Li-Cor IR2 global system. The nuclear extract was bound to oligo containing ERCC6−6530C, which formed one main complex (complex C); the nuclear extract was bound to oligo containing ERCC6−6530G, which formed three main complexes (complex A, B, and D).
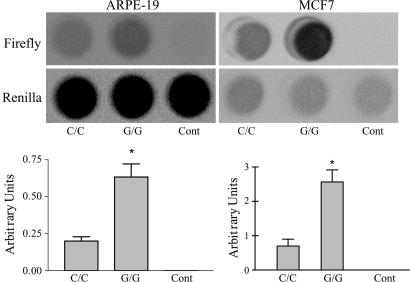
Luciferase reporter constructs possessed 985 bp of the ERCC6 5′ flanking region containing G or C allele, respectively (Fig. 6, which is published as supporting information on the PNAS web site). Transient transfection of allele-specific luciferase reporter constructs into ARPE-19 cells (a human RPE cell line) was performed to elucidate the transcriptional alteration caused by the SNP. As shown in Fig. 2, the G allele caused a three-fold increase in expression activity as compared with the C allele. The same pattern was also observed in transfection of MCF7 cells, a breast cancer cell line (Fig. 2).
Fig. 2.
Correlation between luciferase activities and an SNP in 5′ flanking region of ERCC6. Transcription activity of the −7,025 to −6,040 nucleotide region of ERCC6. (Upper) Firefly luciferase activities were normalized against the internal control Renilla luciferase activity values. (Lower) The data indicate the mean values with the SDs from four independent experiments. The two cell lines (ARPE-19 and MCF7) show similar patterns.
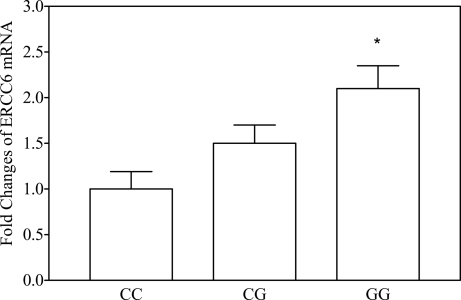
The mRNA level of ERCC6 in various human lymphocyte cells from unrelated individuals (n = 5 in each group, age-matched) differed significantly depending on the ERCC6 C−6530>G SNP type (Fig. 3). Expression was two-fold higher in the cells with the GG homozygozity as compared with the cells with CC homozygozity, which was in agreement with the results of the in vitro experiments.
Fig. 3.
ERCC6 transcript in human lymphocyte (n = 5 in each group) detected by real time RT-PCR. The donors were age-matched. Cells with G allele of ERCC6−6530 express more ERCC6 mRNA than those with the C allele. The fold change was normalized against β-actin. ∗, P < 0.05 vs. CC and CG genotypes.
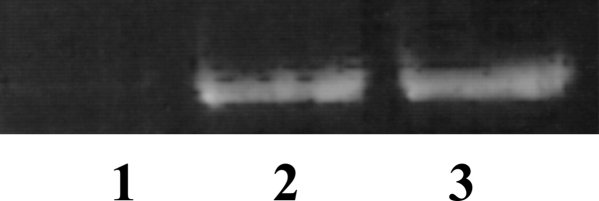
ERCC6 transcript was detected by RT-PCR in human ocular tissue, including RPE cells, by using National Eye Institute (NEI) Bank RPE cDNA library and cDNA synthesized from frozen human retina as templates (Fig. 4).
Fig. 4.
Human ocular ERCC6 transcript detected by RT-PCR. Lane 1, negative control; lane 2, NEI Bank RPE cDNA library; lane 3, human retinal cDNA.
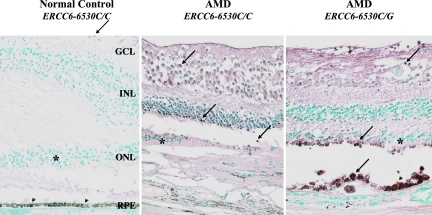
Enhanced expression of ERCC6 protein was observed in the retinal cells at the macula of the AMD eyes as compared with the normal macula. ERCC6 was mildly expressed in 5 of 18 (27.8%) normal eye slides, whereas 15 of 25 (60%) AMD slides were stained strong positive for ERCC6. The representative pictures were shown in Fig. 5. The immunoreactivity was more intense in the AMD eye with CG genotype than the eye with CC genotype. AMD eyes generally exhibited stronger immunoreactions against ERCC6 as compared with normal eyes.
Fig. 5.
