Abstract
Previous studies have narrowed the interval containing PSORS1, the psoriasis-susceptibility locus in the major histocompatibility complex (MHC), to an ∼300-kb region containing HLA-C and at least 10 other genes. In an effort to identify the PSORS1 gene, we cloned and completely sequenced this region from both chromosomes of five individuals. Two of the sequenced haplotypes were associated with psoriasis (risk), and the other eight were clearly unassociated (nonrisk). Comparison of sequence of the two risk haplotypes identified a 298-kb region of homology, extending from just telomeric of HLA-B to the HCG22 gene, which was flanked by clearly nonhomologous regions. Similar haplotypes cloned from unrelated individuals had nearly identical sequence. Combinatorial analysis of exonic variations in the known genes of the candidate interval revealed that HCG27, PSORS1C3, OTF3, TCF19, HCR, STG, and HCG22 bore no alleles unique to risk haplotypes among the 10 sequenced haplotypes. SPR1 and SEEK1 both had messenger RNA alleles specific to risk haplotypes, but only HLA-C and CDSN yielded protein alleles unique to risk. The risk alleles of HLA-C and CDSN (HLA-Cw6 and CDSN*TTC) were genotyped in 678 families with early-onset psoriasis; 620 of these families were also typed for 34 microsatellite markers spanning the PSORS1 interval. Recombinant haplotypes retaining HLA-Cw6 but lacking CDSN*TTC were significantly associated with psoriasis, whereas recombinants retaining CDSN*TTC but lacking HLA-Cw6 were not associated, despite good statistical power. By grouping recombinants with similar breakpoints, the most telomeric quarter of the 298-kb candidate interval could be excluded with high confidence. These results strongly suggest that HLA-Cw6 is the PSORS1 risk allele that confers susceptibility to early-onset psoriasis.
Psoriasis is an inflammatory and hyperproliferative skin disease affecting ∼2% of the United States population.1 The cutaneous manifestations of psoriasis are unpleasant and obvious, with a negative impact on quality of life.2 Moreover, up to 40% of psoriatics develop psoriatic arthritis; in 5% of psoriatics, the arthritis is severe and deforming.3 The disease is characterized by marked hyperplasia and altered differentiation of the epidermis, greatly increased blood flow, leukocytic infiltration of the skin, and a poorly understood relationship with nervous-system function.4 Many observations suggest that psoriasis is mediated by T cells with a Th1-dominated cytokine profile.5 However, the root cause of psoriasis remains unknown.
Twin and family studies clearly demonstrate that psoriasis has a genetic basis and is multifactorial in most if not all cases.6 However, despite numerous genetic linkage studies yielding at least 19 candidate loci, the identities of the genes involved remain unclear.7 Nevertheless, there is general agreement that a major genetic determinant of psoriasis, designated “psoriasis susceptibility 1” (PSORS1 [MIM 177900]), resides in the major histocompatibility complex (MHC).8
The existence of allelic associations between psoriasis and human leukocyte antigen (HLA) genes in the MHC has been appreciated for >30 years.9 There are numerous reports of strong allelic associations with the gene encoding HLA class I antigen HLA-Cw6.6 This association is particularly strong in patients with an early age at onset10 and in patients with the guttate subtype of psoriasis.11 These findings have led many researchers to suggest that HLA-Cw6 is the disease allele at PSORS1. This would be consonant with knowledge that MHC class I molecules play an important role in the function of CD8+ T cells.5 However, proof of this assertion is lacking.
The major obstacle to confirming or refuting the role of HLA-Cw6 in psoriasis has been linkage disequilibrium (LD). The MHC is characterized by extensive and presumably selection-driven variation, as well as the existence of particularly strong extended haplotypes.12 Overall, the recombination rate for the MHC is lower than the genomewide rate determined by sperm typing, and, in addition, the MHC includes defined subregions of low recombination.12 One of these regions, characterized by a 2.3-fold reduction in recombination rate relative to the genomic average, resides just telomeric to HLA-C.12 At least 10 genes have been identified within this region.13 As the allelic variation in these genes has been characterized and tested, it has become clear that many of these genes are also strongly associated with psoriasis. Attempts to stratify associations according to HLA-C status have usually failed to identify any association independent of HLA-Cw6. Against this background, it has become evident that neither the magnitude nor the statistical significance of individual allelic associations can provide definitive insight into the identity of the PSORS1 gene.
To overcome this challenge, we and others have turned to recombinant ancestral haplotype analysis. By accumulating and analyzing sufficient numbers of subjects, it is possible to identify individuals carrying only portions of the ancestral PSORS1 risk haplotype and to assess the risk conferred by those haplotypes. Our laboratory used STRs for this purpose,14 and Veal et al. used SNPs.15 Both studies confirmed that present-day MHC haplotypes have been generated by recombination events involving extended ancestral haplotypes.16 Together with more recent work,17 both efforts have identified a region of ∼300 kb just telomeric of HLA-B as the interval containing PSORS1.
In the present study, we extended our earlier haplotype analysis14 to include 16 polymorphisms in the HLA-C and CDSN genes and 188 additional pedigrees. We also incorporated improved methods for haplotype reconstruction. Finally, we used cloning and shotgun DNA sequencing to determine the complete genomic DNA sequence of the 300-kb PSORS1-risk region for 10 haplotypes falling into seven distinct haplotype clusters. Two of these are associated with psoriasis (risk), and the other five are unassociated (nonrisk). Comparison of sequence for the two risk haplotypes confirmed the existence of a 298-kb region of homology flanked by two clearly nonhomologous regions. Sequences of haplotypes drawn from the same cluster were nearly identical. Comparison of risk and nonrisk haplotypes revealed that only two genes (HLA-C and CDSN) in the 298-kb candidate interval encode variants unique to risk at the level of translated protein. Focusing on those haplotypes that have undergone recombination events between the risk alleles of HLA-C and CDSN, we further report that only those bearing HLA-Cw6 confer risk of psoriasis and that we can exclude the telomeric 25% of the risk interval with high confidence. Although the possibility of regulatory variants or unknown genes in the remainder of this interval cannot be completely excluded, this combined sequencing and haplotype-mapping approach strongly supports HLA-Cw6 as the major PSORS1 disease allele in early-onset psoriasis.
Subjects and Methods
Family Sample
The study sample consisted of 678 pedigrees of various structures (table 1), which either were identified through the dermatology services of the University of Michigan Medical Center, the Ann Arbor Veterans Affairs Hospital, the University of Kiel, and Henry Ford Hospital or were provided by the National Psoriasis Foundation Tissue Bank. Only families in which the proband's age at onset was <40 years were included.10 Individuals were considered to be affected if chronic plaque or guttate psoriasis lesions covered >1% of the total body surface area or if at least two skin, scalp, nail, or joint lesions were clinically diagnostic of psoriasis.18 A total of 2,723 individuals were recruited, 1,432 of whom were affected with psoriasis. Most (642) of the pedigrees were of white European ancestry. Informed consent was obtained from all subjects, under protocols approved by the institutional review boards of the participating institutions.
Table 1.
Composition of the Family Sample
|
No. of Nuclear Familiesa |
No. ofMultigenerationalFamilies by No.of Generations |
|||||||
|
No. of Affected Members |
Dyads | Triads | Sibships | 3 | 4 | 5 | 6 | Total No. of Families |
| 1 | 57 | 218 | 7 | 1 | 0 | 0 | 0 | 283 |
| 2 | 13 | 79 | 107 | 20 | 0 | 0 | 0 | 219 |
| 3 | 0 | 4 | 55 | 34 | 4 | 0 | 0 | 97 |
| 4 | 0 | 0 | 14 | 22 | 2 | 0 | 0 | 38 |
| 5 | 0 | 0 | 1 | 12 | 5 | 0 | 0 | 18 |
| ⩾6 | 0 |
0 |
0 |
7 |
14 |
1 |
1 |
23 |
| Total | 70 | 301 | 184 | 96 | 25 | 1 | 1 | 678 |
Nuclear (2-generation) families are classified as dyads, triads, or sibships (affected child and one or more collected siblings, with or without parents).
Markers and Genotyping
The 34 microsatellite markers originally used for haplotype-clustering analysis of a 490-pedigree subset of the current sample14 were genotyped for an additional 130 pedigrees. HLA-C and HLA-B antigen alleles were genotyped for 157 of the combined sample of 620 pedigrees, with use of DNA-based methods for HLA-C and serological methods for HLA-B, as described elsewhere.19 Seven SNPs in HLA-C and an additional eight SNPs and one indel in CDSN were genotyped for all 678 families in the current analysis (i.e., the 620 families subjected to microsatellite typing plus an additional 58 families). To locate ancestral recombination breakpoints between HLA-C and CDSN, 15 additional markers (12 SNPs and 3 indels) were genotyped for all individuals carrying either of two rare haplotypes that are possibly associated with psoriasis (HLA-Cw6-B45 and HLA-Cw7-B58). Nomenclature, amplification primers, probes, and genomic locations for all markers used in this study are listed in tables 2 and 3.
Table 2.
Nomenclature, Amplification Primers, and Genomic Locations of Microsatellite and Indel Markers
| Amplification Primer(5′→3′) | ||||||
| Marker | Alternate Name | Marker Type | Size Range(bp) | Location on NCBI Build 34 | Forward | Reverse |
| D6S273 | AFM142xh6 | (CA)n | 120–144 | 31787947–31788240 | GCAACTTTTCTGTCAATCCA | ACCAAACTTCAAATTTTCGG |
| M6S124 | D6S2671 | (CA)n | 252–284 | 31681654–31681913 | CCAGCCTGGATAACAGAACGAGAC | GCAACAAACCCCAACCTTAGG |
| TNFB | (CA)n | 101–129 | 31639721–31639831 | GCCTCTAGATTTCATCCAGCCACAG | CCTCTCTCCCCTGCAACACACA | |
| M6S125 | D6S2672 | (CA)n | 102–136 | 31576017–31576137 | ATACAGGGGCTTCAGTTTCTCTACC | CCACTTCAAACCAATCAGGGTG |
| MICA | (CA)n | 179–194 | 31484290–31484472 | CCTTTTTTTCAGGGAAAGTGC | CCTTACCATCTCCAGAAACTGC | |
| M6S166 | D6S2673 | (AC)n, (C)n, 4 nt indel, and 1 nt indel | 180–214 | 31453507–31453708 | TTCTGCGTTTTCAGCCTGCTAG | GAACCACTCTTCGTACCACAGTCTC |
| M6S101 | D6S2674 | (CA)n | 130–172 | 31363826–31363968 | CCTGAAACTTGGGCAATGAGTC | CCAGGCAAAAGTCAAGCATATCTG |
| M6S151 | D6S2675 | (CT)n and 1 nt indel | 200–204 | 31357982–31358181 | GCTGTGGCATGAGCTTCTTGAG | GGGTGGTTTATGACTGAGTAGGACC |
| M6S167 | D6S2676 | (GGAA)n and 1 nt indel | 220–276 | 31315194–31315448 | TGGGTGACAGAACAAGGCTCCATC | CACTTTCTCAGGTACTGGGGGTTAG |
| M6S105 | D6S2677 | (GTT)n and (T)n | 127–157 | 31313804–31313936 | GGGTGCCACAAGAATTGCAG | TCCAGCCTGTGATAGAGTGAGACC |
| M6S168 | D6S2678 | (GAAA)n | 258–326 | 31310300–31310567 | TTGCAGTGAGCCAAGATCGC | CCCCACAAAAAACCCCTGTTTATC |
| M6S178 | D6S2679 | (AG)n, (A)n, and 1 nt indel | 271–278 | 31303903–31304171 | TTGAACCAGGAGGTGGAGGTTG | CAAGAAATGACCACAGCAGGAAC |
| M6S102 | D6S2680 | (A)n, 5 nt indel, and 4 nt indel | 254–263 | 31302286–31302543 | CCTAATCCAAAGGCATGGCTTC | GGTGACAAAGCGAGACACCATC |
| M6S172 | D6S2681 | (ATTTT)n | 163–203 | 31294336–31294495 | CATAAGAAGGCATGGAAATAGGGC | TGGAGGTTGATGGTGAGCTGAG |
| M6S176 | D6S2682 | (T)n and 12 nt indel | 157–171 | 31286564–31286731 | GAAAGTCTTGAAGTCAGGTGTGGC | TGAGCTGAGATTGCACTCCAGC |
| M6S145 | D6S2683 | 4 nt indel | 209 and 213 | 31273045–31273249 | ACAGGTGTGAGCCCAAACTCCTAC | ATTAACCCACAACTGCACTCCGCC |
| M6S143 | D6S2684 | (CA)n, (A)n, and 4 nt indel | 240–261 | 31271236–31271472 | CCAACATGGTGAAGCCTTCTCTAC | CCGCTGGTCTAGTATCATTTCAGTG |
| M6S111 | D6S2685 | (AT)n | 170–192 | 31255818–31256002 | CAAACCTTGACTGTTCTTGCCC | TTACAGGTGCCTGCCACCATAC |
| M6S169 | D6S2686 | (TA)n, (A)n, and 2 nt indel | 156–162 | 31253800–31253953 | GAAAAGGAGCCAATGATAGCAGC | AGATTGCACCACTGCACTCCAG |
| M6S200 | D6S2687 | (AAAAG)n and (A)n | 132–142 | 31223152–31223292 | GCCTGGGTGACAAGAACAAGAC | GAATGTGGAACATGGAACACTGG |
| M6S198 | D6S2688 | (AAC)n | 237–243 | 31218046–31218282 | AGATCATGCCACTGCACTCCAG | CTCAAGCCTGGGTGATAGAGAAAG |
| M6S235 | 46 nt indel and 3 nt indel | 136, 139, and 186 | 31216036–31216175 | TGGACCCTGAGAGCCTACAT | GCCTCTCCCCAGAGTCAGTT | |
| M6S236 | 1 nt indel | 186–187 | 31210869–31211054 | ATGCCCGGCTAATTTTTGTA | AGGACTGTGAAGAGGGCATC | |
| M6S237 | 13 nt indel | 168 and 181 | 31207361–31207541 | GAGCCTCATACAGGGTGCAG | AGGCATCCCTGAGGAGAAAT | |
| M6S190 | D6S2689 | (ATT)n | 127–154 | 31201538–31201686 | CGTGCAGGACTGATCTCCATTC | TGGGGTTTCATCATGTTGGC |
| M6S161 | D6S2690 | (GA)n and 1 nt indel | 250–290 | 31151532–31151779 | TTGAACACAACCATCTCTGCTCC | ATCAGCCTGCTTCTGGGATTCTCC |
| M6S160 | D6S2691 | (TTTC)n | 129–185 | 31144072–31144202 | GTAGCTGTGGAAACAGTGTCCATG | GGCTTGACTTGAAACTCAGAGACC |
| M6S159 | D6S2692 | 4 nt indel and 1 nt indel | 215–221 | 31143530–31143742 | GCTCGGATAACATAGCAAGACTCTG | CAGTGTGCTTGTCCAATAATCCAC |
| M6S162 | D6S2693 | (AGGA)n and (A)n | 154–207 | 31129257–31129416 | CGCTATTGCATTCCAGCCTG | TCTAGTCATCCCTCCCTACTGCTG |
| M6S224 | D6S2694 | (CTTT)n, (CTTC)n, and 14 nt indel | 223–290 | 31126809–31127066 | AACTGTTCCTCTCTTAGAAGGCAGC | ACCTGGGCAATACAGCAAGACC |
| M6S179 | D6S2695 | 4 nt indel | 163 and 167 | 31115915–31116078 | CCAGTTCCTTCATTCGCAGGTC | AGCTTGTAGTGAGCCGAGATCG |
| M6S181 | D6S2696 | 4 nt indel | 241 and 245 | 31107168–31107409 | CGCATTTTGGATGGCTGAAC | GAGCAAGATCCTGTCTCGAAAGG |
| M6S187 | D6S2697 | (ATTTT)n | 150–190 | 31058949–31059110 | AGGTGTAGGGGTAGGAGAGAAATTG | TTGCTCCACTATACTCCAGCCTGG |
| M6S164 | D6S2698 | (GT)n | 212–234 | 30990360–30990573 | TTCCCGACACAGCTCTGACTTC | GCGAGTGGAAGAAGGTGAGATTTAG |
| M6S163 | D6S2699 | (GT)n | 173–201 | 30965475–30965653 | CGACTCCACCTATGACGGACATAC | CCTCTTCTCAGCTCTTCCATCTCAC |
| M6S165 | D6S2700 | (CA)n | 144–168 | 30887321–30887463 | AAAAGGAGGAAGAGCCACGGAG | TCTGCCCAGCATGTTCTTGAC |
| M6S201 | D6S2701 | (GAAA)n | 112–156 | 30513856–30513991 | GCCACTGAGGTCTGTGGTCATAAC | TATTTTCCTGCTGCTGCCCC |
Table 3.