Photomicrographs illustrating positive ERCC6 retina cells (arrows) in the retina. (Left) Only few ganglion cells were positive (arrow) in the macula of a normal eye. (Center and Right) The AMD eyes showed replacement of the photoreceptor cells (asterisk) and retinal pigmented epithelia (arrowheads) by a layer of neovascular fibrous tissue. Increased positive staining is observed in the AMD eyes as compared with the normal eye; the AMD eye with ERCC6−6530C/G shows strongest staining. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigmented epithelium. Avidin–biotin–immunoperoxidase staining. (Magnification: ×200).
The demographic information of the case-control study was summarized in Table 5, which is published as supporting information on the PNAS web site. The age and gender of the groups were appropriately matched between the 460 advanced AMD clinical cases and 269 control subjects, as well as between the 57 archived advanced AMD cases and 18 archived normal eyes. No significant deviation from Hardy–Weinberg equilibrium was noted within the study groups for both the ERCC6 (rs3793784) and CFH (rs380390) SNPs. An increased prevalence of ERCC6−6530G and CFH intron C carriers and alleles was observed in the clinical AMD cases, as compared with the controls even after a Bonferroni correction was introduced (Tables 1 and 2). The association was adjusted for age, gender, and smoking for the clinical cohort. The data from archived AMD and its control group replicated the AMD–ERCC6 association. Both ERCC6−6530G carrier and allele frequency were higher than that of the controls (Table 1).
Table 1.
Distribution of ERCC6 C−6530>G (rs3793784) SNP types among cases and controls
| Genotype | Clinical cases |
Archived cases |
||||||
|---|---|---|---|---|---|---|---|---|
| Control (n = 269) | Case (n = 460) | P value† (χ2) | OR (CI) | Control (n = 18) | Case (n = 57) | P value† (χ2) | OR (CI) | |
| CC | 114 | 149 | Reference | Reference | 9 | 12 | Reference | Reference |
| CG | 120 | 238 | 0.02 (5.50) | 1.51 (1.07–2.11) | 6 | 33 | 0.01 (5.13) | 4.13 (1.47–11.54) |
| GG | 35 | 73 | 0.057 (3.64) | 1.60 (0.99–2.61) | 3 | 12 | 0.08 (1.98) | 3.00 (0.83–10.85) |
| CG + GG | 155 (57.6%) | 311 (67.6%) | 0.02 (5.50)* | 1.47 (1.07–2.02) | 9 (50.0%) | 35 (61.4%) | 0.03 (3.43) | 2.92 (1.13–7.55) |
| C allele | 348 (64.7%) | 536 (58.3%) | Reference | Reference | 24 (65.4%) | 57 (50.0%) | Reference | Reference |
| G allele | 190 (35.3%) | 384 (41.7%) | 0.02 (5.87)** | 1.31 (1.05–1.65) | 12 (34.6%) | 57 (50.0%) | 0.04 (3.00) | 2.00 (1.04–3.86) |
†P values of clinical cases were adjusted by age, gender, and smoking status (ever smoked, never smoked). P values of archived cases were obtained by one-tailed χ2 test because the clinical cases indicated the SNP rates are higher in the cases than the control. CI, 95% confidence interval. Reference, cell was taken as the baseline for the indicated statistical calculation: CC was taken as reference when comparing the genotypes, and C was taken as reference when comparing alleles. ∗, Bonferroni corrected P = 0.04; ∗∗, Bonferroni corrected P = 0.03.
To investigate the possible haplotype block which might have LD with ERCC6 C−6530>G, we selected four SNPs (rs1012554, rs11101153, rs4250004, and rs1964145) with minor allele frequency >10% within the 20-kb region flanking ERCC6 C−6530>G to determine the possible LD with ERCC6 C−6530>G. Ninety samples from the controls were analyzed. The results showed no LD among those SNPs (data not shown).
Although the magnitude of the AMD association of ERCC6 C−6530>G was moderate, logistic analysis on the joint contribution of these two SNPs unveiled a strong synergic effect of the G allele at the ERCC6 locus on C allele at the CFH locus toward AMD susceptibility (Table 3). The order of disease odds ratios (ORs) of individuals with various combinations of two SNP loci in comparison with CC/GG (ERCC6 locus/CFH locus) was CC/CC (OR = 4.29), CG/CC (OR = 7.70), and GG/CC (OR = 23.05). Although the combination analysis highly stratified the samples, a significant difference was found by comparing GG/CC verses CC/CC or CG/CC between the case and control groups (Table 3). Individuals homozygous for both the ERCC6−6530 risk alleles GG and the CFH intron risk alleles CC had dramatically increased disease risk compared with the CC/GG homozygote (OR = 23.05; Table 3). This OR of 23.05 is two-fold larger than the product of the OR of the homozygous ERCC6 risk allele (GG) times the OR of homozygous CFH risk allele (CC) (1.60 × 6.77 = 10.83) (Tables 1–3).