Nomenclature, Amplification Primers, Probes, and Genomic Locations of SNPs Used in the Present Study
| Amplification Primer(5′→3′) | |||||||
| Marker | dbSNP ID | Forward | Reverse | Variation | Single Base Extension Primer | Probe Stranda | NCBI Build 34 Position |
| SNP25 | AGCGAGGKGCCCGCCCGGCGA | GGAGATGGGGAAGGCTCCCCACT | C/G | CGACGCCGCGAGTCCRAGAGGGGAGCC | F | 31344254 | |
| SNP38 | AGCGAGGKGCCCGCCCGGCGA | GGAGATGGGGAAGGCTCCCCACT | C/A | CCGCGAGTCCRAGAGGGGAGCCSCGGG | F | 31344249 | |
| SNP39 | rs1131123 | AGCGAGGKGCCCGCCCGGCGA | GGAGATGGGGAAGGCTCCCCACT | C/A | CGCGGCTACTAYAACCAGAGCGAGG | F | 31344128 |
| SNP26 | rs1131118 | AGCGAGGKGCCCGCCCGGCGA | GGAGATGGGGAAGGCTCCCCACT | T/A | CGGGGCCAGGKTCTCACAYCMTCCAG | F | 31343856 |
| SNP36 | rs17416863 | AGCGAGGKGCCCGCCCGGCGA | GGAGATGGGGAAGGCTCCCCACT | C/G | GWGGATGTNYGGCTGCGACSTGGGGCC | F | 31343830 |
| SNP40 | rs17839985 | AGCGAGGKGCCCGCCCGGCGA | GGAGATGGGGAAGGCTCCCCACT | C/T | CAAGGATTACATCGCCCTGAAYGAGGA | F | 31343758 |
| SNP37 | AGCGAGGKGCCCGCCCGGCGA | GGAGATGGGGAAGGCTCCCCACT | C/G | CACGCACYYKCCCTCCAGGTAGGCTCT | R | 31343677 | |
| SNP66 | rs887464 | GTAAGCAGGCAGGTGAGAGG | AGGGCCTCATCTAGGCTCAT | G/A | AAATCTCTTCATCCCTGTTCTTCACAT | R | 31252187 |
| SNP67 | rs3132514 | GATCCCTTTGACGTGAGCAT | CTTGAACTCAGGCAGCAGAG | G/A | GGGTGCAGTGAGCTGAGATCGAGCCAC | R | 31249026 |
| SNP68 | rs3094191 | AGGTGTGGGAGTGATTCCAG | GATGGGGCCTTGCTATGTTA | G/A | TGGTCTTGAATTCCTGTCCTCAAGAAA | R | 31245179 |
| SNP69 | TGCTGGTCTTGAACTCCTGA | CATTCACCCATTCCCTGTTC | G/T | TTCTAACCTTCATAACCTGACAGGTGT | F | 31240385 | |
| SNP70 | rs7750641 | AGCAAAGTCCTGAGGCTTGA | CCCCACCATCAACAGGTACT | G/A | GACCTCCTGACCTTTGGCCCTGAAGGG | R | 31235557 |
| SNP71 | rs3130455 | CCTCCTCCCAACAGAGTGAA | CGAACGTCCCTTACCAACC | T/A | GCGGGGCCGGGAGCGAGGCGTAGGGGG | F | 31232260 |
| SNP72 | rs3094662 | GCCCTGCTTCCCTTTCTAGT | AGGCTGTCAGTTCCATCCAC | G/T | GGGTATTGTGGAGGGCAGTGAGGAAGG | F | 31228208 |
| SNP74 | rs6929464 | AGGTCTGGACCCCTCAAACT | GGTAGGAGGATCCCTTGAGC | G/A | AACTGACCCTCAAGCCAATAACGTGGA | F | 31204184 |
| SNP75 | rs6909321 | TCTTGGTCACCAGAGGAACC | CTTCCCTGTGGACTCACCTC | C/T | GCACAGGAAAAACAACTCAGAGGACAT | F | 31199434 |
| SNP1 | rs3095318 | CACCCAAGTGTGGAAGAAAAAGC | TACCTGAGGCGACCATACAGTGAG | T/A | GTGTGGGTGGGCACGGGATG | F | 31194387 |
| SNP34 | rs7742033 | CCGATGACTGAGATAAGGCAGAAAG | GTTCCTGGCTTAAAAGATCCTGC | C/T | AGCCGCTGGAGTCACCCTTCCCAGTGA | R | 31191469 |
| SNP2 | rs3130984 | GCTCCATTTCCAGTGCCAGAAG | TTGGTGAAGTAGCCCACAGGRTAG | G/A | GGGAGCAGCAGCTCTCACTCGGGAA | R | 31191207 |
| SNP9b | GCTCCATTTCCAGTGCCAGAAG | TTGGTGAAGTAGCCCACAGGRTAG | C/A | TCACTCGGGAARCAGCGGCTCTCACTCGGG | F | 31191186 | |
| SNP3 | rs707913 | GCTCCATTTCCAGTGCCAGAAG | TTGGTGAAGTAGCCCACAGGRTAG | C/T | CTGGACAAAGCTCTTCCTCTTCCCARACCT | F | 31191030 |
| SNP35 | GCAAATACTTCTCCAGCAACCCC | AGTGTATGTGCTTGTTTGTGCCC | G/A | TCTCCCTCCAGTTCTCGAGTCCCCAGC | F | 31190434 | |
| SNP4 | rs1042127 | GGCAAATACTTCTCCAGCAACCC | ACAGACATGCAAGGGTGACCAG | G/T | GTTCTAGCATTTCCAGCAGC | F | 31190413 |
| SNP5 | rs3132554 | GGCAAATACTTCTCCAGCAACCC | ACAGACATGCAAGGGTGACCAG | C/T | AGCACTGCCGCAGGGATGGTAGGGT | R | 31190406 |
| SNP6 | rs3130981 | GCAAATACTTCTCCAGCAACCCC | AGTGTATGTGCTTGTTTGTGCCC | G/A | GAAGTTTTCCTACCCCAAGGAGAGTTACTC | F | 31190056 |
| SNP76 | rs3734852 | CTTGCTATCCCCATCCTTCA | TGCAGGTGCCATCTCAATTA | C/T | CTGGAACAGCTCTCCCACCTGGCCGGG | R | 31185050 |
| SNP77 | rs7756290 | GATCTGACCCAGTGCAGTCC | TGCTGTGGTTTTGCAGACTC | T/A | CCAGTGACGGCCACAGGGGTGCTTGTG | F | 31176558 |
| SNP78 | rs12214039 | AGAAGAGACGGGGGTCTCAC | CCTTGGAAGGATGTTTGGAA | C/A | CAAGGTGGKCAGATCACCTGAGATCAG | R | 31166604 |
F = forward; R = reverse.
This marker is an AAG indel in exon 2 of CDSN but was typed as a SNP.
The SNPs typed for HLA-C (SNP25, SNP38, SNP39, SNP26, SNP36, SNP40, and SNP37) (table 3) are located in exons 2 and 3 of the gene at positions 213, 218, 341, 361, 387, 459, and 540 of GenBank mRNA reference sequence NM_002117.4 (unless otherwise noted, mRNA numberings denote the A of the initiator codon as nt +1). When haplotype phase could be unambiguously determined, the two SNPs at positions 213 and 361 allowed HLA-C to be typed to a biallelic level by distinguishing all HLA-Cw6 alleles (HLA-Cw0602–Cw0613) from the other 186 known alleles of the gene (July 2005 release 2.10.0 of the IMGT/HLA Sequence Database maintained by the HLA Informatics Group20 [see the Anthony Nolan Trust Web site]). Typing all seven SNPs provided absolute discrimination of HLA-Cw6 alleles, even when phase could not be determined with certainty, and allowed classification of HLA-C types into 15 allele groups when phasing was unambiguous. The nine polymorphisms typed for CDSN (SNP1, SNP34, SNP2, SNP9, SNP3, SNP35, SNP4, SNP5, and SNP6) (table 3) are all protein-altering variations and are located at positions 52, 166, 428, 447–449, 605, 1201, 1222, 1229, and 1579 of GenBank mRNA reference sequence NM_001264.2. Seven of these polymorphisms constitute the five different combinations that make the psoriasis-associated risk haplotypes unique among the 10 sequenced haplotypes of this study, and the other two (SNP34 and SNP35) define two known CDSN protein variants among psoriasis-risk haplotypes (see the “Results” section for more details).