Table 3.
Distribution of the ERCC6 (rs3793784) and CFH (rs380390) SNP types and the combined effect on the disease risk
| ERCC6–C6530>G | CFH rs380390 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Control (n = 268) |
Case (n = 460) |
OR (CI) |
|||||||
| GG | GC | CC | GG | GC | CC | GG | GC | CC | |
| CC | 45 (16.8%) | 53 (19.8%) | 16 (6.0%) | 33 (7.2%) | 66 (14.4%) | 50 (10.9%) | Reference | 1.65 (0.90–3.03) | 4.29* (2.02–9.09) |
| CG | 43 (16.0%) | 56 (20.9%) | 20 (7.5%) | 42 (9.1%) | 94 (20.4%) | 102 (22.2%) | 1.24 (0.65–2.38) | 2.17 (1.20–3.90) | 7.70* (3.86–15.34) |
| GG | 16 (6.0%) | 17 (6.3%) | 2 (0.8%) | 8 (1.7%) | 31 (6.7%) | 34 (7.4%) | 0.62 (0.23–1.69) | 2.64 (1.22–5.75) | 23.05** (5.07–104.78) |
CI, 95% confidence interval. Reference, cell was taken as the baseline for the indicated statistical calculation. ∗, P < 0.0001, using CC/GG as the reference. ∗∗, P < 0.0001, using CC/GG as reference or P < 0.05, using CC/CC (two-tailed) or CG/CC (one-tailed) as the references.
Discussion
To our knowledge, this study identifies for the first time a functional SNP in a regulatory region of ERCC6. By means of EMSA and transient transfection reporter assays, we have provided in vitro evidence that the ERCC6 SNP is functional with respect to the transcriptional regulation of the gene. We have also provided in vivo evidence that ERCC6−6530G induces higher gene expression in comparison with the reference sequence. The results from immunohistochemistry on archived ocular tissue of AMD and normal eyes showed increasing expression of ERCC6 protein in the AMD eyes, especially in the individuals who carried ERCC6−6530G allele, suggesting that high expression of this gene is predisposing toward the disease. We reported, for the first time to our knowledge, that an SNP C−6530>G in the 5′-flanking region of ERCC6 was associated with AMD, both independently and through interaction with an SNP in the CFH intron, which has been reported to be highly associated with AMD (20).
Current knowledge on ERCC6 function does not readily provide a clear underlying mechanism for why overexpression of this gene induces AMD susceptibility. Moreover, because ERCC6 and CFH are located at chromosome 10q11 and 1q, respectively, and function in different pathways, we also do not have a biological model to elucidate this statistical epistasis of the two AMD loci. However, the effect of ERCC6 alone is likely related to ERCC6’s role as a common player in DNA metabolism (28, 29). ERCC6 functions in DNA repair, regardless of whether the repair is transcription coupled or base incision, as a partner in protein complexes (30, 40). It has been reported that overexpression of just one element in an enzyme complex can adversely affect the desired end result (41–45). Even though a statistically identified epistasis does not necessarily mean a biological epistasis (24), prolonged enhanced expression of ERCC6 could result in significant biological epistasis because ERCC6 is known to play an important role in universal transcription by functioning as a component of RNA pol I transcription complex (40). Based on the fact that CFH is the first major disease-related gene for AMD and that the CFH intron SNP is in absolute LD with CFH-tyr402his, it is worth exploring whether the complement cascade could be affected by elevated ERCC6.
An association of CFH SNPs and AMD has been documented (17–20). We not only replicated the CFH association (20) but also discovered a strong interaction between the ERCC6 and the CFH SNP. Functional studies on ERCC6 C−6530>G implicate this polymorphism’s role in gene regulation. Although the SNP is positioned 6,530 bp away from the translation start codon, it is only 492 bp away from the transcriptional start site. EMSA experiments have clearly revealed that nuclear extracts from human RPE cell lines distinctively bind to this SNP adjacent sequence with different affinities as well as binding patterns, which is in agreement with the in silico analysis. Cells having CC, CG, and GG ERCC6−6530 SNP types also produce different levels of transcripts accordingly. This effect is likely because of altered transcriptional regulation elements binding with unidentified transcription factors. Although a supershift experiment with specific antibodies might be helpful to define which transcription factors are involved in this alteration, our goal in this study is to demonstrate the different biochemical effects caused by a C>G transition at the −6,530 nucleotide of ERCC6.