DNA was prepared from peripheral-blood mononuclear cells or Epstein-Barr virus–immortalized lymphoblastoid cell lines, as described elsewhere.21 Microsatellite markers were genotyped either by 32P-labeled primers and polyacrylamide gel electrophoresis, as described elsewhere,21 or by fluorescent labeling followed by capillary electrophoresis on ABI Prism 3100 Genetic Analyzers (Applied Biosystems). Indels were typed by the same method. SNPs were typed by single-base primer extension, as implemented in the SnapShot assay protocol (Applied Biosystems), per the manufacturer’s instructions. Genotypes were checked for Mendelian inheritance errors by use of Pedstats22 and Pedmanager and, for unlikely genotypes, by use of the “error” option of version 1.0-alpha of Merlin.23
Genomic DNA Cloning and Sequencing
High–molecular weight DNA was isolated by sucrose density gradient centrifugation, and large-insert (∼35–45 kb) libraries were prepared in either a SuperCos1 cosmid vector (Stratagene) or a pEPIFOS-5 fosmid vector (Epicentre Biotechnologies), per the manufacturers' instructions. Each library had at least 12-fold haploid genome coverage. Libraries were screened using gel-purified probes derived by PCR amplification from the region of interest, and the probes were labeled with 32P by random priming. To map clones, we sequenced clone termini and used microsatellite and SNP genotyping to determine whether each clone was derived from the maternally or paternally inherited chromosome. Overlapping clones were selected for sequencing, to provide complete coverage of a 350-kb target region on both chromosomes.
For sequencing, the large insert fosmid or cosmid clones were sheared into ∼1-kb fragments by use of a nebulizer (Invitrogen), were gel purified, were blunt ended, were cloned into a pGEM3-ZF(−) plasmid vector (Promega), and were sequenced in both directions with the vector primers SP6 and T7. Raw sequences were assembled into contigs by use of SeqMan (DNAStar, version 6.0), assisted by alignment of shotgun sequence against an HLA-Cw7-B8 reference sequence from the COX homozygous cell line.24 The coordinate system used in the tables and figures of the present study was based on this reference sequence, starting with the first base of the 5′ primer (GCAACTTTTCTGTCAATCCA) used to amplify microsatellite marker D6S273 and extending in the telomeric direction (see table 4 for a listing of accession numbers for the reference sequence). High-quality sequence coverage from at least two different plasmid subclones—and from both strands, whenever possible—was required for the entire cosmid or fosmid insert. Examination of overlapping regions of sequenced clones for the same haplotype of the same individual yielded an estimated total error rate of well under one error per 100,000 bases, which is consistent with the error rate seen for the Human Genome Project.25
Table 4.
GenBank Accession Numbers and HLA-C and HLA-B Alleles for Haplotype Sequences
| Haplotype Clustera and Sourceb |
HLA-Cc | HLA-Bc | GenBank Accession Number(s) |
| 7 (66): | |||
| Patient 144 | 120301 | 3801 | DQ249175 |
| Patient 495 | 120301 | 3801 | DQ249179 |
| 37 (14): | |||
| Patient 541 | 07020103 | 070201 | DQ249181 |
| PGF cell line | 07020103 | 070201 | AL671883, AL662850, AL662844, and AL669830 |
| 41 (19): | |||
| Patient 541 | 0602 | 5001 | DQ249182 |
| 44 (25): | |||
| Patient 495 | 0602 | 570101 | DQ249180 |
| Patient 388 | 0602 | 570101 | DQ249178 |
| DBB cell line | 0602 | 5701 | CR759814, CR38829, CR753819, CR847993, and CR753812 |
| 49 (26): | |||
| Patient 135 | 070101 | 080101 | DQ249172 |
| Patient 144 | 070101 | 080101 | DQ249174 |
| COX cell line | 070101 | 080101 | AL670886, AL662847, AL662866, AL669854, AL845556, AL662833, AL662867, and AL663093 |
| 51 (17): | |||
| Patient 135 | 0802 | 140201 (B65) | DQ249173 |
| Patient 218 | 0802 | 140201 (B65) | DQ249176 |
| 60 (48): | |||
| Patient 218 | 030401 | 15010101 (B62) | DQ249177 |
| MCF cell line | 030401 | 15010101 (B62) | CR759828, CR933862, CR759815, CR938714, CR936880, CR759805, and CR759772 |
Haplotype-cluster number for the new analysis of the present study (cluster number from our original study14).
Patient numbers refer to haplotypes sequenced for the present study; cell lines refer to HLA-homozygous cell lines sequenced by the Sanger Institute for the MHC Haplotype Project.
HLA-C and HLA-B allele designations follow the nomenclature of the IMGT/HLA Sequence Database, release 2.10.0, maintained by the HLA Informatics Group.20 HLA-B alleles in parentheses are serological equivalents that are used elsewhere in this study in preference to the DNA allele designations listed here.
Comparison of Sequenced Haplotypes
Overlapping cosmid and fosmid clone sequences were assembled into a single contig for each of the 10 haplotypes (GenBank accession numbers are provided in table 4). Haplotype contigs were then aligned with each other in SeqMan. Whenever necessary, sequence alignments were manually adjusted to yield the minimum possible number of polymorphisms. For each polymorphism, its location in the COX HLA-Cw7-B8 reference sequence and its alleles for the 10 haplotypes were recorded. Haplotype contig sequences were then compared with that of the HLA-Cw6-B57 contig by determining a weighted percentage difference of polymorphic alleles over 2.5-kb intervals. Weighting each polymorphism by the inverse of the number of variations found within 1.25 kb of the polymorphism allowed the percentage-difference metric to incorporate a true local density of polymorphisms.
Analysis of Candidate Genes
All expressed and transcript genes within the 300-kb PSORS1 interval that are listed in a recently published gene map for the human MHC13 were selected as candidates for PSORS1 (table 5). With the exception of a computer-predicted locus (LOC442199), the information in this table corresponds to all genes shown in build 35.1 of National Center for Biotechnology Information (NCBI) Map Viewer for the candidate interval. For each gene, all mRNA sequences available in build 186 of NCBI Unigene were considered. Two-sequence BLAST was used to align each mRNA sequence with the relevant portion of the COX HLA-Cw7-B8 genomic reference sequence, and all exon boundaries were mapped. The mRNA sequences for each gene were then sorted into groups with similar splicing patterns. When more than one sequence existed for a given splice variant, that sequence with the most-complete 5′ and 3′ UTRs was used as a reference (reference mRNAs are provided in table 5). All variations among the 10 sequenced haplotypes that occurred in the spliced transcript of the reference mRNA sequence were located, and their effect on the predicted protein sequence was determined. The predicted coding sequence included as an annotation within the GenBank sequence record was used whenever available; the longest ORF predicted by MacVector (version 6.5.3 [Accelrys]) was used instead for those two reference mRNA sequences without this information (BX647174.1 for TCF19 and AK094433.1 for HCG22). Version 0.97-600-1000 of the MINCOV program (Stanford Center for Tuberculosis Research Web site)26 was used to search for minimal sets of polymorphism alleles that distinguished the two risk haplotypes (Cw6-B57 and Cw6-B50) from the eight nonrisk haplotypes. Each splice variant of each gene was analyzed; separate analyses were conducted for all polymorphisms within the mRNA transcript, as well as for the subset of protein-altering mutations within the coding sequence.
Table 5.