Although we have not identified SNP(s) that are in LD with ERCC6 C−6530>G SNPs with the selection of several common nearby SNPs, we cannot exclude that other variation(s) in LD with ERCC6 C−6530>G might be the true contributor to AMD risk.
In conclusion, our data suggest that the G allele of ERCC6 C−6530>G is associated with a risk of AMD development and possibly interacts with an SNP in CFH to influence AMD susceptibility. We have also presented evidence that ERCC6 C−6530>G, which is located in the regulatory region, up-regulates the transcript and protein expression. These data also support the hypothesis that DNA repair mechanisms may play a role in AMD pathogenesis. Additional analysis of polymorphic ERCC6 transcription, and the relationship of ERCC6 protein with CFH, especially in ocular tissue, will further elucidate the nature of the observed AMD association and the SNP-SNP interaction. These findings expand the spectrum of known AMD risk factors and can hopefully suggest appropriate and specific candidate target molecules in AMD treatment and prevention.
Methods
In Silico Analysis.
The sequence flanking the SNP was screened for transcription factor binding sites. Web-based alibaba2.1 (www.alibaba2.com) software was used for the analysis.
EMSA.
ARPE-19 cells, a spontaneously arising human RPE cell line established by the selective trypsinization of a primary RPE culture (46), were maintained in Dulbecco’s modified Eagle’s medium (DMEM) F12 supplemented with 10% FBS and 100 units/ml each of penicillin and streptomycin. Nuclear extracts from the RPE cells were isolated from confluent cultures by using a nuclear extraction kit (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions. Infrared dye (IRD-800)-labeled DNA probes were purchased from Li-Cor (Lincoln, NE). The top strand sequences for the DNA probes were: C allele, 5′-GAC AGC TCT CCA TCC TTC CCG-3′ and G allele, 5′-GAC AGC TCT GCA TCC TTC CCG-3′. Unlabeled oligos and the corresponding complementary oligos were ordered from Integrated DNA Technologies (Coralville, IA). The binding reactions were carried out for 20 min at room temperature. Each 5-μl binding reaction contained 50 mM Tris·HCl (pH 7.5), 25 mM KCl, 5 mM MgCl2, 100 ng Poly(dI)·(dC), 1 mM DTT, and 0.1-nmol-labeled probe, with or without 4-μg nuclear extracts and different amounts (0–25 nmols) of unlabeled probes. Next, 2.5 μl of reaction mixture was loaded onto a native 6% polyacrylamide/bis gel (18 cm) by using 0.5 μl 6× loading buffer [60 mM Tris·HCl (pH 7.5) with 0.015% Orange G and 60% glycerol] and subjected to a Li-Cor IR2 global system for electrophoresis. Electrophoresis was carried out in 0.5× TBE buffer with the parameters: 200–400 V, 30 mA, and 25°C for 5 h. The raw image from the Li-Cor IR2 global system was compressed by using Adobe Photoshop 7.0.
Construction of Reporter Plasmids.
The ERCC6 promoter-luciferase reporter plasmids containing either the 6530C or 6530G sequence were constructed by using gene-specific PCR to amplify a 985-bp fragment of DNA from the 5′-flanking region of ERCC6 (ERCC6−6040 to ERCC6−7025, referring to accession no. AY204752) by using primers with restriction sites (Fig. 1). The forward primer was 5′-GAA TAC GCG TTG GGT TGG GGC CGC TGA CAG GAG-3′ and the reverse primer was 5′-GCC TAA GCT TGC CGC CAG CCT TGG AAC C-3′. Here the bold bases contain the cutting sites for the restriction enzymes MluI and HindIII, respectively. An advantage-GC genomic PCR kit (BD Biosciences, San Jose, CA) was used to perform DNA amplification following the manufacturer’s instructions. Two sets of human genomic DNA, confirmed by direct sequencing as CC homozygous and GG homozygous, were used as the templates for making reporter constructs. The PCR products were digested with MluI and HindIII (New England BioLabs), respectively. Restriction enzyme-digested PCR products were gel-purified by using a Qiagen kit and subsequently ligated to the pGL3-basic luciferase reporter vector (Promega) by using a DNA ligation kit (Stratagene). The clones containing either the C allele or G allele were confirmed by sequencing the full insert.
Transient Transfections and Luciferase Assays.