Nomenclature and mRNA Reference Sequences for All Known Genes in the PSORS1 Candidate Interval
| Gene Symbola | HGNC Symbolb |
HGNC IDc | Entrez IDd | HGNC Namee | mRNA Reference Sequence(s)f |
| HLA-C | HLA-C | 4933 | 3107 | Major histocompatibility complex, class I, C | NM_002117.4 |
| HCG27 | HCG27 | 27366 | 253018 | HLA complex group 27 | NM_181717.1 and BC041465.2 |
| PSORS1C3 | PSORS1C3 | 17203 | 170681 | Psoriasis susceptibility 1 candidate 3 | AY484516.1 |
| OTF3 | POU5F1 | 9221 | 5460 | POU domain, class 5, transcription factor 1 | NM_002701.3 and NM_203289.2 |
| TCF19 | TCF19 | 11629 | 6941 | Transcription factor 19 (SC1) | S53374.1, BC033086.1, BC044632.1, and BX647174.1 |
| HCR | CCHCR1 | 13930 | 54535 | Coiled-coil alpha-helical protein 1 | NM_019052.2, AB112474.1, AB029331.1, and AK000533.1 |
| SPR1 | PSORS1C2 | 17199 | 170680 | Psoriasis susceptibility 1 candidate 2 | NM_014069.1 |
| SEEK1 | PSORS1C1 | 17202 | 170679 | Psoriasis susceptibility 1 candidate 1 | NM_014068.1 and AF484419.1 |
| CDSN | CDSN | 1802 | 1041 | Corneodesmosin | NM_001264.2 |
| STG | C6orf15 | 13927 | 29113 | Chromosome 6 open reading frame 15 | AY358438.1 |
| HCG22 | HCG22 | 27780 | 285834 | HLA complex group 22 | AK094433.1 |
Gene symbol used in the present study.
Official gene symbol that has been approved by the HUGO Gene Nomenclature Committee (HGNC).
A unique gene ID provided by the HGNC.
Gene ID in the Entrez Gene database curated by the NCBI.
Official gene name that has been approved by the HGNC.
GenBank entry number(s) for mRNA sequences used as references in the present study. Multiple mRNA sequences for a gene correspond to multiple known splicing variants.
Haplotype Reconstruction
Haplotypes were reconstructed by three different methods. The first method generated maximum-likelihood haplotypes with Merlin,23 with a new version that models LD among clusters of tightly linked markers.27 Population-haplotype frequencies within the cluster were computed by Merlin from our pedigree data with an expectation-maximization algorithm. The second method used a combination of Merlin and PHASE,28,29 as described in detail elsewhere.30 In brief, maximum-likelihood haplotypes were created with an implicit assumption of linkage equilibrium between markers by use of the “best” option of Merlin (version 0.10.2). Phase ambiguities in the most-likely Merlin haplotypes were then resolved by PHASE (version 2.1.1) whenever the confidence of the phase call was at least 99%. The third method used version 1.5.5 of the family-based association test (FBAT)31,32 (see the Harvard School of Public Health Web site), which reconstructs haplotypes in a probabilistic manner, with a conditioning approach that allows use of haplotypes with missing genotype or phase information without introducing bias.33
Haplotypes for the nine genotyped polymorphisms of CDSN and the seven typed SNPs of HLA-C were reconstructed by method 1. Haplotypes for HLA-C and CDSN in the 58 families typed only for these two genes were also generated by method 1; the entire pedigree sample was used to improve estimation of haplotype frequencies in the underlying population. Thirty-six marker haplotypes (HLA-Cw6, CDSN, and 34 microsatellites) in 620 families were generated by method 2 because of computational constraints of methods 1 and 3.
Accuracy of HLA-C and CDSN typing was confirmed by several methods. For CDSN, identical haplotypes were obtained with methods 1 and 2, and the founder-haplotype frequencies produced by method 3 were very similar to those yielded by the first two methods. Accuracy of haplotypes for the seven typed SNPs of HLA-C generated by method 1 was assessed by comparison of the inferred haplotype configuration for each person with the predicted configurations for all pairwise combinations of the 198 known HLA-C alleles. Haplotypes were retained if they were the only possible outcome given the genotypes of that person and other family members or if the only other choices involved HLA-C haplotypes known to be very rare in the study population. Moreover, method 3 frequencies for 7-SNP HLA-C haplotypes were very similar to those of method 1. As mentioned before, HLA-C haplotypes could be absolutely discriminated to the level of HLA-Cw6 versus non–HLA-Cw6 in the absence of external phase information from the population or other family members.
Accuracy of HLA-C–CDSN haplotypes was assessed by comparison of the results of methods 1 and 3, which yielded very similar haplotype frequencies. Merlin, as used by method 2 for 36-marker haplotype construction, incorrectly assumes linkage equilibrium among markers, so the reliability of the most-likely haplotype vectors of Merlin was assessed by comparison of them with all possible haplotype vectors (under the assumption of no recombination) for each member of the pedigree sample. Only 0.006% (5 of 88,340) of the alleles were phased differently between the two sets. For 1,584 alleles, phase was ambiguous in one or more of the possible haplotypes for a person but resolved in the most likely haplotype; when method 2 was applied to that set of possible haplotypes for each pedigree with the most such discrepancies, all but 51 of these ambiguous phases were resolved identically for the possible and most likely haplotype.
For all haplotypes in this study, inferred haplotypes of uncollected family members were discarded for purposes of analysis, because their use by the transmission/disequilibrium test (TDT) and pedigree disequilibrium test (PDT) can lead to bias.34,35 Because these two tests can handle only one possible haplotype configuration per individual, haplotypes with ambiguous or missing phase information were also discarded, even though use of only phase-known haplotypes can also lead to bias.36 Bias of this sort should be minimal—none of the CDSN haplotypes and only 0.7% of the multiallelic HLA-C, 0.2% of the biallelic HLA-C, and 0.3% of 36-marker haplotypes needed to be discarded because of missing or unresolved phase information for those people with genotypes for at least one marker of the haplotype.
Haplotype Clustering
For founder chromosomes of the pedigree sample, 36-marker haplotypes were clustered using an average-distance agglomerative hierarchical method with a percentage-difference metric. To qualify for clustering, at least half of the alleles of the haplotype were required to be typed and of known phase. A total of 2,710 founder haplotypes either were directly typed or were potentially inferable from other typed members of the pedigrees; of these, 2,700 qualified for clustering. Untyped or unphased alleles—which comprised 0.10% and 0.14%, respectively, of the alleles of clustered founder haplotypes—were labeled as “missing” rather than as “zero,” to avoid false matches and mismatches. The criteria for assignment of haplotypes to a cluster were ⩾80% homogeneity of marker alleles and a minimum of five founders. All other haplotypes were lumped together into a single cluster. Clusters were then numbered and assigned to all nonfounders in the pedigrees, according to the segregation of founder chromosomes. Recombinations between founder haplotypes occurred six times among ∼1,400 informative meioses in the pedigree sample; the resulting recombinant haplotypes were discarded for all individuals inheriting them. Genotypes for HLA-C and HLA-B antigen alleles, which were available for only a subset of the pedigree sample, were assigned to haplotype clusters by inspection of informative pedigrees.
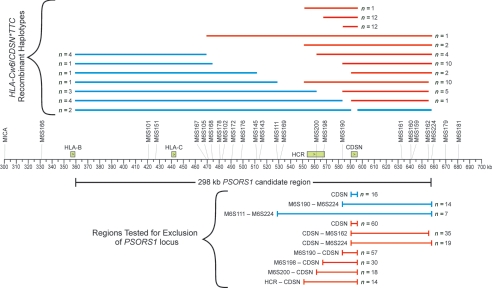
Mapping Breakpoints of Recombinant Haplotypes
Breakpoints for haplotypes with an ancestral recombination between the psoriasis-risk alleles of CDSN and HLA-C were mapped using the 34 microsatellite markers. Identity with the risk alleles of the PSORS1 candidate region was assessed, allowing for mutable markers for which two or three alleles (differing in size by one or two tandem repeats) occur on risk haplotypes that are otherwise similar. Of the 58 pedigrees that were typed only for CDSN and HLA-C, 2 carried a recombinant haplotype; all members of these two families were typed for additional markers, to map the breakpoints to the same resolution as the other recombinant haplotypes. Because association testing was used to exclude portions of the PSORS1 candidate region, breakpoints of HLA-Cw6−/CDSN*TTC+ recombinants (which were found to be unassociated with psoriasis) were mapped to the last marker on both sides of CDSN that bears a risk allele, to ensure that these haplotypes fully retained the portion of the candidate interval being tested for exclusion. Conversely, breakpoints of HLA-Cw6+/CDSN*TTC− recombinants (which were found to be positively associated with psoriasis) were mapped to the first marker on both sides of HLA-C that does not carry a risk allele, to ensure that these haplotypes had not retained any portion of the PSORS1-risk interval being tested for exclusion. To better compare two rare putative risk haplotypes (HLA-Cw7-B58 and HLA-Cw6-B45) with the reduced PSORS1-candidate region, 15 additional markers (M6S235–M6S237, SNP66–SNP72, and SNP74–SNP78) were used to fine map their breakpoints. These breakpoints were mapped to the first markers with a nonrisk allele, to delineate the largest possible portions of these haplotypes that are homologous to the ancestral HLA-Cw6-B57 risk haplotype.
Family-Based Association Tests
Pedigrees were analyzed for putative disease-associated alleles and haplotypes with three different family-based association tests: the TDT,37 the PDT,38,39 and the FBAT.31,32 All three methods were implemented as biallelic two-sided tests of the null hypothesis of no association in the presence of linkage. Both the TDT and PDT were extended to include dyads (affected child and one collected parent) when triads (affected child and two collected parents) were not available, with appropriate precautions.40 For the TDT, a single triad or dyad was randomly extracted from each pedigree. Because results vary depending on the particular random selection, the analysis was repeated 999 times with different random number seeds, and the median result was reported. Exact binomial P values were used. For the PDT, we used the PDT-avg test, which gives equal weighting to all families. All triads, dyads, and discordant sib pairs in a family contributed to the test. We also computed 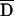 , a standardized measure of association between the disease and marker loci, as assessed by the PDT, which has a range of −1 to 1.17 For the FBAT, version 1.5.5 of the software41 was used with an additive model, the empirical variance, and an offset of 0. FBAT does not output a measure of LD that is independent of sample size; for this purpose, we calculated a statistic S*, which is the mean deviation of the FBAT test statistic S from its expected value over all families that are genotypically informative for the test allele. The number of families informative for association testing varied, depending on both the locus and test (table 6).
, a standardized measure of association between the disease and marker loci, as assessed by the PDT, which has a range of −1 to 1.17 For the FBAT, version 1.5.5 of the software41 was used with an additive model, the empirical variance, and an offset of 0. FBAT does not output a measure of LD that is independent of sample size; for this purpose, we calculated a statistic S*, which is the mean deviation of the FBAT test statistic S from its expected value over all families that are genotypically informative for the test allele. The number of families informative for association testing varied, depending on both the locus and test (table 6).
Table 6.
Number of Families Informative for Association Testing[Note]
|
No. ofInformativeFamilies |
||||
| Gene or Haplotype | TDT | PDT | FBATa | No. of Typed Families |
| HLA-C (biallelic) | 492 | 530 | 544 | 677 |
| HLA-C (multiallelic) | 623 | 634 | 642 | 670 |
| CDSN | 629 | 640 | 649 | 678 |
| HLA-C (biallelic) and CDSN haplotypes | 625 | 636 | 648 | 677 |
| 36-Marker haplotypes | 576 | 584 | … | 620 |
Note.— Although typing success was uniformly high (>99%) for all markers and haplotypes of the present study, the number of informative families varies because some families were not typed for the 34 microsatellite markers, the three association tests differ in what sort of pedigrees they can use, and dyads cannot be tested for biallelic markers. The numbers shown for each association test count only those families with at least one typed and phenotypically informative unit. For the TDT, this unit is a triad or dyad; for the PDT, it is a triad, dyad, or discordant sib pair (an affected child and unaffected sibling); and, for the FBAT with the settings used, it is a triad, dyad, discordant sib pair, or a sibship with three or more affected siblings.
As indicated by the ellipses (…), the FBAT cannot be applied to the 36-marker haplotypes, because the computational demands are too great; furthermore, FBAT can test only groups of haplotypes with exact matches for all alleles, and these constitute only a small fraction of all haplotypes in the sample.
The equality of TDT percentage transmission (%T) values for two risk alleles of the same locus or haplotype was tested by generating 100,000 simulated data sets. In each simulation, gentoypes were randomly assigned to the parents on the basis of observed allele frequencies in founders and an assumption of Hardy-Weinberg equilibrium. The probability of segregation from parents carrying exactly one risk allele was based on the observed %T for all risk alleles combined; for homozygous parents, alleles were assumed to segregate in a Mendelian fashion. Significance levels were determined by comparison of the difference of %T values in our original pedigree sample with the difference of %T values in simulated pedigrees.
Recombinant haplotypes separating the risk alleles of HLA-C and CDSN were tested for their association with psoriasis. Correction for multiple correlated tests was determined using a Monte Carlo method.42 In brief, founder haplotypes were randomly assigned on the basis of observed allele frequencies and an assumption of Hardy-Weinberg equilibrium, and Mendelian segregation of founder haplotypes to nonfounders was then simulated. Association tests were repeated in the simulated data, and the most significant P value among all association tests was stored for each of 100,000 simulated samples, to create a reference distribution. The corrected P value for each test of the actual data was then simply the rank of its nominal P value within the sorted reference distribution.
TDT Power Analysis
Power to detect association with various recombinant haplotypes was determined by simulation, under the alternative hypothesis of LD between psoriasis and the marker locus. We generated 100,000 samples of triads and dyads that were equivalent in size and parental haplotype-cluster frequencies to those extracted from the observed pedigrees for the TDT, using a gene-drop algorithm with rejection sampling.30 We assumed a population prevalence of 2% for psoriasis. Estimates of genotype relative risk (GRR) for carriers of a recombinant haplotype presumed to contain PSORS1 were based on estimates of GRR for HLA-Cw6—10.4 for homozygotes and 5.2 for heterozygotes.17 Because only a small fraction of haplotypes carrying HLA-Cw6 exhibit an ancestral recombination between it and the CDSN risk allele, the GRR values for the relatively rare recombinants between these two alleles must be adjusted accordingly. The penetrance of noncarriers of recombinant haplotypes will be nearly double that of non–HLA-Cw6 carriers, so GRR values need to be halved to ∼5 for homozygotes and ∼3 for heterozygotes. Nominal power was then estimated using a type I error rate of 0.05 for each individual test. Estimates of corrected power utilized a type I error of 0.016, which was determined by simulation (see above) to ensure a global type I error rate of 0.05 for all 10 tests of recombinant haplotypes.
Results
Delineation of the PSORS1 Interval
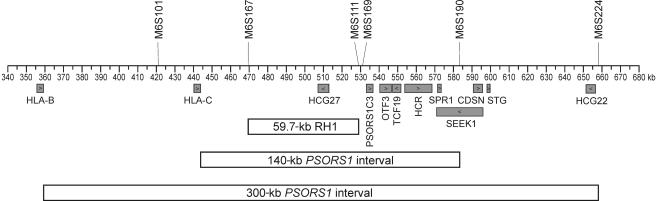
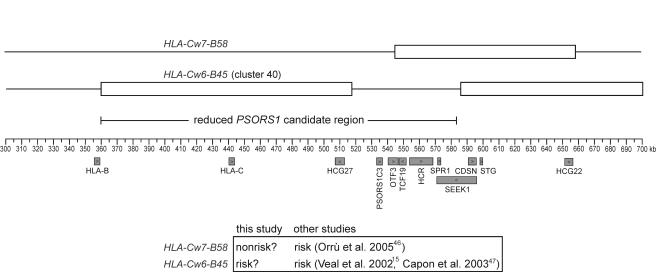
Our original localization study analyzed association between psoriasis and 62 microsatellite markers situated throughout the MHC.14 The strongest associations were found for markers lying within a 1.2-Mb region of the central MHC. A comparison of clusters of 34-marker haplotypes spanning this region narrowed the candidate interval for PSORS1 to a 59.4-kb region designated “RH1” (fig. 1), which was the shortest microsatellite haplotype segment common to all identifiable risk haplotypes.14 RH1 was therefore our original target for DNA sequencing. However, analysis of sequences flanking RH1 and evidence that an uncommon haplotype showing association with psoriasis in our original sample is probably not associated17 prompted us to extend the PSORS1 interval to 300 kb, bounded centromerically by HLA-B and telomerically by M6S224 (fig. 1).
Figure 1.
Delineation of the PSORS1 candidate interval. Known genes are shown below the coordinate axis, and key microsatellite markers are shown above. The candidate interval is depicted as originally defined (59.7-kb RH1 interval14), after its initial expansion on the basis of sequencing the regions immediately flanking RH1 (140-kb PSORS1 interval) and after incorporating results with reassessment of the association of an HLA-Cw8-B65 haplotype with psoriasis (300-kb PSORS1 interval17).