ARPE-19 cell is an established human RPE cell line that has been shown to express many of the characteristics of RPE cells in culture (46). The cell line was originally obtained from American Type Culture Collection. The cells were maintained in DMEM/F12 supplemented with 10% FBS and 100 μl/ml each of penicillin and streptomycin in a humidified, 5% CO2 incubator at 37°C. Cells were seeded into 96-well tissue-culture plates (F96 MicroWell plates; Nalge; Nunc) at a density of 20,000 cells per well in 200 μl and allowed to grow overnight. Transfection was performed by using Lipofectamine reagent (Lipofectamine 2000 transfection reagent; Invitrogen) according to the manufacturer’s protocol. Cells were cotransfected with 500 ng of reporter plasmid and pRL-TK plasmid. The latter, containing Renilla reniformis luciferase, was used to serve as a reference for transfection efficiency. DNA luciferase activity was determined according to the manufacturer’s protocol by using a dual-luciferase assay system (Promega). Briefly, cells were lysed with 50 μl of lysis buffer. Luciferase activities were immediately captured by an imagine document system with BioChemi camera (Ultraviolet Products, San Gabriel, CA) after mixing 100 μl of substrates of firefly luciferase and Renilla luciferase with the cell lysate. For each plasmid construct, three independent transfection experiments were performed, and each was done in triplicate. The empty pGL3 basic vector cotransfected with pRL-TK plasmid served as a control. Fold increase of the image densities was calculated by defining the activity of empty pGL3 basic vector as 1. Differences were determined by t test, and P < 0.01 was considered significant.
Detection of ERCC6 Transcripts by RT-PCR.
To detect the correlation of ERCC6 mRNA level with ERCC6 C−6530>G genotype, normal human lymphocytes with different genotypes (n = 5 in each genotype) were subjected to RNA extraction. The donors were age-matched. Real-time RT-PCR was performed by using the validated and inventoried ERCC6 and β-actin Taqman gene expression kit (Applied Biosystems). The assay was performed according to the manufacture’s instruction. The comparative Ct method was used to establish relative quantification of the fold changes in gene expression according to Applied Biosystems Prism 7700 sequence detection system. Fold changes were normalized first by the level of β-actin. The average fold change due to the treatment was again normalized to the level of the naïve cell and presented graphically. To detect whether ERCC6 is expressed in ocular tissue, RNA was extracted from frozen human ocular tissue. cDNA was synthesized by using a reverse transcription kit (RETROscript; Ambion, Austin, TX). cDNA from human RPE was obtained from the NEI Bank RPE cDNA library. RT-PCR was performed as described (47).
Immunohistochemical Detection of ERCC6 on Paraffin-Embedded Ocular Section.
Archived ocular sections from 25 AMD cases and 18 normal eyes were subjected to avidin–biotin–immunoperoxidase staining. Endogenous peroxide activity was blocked in hydrogen peroxide. Polyclonal anti-human goat IgG antibodies against a peptide mapping within an internal region of ERCC6 (CSB) (Santa Cruz Biotechnology) were applied in 1:50 dilution for 60 min at room temperature. Biotinylated rabbit anti-goat IgG was used as the linker molecule between the ERCC6 antibody and avidin–biotin–peroxidase complex for 60 min before application of the abc complex (Vector Laboratories). Diaminobenzidine-hydrogen peroxide was used as the chromogen. The sections were counterstained with 1% methyl green. The staining was quantified and arbitrarily graded based on the positive number of cells and the intensity (brown-blackish color) of the stained cells.
Study Subjects.
All subjects in this study were self-identified as Caucasians of non-Hispanic descent. Each participant signed the informed consent that was part of the protocol approved by the NEI Institutional Review Board.
Clinical diagnoses and categorization followed the guideline of NEI age-related eye disease study (AREDS) (48). All AMD patients had advanced stage, e.g., category >3, as defined by the guidelines of the AREDS. Advanced AMD patients had geographic atrophy involving the macula and/or choroidal neovascularization and large drusen in at least one eye (48). The unrelated control subjects were clinically evaluated and showed an absence of drusen or <5 small drusen (<63 μm), no evidence of significant extra-macular drusen, and an absence of all other retinal disease affecting the photoreceptors and/or outer retinal layers such as high myopia, retinal dystrophies, central serous retinopathy, vein occlusion, diabetic retinopathy, uveitis, and other retinal diseases. Fundoscopic photographs were taken for all patients and controls. The cases and controls were matched as closely as possible for age and gender (Table 5).