Sequencing Strategy
Rather than sequence genomic DNA directly, we chose to clone the region first. In this way, we could separate maternal and paternal chromosomes, avoid the difficulty of specifically amplifying regions with genomic repeats, and still sequence haplotypes originating in affected individuals from our sample. We selected five individuals for sequencing, which yielded a total of 10 haplotypes from seven of the distinct haplotype clusters of our original study.14 Two of these haplotype clusters were associated with psoriasis, and the other five were unassociated. Among risk haplotypes, HLA-Cw6-B57 was selected because it was the most common in our sample, and HLA-Cw6-B50 was chosen because our earlier studies showed that it limited the candidate interval on its telomeric end. The chosen nonrisk haplotypes (Cw12-B38, Cw7-B7, Cw7-B8, Cw8-B65, and Cw3-B62) represent five of the eight most common nonrisk-haplotype clusters in our original study.14 The Cw8-B65 cluster was originally selected as a risk haplotype, but subsequent analysis17 led us to reassign it to the nonrisk category. We deliberately selected three haplotypes (Cw12-B38, Cw7-B8, and Cw8-B65) to be cloned and sequenced twice, to gain a better understanding of the similarity of haplotypes belonging to the same cluster that were cloned from unrelated individuals. The seven distinct haplotypes selected for sequencing were carried by 34% of the founder chromosomes in our original study.14
Homogeneity of Haplotypes from the Same Cluster
Our original study used a threshold of 80% identity among marker alleles to define clusters for 34-marker haplotypes spanning the PSORS1 candidate interval.14 If the clustered sequences are not much more similar than this for all polymorphisms in the interval, then sequencing one member as a representative of the entire cluster would be of little value. Therefore, as a prelude to DNA sequencing, we assessed the homogeneity of the haplotype clusters to be sequenced by determining the percentage of founder haplotypes in each cluster that carry the consensus allele for the marker. This analysis, shown in table 7, revealed that the mean homogeneity of the seven sequenced haplotype clusters has a range of 91%–95% (grand mean 93%) when averaged over all 34 markers, far in excess of the 80% threshold used to define clusters. Homogeneity was even greater for the 24 markers within the 362-kb sequenced region and when the least stable markers (M6S167, M6S168, and M6S224) were omitted.
Table 7.
Homogeneity of Consensus Alleles for the Sequenced Haplotype Clusters[Note]
|
Percentage of Haplotypes in Cluster Carrying Consensus Allele at |
Mean Homogeneity forb |
||||||||||||||||||||||||||||||||||||
| Clustera | D6S273 | M6S124 | TNFB | M6S125 | MICA | M6S166 | M6S101 | M6S151 | M6S167 | M6S105 | M6S168 | M6S178 | M6S102 | M6S172 | M6S176 | M6S145 | M6S143 | M6S111 | M6S169 | M6S200 | M6S198 | M6S190 | M6S161 | M6S160 | M6S159 | M6S162 | M6S224 | M6S179 | M6S181 | M6S187 | M6S164 | M6S163 | M6S165 | M6S201 | All 34 Markers | 24 Markers | 21 Markers |
| 7 (66) | 82 | 84 | 87 | 79 | 97 | 97 | 95 | 100 | 97 | 100 | 87 | 100 | 100 | 100 | 100 | 100 | 97 | 100 | 92 | 100 | 100 | 97 | 64 | 92 | 100 | 97 | 72 | 100 | 100 | 100 | 100 | 97 | 72 | 45 | 92 | 95 | 97 |
| 37 (14) | 72 | 78 | 86 | 93 | 94 | 95 | 90 | 99 | 98 | 59 | 46 | 99 | 100 | 99 | 99 | 100 | 100 | 99 | 100 | 98 | 99 | 100 | 100 | 100 | 99 | 100 | 74 | 100 | 99 | 92 | 99 | 77 | 78 | 58 | 91 | 94 | 97 |
| 41 (19) | 81 | 86 | 86 | 94 | 97 | 90 | 93 | 100 | 84 | 100 | 97 | 97 | 100 | 100 | 100 | 100 | 100 | 90 | 100 | 97 | 97 | 100 | 97 | 100 | 100 | 100 | 77 | 96 | 92 | 90 | 87 | 76 | 86 | 61 | 93 | 96 | 98 |
| 44 (25) | 90 | 90 | 97 | 95 | 100 | 98 | 92 | 100 | 88 | 100 | 99 | 99 | 100 | 100 | 100 | 99 | 100 | 95 | 100 | 100 | 100 | 99 | 99 | 98 | 99 | 100 | 76 | 100 | 100 | 95 | 87 | 87 | 86 | 74 | 95 | 98 | 99 |
| 49 (26) | 79 | 84 | 97 | 89 | 100 | 91 | 97 | 99 | 85 | 100 | 99 | 99 | 100 | 100 | 99 | 99 | 99 | 92 | 100 | 99 | 100 | 90 | 100 | 100 | 100 | 99 | 95 | 100 | 100 | 88 | 99 | 98 | 93 | 71 | 95 | 98 | 98 |
| 51 (17) | 97 | 95 | 97 | 100 | 100 | 100 | 97 | 97 | 65 | 100 | 100 | 95 | 100 | 100 | 100 | 100 | 97 | 73 | 100 | 100 | 100 | 88 | 92 | 76 | 100 | 84 | 39 | 100 | 100 | 100 | 100 | 95 | 84 | 58 | 92 | 92 | 95 |
| 60 (48) | 88 | 88 | 93 | 72 | 100 | 98 | 76 | 100 | 86 | 100 | 91 | 100 | 98 | 98 | 97 | 100 | 100 | 100 | 98 | 98 | 100 | 50 | 100 | 100 | 100 | 100 | 81 | 100 | 97 | 100 | 98 | 90 | 86 | 65 | 93 | 95 | 96 |
| Meanc | 84 | 86 | 92 | 89 | 98 | 96 | 91 | 99 | 86 | 94 | 88 | 98 | 100 | 100 | 99 | 100 | 99 | 93 | 99 | 99 | 99 | 89 | 93 | 95 | 100 | 97 | 73 | 99 | 98 | 95 | 96 | 89 | 84 | 62 | 93 | 95 | 97 |
Note.— The table displays all 34 markers subjected to long haplotype clustering in our original study,14 in centromeric→telomeric order, from left to right. Numbers indicate the percentage of founder haplotypes in the cluster that carry the consensus allele for the marker. The sequenced portion of the haplotype clusters is shaded.
Haplotype-cluster number for the new analysis of the present study (cluster number from our original study14).
The mean homogeneity among founder haplotypes for each cluster for all 34 markers, for the 24 markers within the sequenced portion of the cluster haplotypes, and for the 21 most stable markers (excludes M6S167, M6S168, and M6S224) within the sequenced portion of the haplotypes.
The mean homogeneity of haplotypes among all sequenced clusters for each marker.
Clusters appeared to be even more homogeneous when SNP and indel variants were considered. Specifically, we compared our own three duplicate haplotype sequences, as well as four haplotypes derived from homozygous MHC typing cell lines sequenced by the Sanger Institute for the MHC Haplotype Project. With these sequences, we were able to make eight pairwise comparisons across the PSORS1-candidate interval, comprising six of the seven sequenced haplotype clusters. These comparisons revealed 99.996%–99.999% sequence similarity for haplotypes assigned to the same cluster (table 8). When expressed as a percentage of divergent alleles for those polymorphisms identified among the 10 sequenced haplotypes, these differences had a range of 0.047%–0.17%. These data demonstrate that haplotypes assigned to the same cluster by our methods are indeed nearly identical.
Table 8.