A set of archived paraffin-embedded ocular sections were obtained from 57 advanced-stage Caucasian AMD patients and used as an independent sample set for replication of possible association (49). All cases were diagnosed as AMD category = 4 (geographic AMD and/or neovascular AMD) based on the guidelines of the AREDS. Thirty of the 57 cases had neovascular AMD characterized by subretinal neovascular fibrous tissue with or without hemorrhage, exudate, and/or disciform scar. In addition, with these cases, there was a loss of photoreceptors and RPE alteration within the macular region. The remaining 27 cases were classified as areolar or geographic AMD cases without neovascularization and characterized by a loss of photoreceptors, RPE atrophy or hypotrophy, the presence of diffuse confluent drusen and/or large drusen, calcification, and fibrous glial scar in the macula. Eighteen archived normal eye slides from subjects >65 years old were used as the control.
Construction of DNA Standard.
Standard DNA SNP templates were made to serve as genotyping assay references. Genomic DNA of heterozygous ERCC6 C−6530>G (rs3793784) was PCR-amplified by using the primers 5′-GGG GGG AAC AGA GAA GCA GGA CAG CTA T-3′ and 5′-GCC ATG CGA ATG TAA ATC CT-3′. A mismatch was introduced into the forward primer (bold A) to generate an Nsi restriction site. The 157-bp PCR fragment was inserted into the pGEM-T Easy vector (Promega). The ligation product was transformed to JM109 high efficiency competent cells (Promega). Twenty colonies were collected to determine the specific SNP types by using the restriction fragment length polymorphism (RFLP) assay. RFLP-selected colonies containing the corresponding SNP types were further confirmed by direct sequencing. The colonies corresponding to each allele were propagated in LB broth for extraction of plasmid DNA that later served as the genotype specific DNA standard. The mixture of the plasmids containing two different alleles served as the heterozygous control.
Standard DNA for the CFH intron (rs380390) was cloned in the same fashion by using the primers 5′-CGC GGA TCA AAT TAT CAA AAG TTA TGG AC-3′ and 5′-CCT TAT GGC CTG CAA GGT T-3′ with a mismatch in the forward primer (bold G) to generate an AvaII restriction site. The polymorphic site was confirmed by direct sequence.
SNP Typing.
DNA from NEI AREDS Genetic Repository and DNA extracted from archived paraffin-embedded ocular sections were used for SNP typing (21). A detailed description of the use of PCR-restriction fragment length polymorphism and Taqman assays for SNP typing can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Statistical Analysis.
Hardy–Weinberg equilibrium for each SNP was tested by the χ2 procedure. Logistic regression was performed by using SAS (Release 9.1; SAS Institute, Cary, NC) to compare genotype and allele frequencies in cases and controls and to estimate ORs. Hypothesis testing was at P < 0.05 level. LD analysis was performed by using web-based software SNP analyzer.
Supplementary Material
Table 2.
Distribution of the CFH intron (rs380390) SNP types among cases and controls and its association with AMD
| Genotypes | Control (n = 268) | Case (n = 460) | χ2 (P value) | OR (CI) |
|---|---|---|---|---|
| GG | 104 | 83 | Reference | Reference |
| GC | 126 | 191 | 11.43 (0.0007) | 1.93 (1.32–2.83) |
| CC | 38 | 186 | 63.0 (<0.0001) | 6.77 (4.22–10.86) |
| GC + CC | 164 (61.2%) | 377 (82.0%) | 34.68 (<0.0001) | 2.80 (1.99–3.95) |
| G allele | 334 (62.3%) | 357 (38.8%) | Reference | Reference |
| C allele | 202 (37.7%) | 563 (61.2%) | 74.9 (<0.0001) | 2.74 (2.18–3.44) |
P value was adjusted by age, gender, and smoking status (ever smoked, never smoked). CI, 95% confidence interval. Reference, cell was taken as the baseline for the indicated statistical calculation: GG was taken as reference when comparing the genotypes, and G was taken as reference when comparing the alleles.
Acknowledgments
We thank the Age-Related Eye Disease Study (AREDS) participants, the AREDS Research Group, and National Eye Institute (NEI)–AREDS Genetic Repository for their valuable contribution to this research. We also thank Ms. Katherine Shimel, R.N., and Young Kim, R.N., for patient care and sample collection; Dr. Congxiao Zhang (NEI) for assistance in culturing the ARPE-19 cells; Dr. Wenmei Li (National Institute of Diabetes and Digestive and Kidney Diseases) for providing the MCF cell line used in the functional studies; and Dr. John Paul SanGiovanni (NEI) and David Y. He (Analytical Solution Group, Gaithersburg, MD) for statistical consultation. In addition, we acknowledge the study participants and their families for enrolling in this study. This research was supported fully by the Intramural Research Program of NEI at the National Institutes of Health.
Abbreviations
- AMD
age-related macular degeneration
- AREDS
age-related eye disease study
- CFH
complement factor H
- LD
linkage disequilibrium
- NEI
National Eye Institute
- OR
odds ratio
- RPE
retinal pigment epithelial
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Klein R., Peto T., Bird A., Vannewkirk M. R. Am. J. Ophthalmol. 2004;137:486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 2.Friedman D. S., O’Colmain B. J., Munoz B., Tomany S. C., McCarty C., de Jong P. T., Nemesure B., Mitchell P., Kempen J. Eye Diseases Prevalence Research Group. Arch. Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 3.Age-Related Eye Disease Study Research Group. Arch. Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Age-Related Eye Disease Study Research Group. Ophthalmology. 2000;107:2224–2232. doi: 10.1016/s0161-6420(00)00409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyman L., Neborsky R. Curr. Opin. Ophthalmol. 2002;13:171–175. doi: 10.1097/00055735-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Seddon J. M., Rosner B., Sperduto R. D., Yannuzzi L., Haller J. A., Blair N. P., Willett W. Arch. Ophthalmol. 2001;119:1191–1199. doi: 10.1001/archopht.119.8.1191. [DOI] [PubMed] [Google Scholar]
- 7.Clemons T. E, Milton R. C, Klein R, Seddon J. M, Ferris F. L., III Ophthalmology. 2005;112:533–539. doi: 10.1016/j.ophtha.2004.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans J. R., Fletcher A. E., Wormald R. P. Br. J. Ophthalmol. 2005;89:550–553. doi: 10.1136/bjo.2004.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuo J., Bojanowski C. M., Chan C.-C. Prog. Retin. Eye Res. 2004;23:229–249. doi: 10.1016/j.preteyeres.2004.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman D. S., Katz J., Bressler N. M., Rahmani B., Tielsch J. M. Ophthalmology. 1999;106:1049–1055. doi: 10.1016/S0161-6420(99)90267-1. [DOI] [PubMed] [Google Scholar]
- 11.de Jong P. T., Bergen A. A., Klaver C. C., van Duijn C. M., Assink J. M. Eye. 2001;15:396–400. doi: 10.1038/eye.2001.143. [DOI] [PubMed] [Google Scholar]
- 12.Klaver C. C., Wolfs R. C., Assink J. J., van Duijn C. M., Hofman A., de Jong P. T. Arch. Ophthalmol. 1998;116:1646–1651. doi: 10.1001/archopht.116.12.1646. [DOI] [PubMed] [Google Scholar]
- 13.Seddon J. M., Ajani U. A., Mitchell B. D. Am. J. Ophthalmol. 1997;123:199–206. doi: 10.1016/s0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- 14.Seddon J. M., Cote J., Page W. F., Aggen S. H., Neale M. C. Arch. Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 15.Meyers S. M., Greene T., Gutman F. A. Am. J. Ophthalmol. 1995;120:757–766. doi: 10.1016/s0002-9394(14)72729-1. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsdottir J., Conley Y. P., Weeks D. E., Mah T. S., Ferrell R. E., Gorin M. B. Am. J. Hum. Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conley Y. P., Thalamuthu A., Jakobsdottir J., Weeks D. E., Mah T., Ferrell R. E., Gorin M. B. Hum. Mol. Genet. 2005;14:1991–2002. doi: 10.1093/hmg/ddi204. [DOI] [PubMed] [Google Scholar]
- 18.Haines J. L., Hauser M. A., Schmidt S., Scott W. K., Olson L. M., Gallins P., Spencer K. L., Kwan S. Y., Noureddine M., Gilbert J. R., et al. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 19.Edwards A. O., Ritter I. R., Abel K. J., Manning A., Panhuysen C., Farrer L. A. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 20.Klein R. J., Zeiss C., Chew E. Y., Tsai J. Y., Sackler R. S., Haynes C., Henning A. K., Sangiovanni J. P., Mane S. M., Mayne S. T., et al. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuo J., Smith B., Bojanowski C. M., Meleth A. D., Gery I., Csaky K., Chew E., Chan C.-C. FASEB J. 2004;18:1297–1299. doi: 10.1096/fj.04-1862fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beatty S., Koh H., Phil M., Henson D., Boulton M. Surv. Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 23.Liang F. Q., Godley B. F. Exp. Eye Res. 2003;76:397–403. doi: 10.1016/s0014-4835(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 24.Moore J. H. Nat. Genet. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- 25.Winkler B. S., Boulton M. E., Gottsch J. D., Sternberg P. Mol. Vis. 1999;5:32. [PMC free article] [PubMed] [Google Scholar]
- 26.Ames B. N., Gold L. S. Mutat. Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 27.Conaway J. W., Conaway R. C. Annu. Rev. Biochem. 1999;68:301–319. doi: 10.1146/annurev.biochem.68.1.301. [DOI] [PubMed] [Google Scholar]
- 28.Tornaletti S., Hanawalt P. C. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 29.Hanawalt P. C. Nature. 2000;405:415–416. doi: 10.1038/35013197. [DOI] [PubMed] [Google Scholar]
- 30.Tuo J., Chen C., Zeng X., Christiansen M., Bohr V. A. DNA Repair (Amst.) 2002;1:913–927. doi: 10.1016/s1568-7864(02)00116-7. [DOI] [PubMed] [Google Scholar]
- 31.Boraz R. A. Pediatr. Dent. 1991;13:227–230. [PubMed] [Google Scholar]
- 32.Dollfus H., Porto F., Caussade P., Speeg-Schatz C., Sahel J., Grosshans E., Flament J., Sarasin A. Surv. Ophthalmol. 2003;48:107–122. doi: 10.1016/s0039-6257(02)00400-9. [DOI] [PubMed] [Google Scholar]
- 33.Traboulsi E. I., De Becker I., Maumenee I. H. Am. J. Ophthalmol. 1992;114:579–583. doi: 10.1016/s0002-9394(14)74486-1. [DOI] [PubMed] [Google Scholar]
- 34.Levin P. S., Green W. R., Victor D. I., MacLean A. L. Arch. Ophthalmol. 1983;101:1093–1097. doi: 10.1001/archopht.1983.01040020095016. [DOI] [PubMed] [Google Scholar]
- 35.Soames C. J., Sim R. B. Biochem. J. 1997;326:553–561. doi: 10.1042/bj3260553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein M. L., Schultz D. W., Edwards A., Matise T. C., Rust K., Berselli C. B., Trzupek K., Weleber R. G., Ott J., Wirtz M. K., et al. Arch. Ophthalmol. 1998;116:1082–1088. doi: 10.1001/archopht.116.8.1082. [DOI] [PubMed] [Google Scholar]
- 37.Weeks D. E., Conley Y. P., Mah T. S., Paul T. O., Morse L., Ngo-Chang J., Dailey J. P., Ferrell R. E., Gorin M. B. Hum. Mol. Gen. 2000;9:1329–1349. doi: 10.1093/hmg/9.9.1329. [DOI] [PubMed] [Google Scholar]
- 38.Zareparsi S., Branham K. E., Li M., Shah S., Klein R. J., Ott J., Hoh J., Abecasis G. R., Swaroop A. Am. J. Hum. Genet. 2005;77:149–153. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hageman G. S., Anderson D. H., Johnson L. V., Hancox L. S., Taiber A. J., Hardisty L. I., Hageman J. L., Stockman H. A., Borchardt J. D., Gehrs K. M., et al. Proc. Natl. Acad. Sci. USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradsher J., Auriol J., Proietti d. S., Iben S., Vonesch J. L., Grummt I., Egly J. M. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 41.Frosina G. Carcinogenesis. 2001;22:1335–1341. doi: 10.1093/carcin/22.9.1335. [DOI] [PubMed] [Google Scholar]
- 42.Frosina G. Eur. J. Biochem. 2000;267:2135–2149. doi: 10.1046/j.1432-1327.2000.01266.x. [DOI] [PubMed] [Google Scholar]
- 43.Kim P. M., Allen C., Wagener B. M., Shen Z., Nickoloff J. A. Nucleic Acids Res. 2001;29:4352–4360. doi: 10.1093/nar/29.21.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishel M. L., Seo Y. R., Smith M. L., Kelley M. R. Cancer Res. 2003;63:608–615. [PubMed] [Google Scholar]
- 45.Rinne M., Caldwell D., Kelley M. R. Mol. Cancer Ther. 2004;3:955–967. [PubMed] [Google Scholar]
- 46.Dunn K. C., Aotaki-Keen A. E., Putkey F. R., Hjelmeland L. M. Exp. Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 47.Tuo J., Muftuoglu M., Chen C., Jaruga P., Selzer R. R., Brosh R. M., Jr., Rodriguez H., Dizdaroglu M., Bohr V. A. J. Biol. Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 48.Age-Related Eye Disease Study Research Group. Control Clin. Trials. 1999;20:573–600. doi: 10.1016/s0197-2456(99)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan C.-C., Tuo J., Bojanowski C. M., Csaky K. G., Green W. R. Histol. Histopathol. 2005;20:857–863. doi: 10.14670/hh-20.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.