Sequence Comparison of Haplotypes That Belong to the Same Cluster
| Clustera, HLA-CB Haplotype, and Pairwise Haplotype Comparison |
Shared Sequence Length (bp) |
No. of Differences |
Fraction of Divergent Sequences |
No. of Known Polymorphismsb | Fraction of Divergent Polymorphisms |
| 7 (66): | |||||
| Cw12-B38: | |||||
| Patient 144 vs. patient 495 | 327,895 | 11 | 1/29,809 | 6,639 | 1/604 |
| 37 (14): | |||||
| Cw7-B7: | |||||
| Sanger PGF cell line vs. patient 541 | 341,361 | 3 | 1/113,787 | 6,415 | 1/2,138 |
| 44 (25): | |||||
| Cw6-B57: | |||||
| Sanger DBB cell line vs. patients 495 and 388 | 313,596 | 5 | 1/62,719 | 5,681 | 1/1,136 |
| Sanger COX cell line vs. patient 135 | 342,745 | 3 | 1/114,248 | 6,235 | 1/2,078 |
| 49 (26): | |||||
| Cw7-B8: | |||||
| Sanger COX cell line vs. patient 144 | 338,544 | 4 | 1/84,636 | 6,684 | 1/1,671 |
| Patient 135 vs. patient 144 | 319,157 | 6 | 1/53,193 | 6,135 | 1/1,023 |
| 51 (17): | |||||
| Cw8-B65: | |||||
| Patient 135 vs. patient 218 | 336,786 | 10 | 1/33,679 | 6,247 | 1/625 |
| 60 (48): | |||||
| Cw3-B62: | |||||
| Sanger MCF cell line vs. patient 218 | 323,140 | 6 | 1/53,857 | 6,078 | 1/1,013 |
Haplotype-cluster number for the new analysis of the present study (cluster number from our original study14).
Results are based on polymorphisms observed among the 10 sequenced haplotypes of the present study.
Sequence Characterization of the PSORS1 Interval
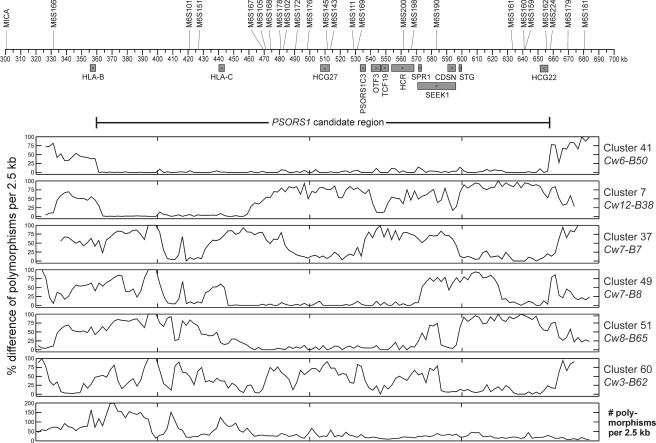
Figure 2 depicts the sequence divergence of one example of each of the sequenced haplotypes relative to the HLA-Cw6-B57 haplotype. Note that the divergence between the two risk haplotypes is very low across an interval extending from just telomeric of HLA-B (359.8 kb) to just telomeric of the HCG22 gene (657.6 kb). As defined by the first points at which the two risk-haplotype sequences diverge by >20% over a span of 2.5 kb, the PSORS1-risk interval is 297.8 kb in length. The two risk haplotypes diverge at 0%–17% (mean 2%; median 0%) of polymorphic sites within the 298-kb region of homology, at 39%–82% (median 43%) of sites in the 35 kb of sequenced centromeric flanking region, and at 29%–100% (median 75%) of sites in the sequenced 25 kb of telomeric flanking region. Thus, sequence comparison of the two risk haplotypes allowed us to more accurately define the risk interval than when the boundaries were defined using microsatellite markers14 or SNPs15 and to conclusively demonstrate continuous homology between risk haplotypes throughout the interval.
Figure 2.
Divergence of sequenced haplotypes from the HLA-Cw6-B57 risk haplotype. Known genes and their direction of transcription are shown below the coordinate axis, and microsatellite markers are shown above. The 298-kb PSORS1 candidate region is also depicted. The weighted percentage difference of polymorphism alleles, when compared with the sequence of the HLA-Cw6-B57 haplotype, is plotted by 2.5-kb intervals for each of the remaining six distinct haplotype clusters that were sequenced. Only those polymorphisms observed among the 10 sequenced haplotypes were considered when computing percentage difference. A plot for the second risk haplotype is shown first, followed by plots for the five nonrisk haplotypes. The bottom panel plots the number of polymorphisms observed among the 10 sequenced haplotypes, for 2.5-kb intervals of the sequenced region. The Cw6-B57 haplotype was derived from two individuals, because the region telomeric of CDSN on the Cw6-B57 chromosome of the person originally selected for cloning is actually derived from a Cw7-B8 haplotype by ancestral recombination.
Comparison of Risk and Nonrisk Haplotypes
In contrast to the extended region of sequence similarity between the two risk haplotypes, comparisons of nonrisk haplotypes with the HLA-Cw6-B57 risk haplotype demonstrate extensive variation. However, some portions of the nonrisk haplotypes HLA-Cw7-B8, HLA-Cw8-B65, and HLA-Cw12-B38 manifest marked similarity to risk haplotypes, with shorter regions of lesser similarity on nonrisk haplotypes HLA-Cw7-B7 and HLA-Cw3-B62 (fig. 2).
The bottom panel of figure 2 depicts the total number of polymorphisms observed among the 10 haplotypes, per 2.5-kb intervals. This panel reveals a highly uneven distribution of sequence variation, ranging from a low of 2.4 polymorphisms per kb in the telomeric portion of the PSORS1 interval to a peak of nearly 80 polymorphisms per kb in the interval between HLA-B and HLA-C. The 6,856 variations observed within the 362-kb sequenced interval include SNPs (85.0%), indels (8.5%), SNPs within indels (4.8%), poly A/T tracts (1.3%), and STRs (0.5%). Eleven of the variations are indels and inversions >100 bp in size; the largest, a 9.7-kb indel halfway between HLA-B and HLA-C. The mean density of 16.1 SNPs per kb observed for the sequenced region is much higher than the mean density of 2.9 SNPs per kb seen for the entire human genome (build 124 of dbSNP).
Analysis of Candidate Genes
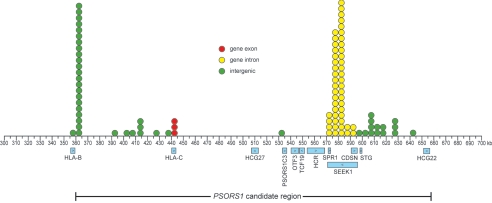
In all, 5,545 polymorphisms were found in the 297.8-kb candidate interval, 205 of them within spliced gene transcripts. We considered a candidate for PSORS1 to be either an allele at a single polymorphism or a combination of alleles at multiple polymorphisms that is unique to risk; that is, present on both risk chromosomes but absent on all five nonrisk chromosomes. Of the 5,545 polymorphisms, 98 (1.8%) met this criterion when considered individually. Three of these are SNPs in the second exon of HLA-C. None of the other 95 polymorphisms unique to risk haplotypes occur within spliced gene transcripts. However, 49 of the 95 occur within SEEK1 introns, and 2 of these 49 also occur in the first intron of CDSN, because of the overlapping nature of these two genes. The distribution of unique individual SNPs within the PSORS1 candidate region is depicted in figure 3.
Figure 3.
Locations of polymorphisms unique to risk haplotypes within the PSORS1 candidate region. Polymorphisms with alleles borne by the two sequenced risk haplotypes that differ from the alleles borne by all eight sequenced nonrisk haplotypes are plotted, with circles, to the nearest 5-kb interval. Fill color of the circle indicates whether the polymorphism occurs within a gene exon (red), gene intron (yellow), or intergenic region (green). Known genes and their direction of transcription and the PSORS1 candidate interval are shown below the coordinate axis.
Given the fact that alleles for HLA and non-HLA genes within the MHC differ by varying combinations of SNPs and indels, it is important to consider the possibility that the PSORS1-disease allele differs from other nonrisk alleles only when multiple polymorphisms within a functional unit are considered in combination. On the basis of our sequence data, thousands of two-way and millions of three-way combinations of polymorphisms within the candidate interval are unique to risk. Therefore, our analysis focused on variations found in spliced transcripts of all known genes in the area, with a particular focus on variations leading to alteration of amino acid sequences. A summary of the genetic variations, for each candidate gene, that are unique to risk haplotypes HLA-Cw6-B57 and HLA-Cw6-B50 is given in table 9. In the detailed analysis below, gene polymorphisms are named per the recommendations of Antonarakis et al.,43 with use of the mRNA reference sequences in table 5.
Table 9.
Polymorphisms in Spliced Transcripts of Known Genes of the PSORS1 Candidate Interval
|
No. of Qualifying Polymorphisms or Polymorphism Combinations ata |
|||||||||||
| Type of mRNA Polymorphism or Polymorphism Combination | HLA-C | HCG27 | PSORS1C3 | OTF3 | TCF19 | HCR | SPR1 | CDSN | SEEK1 | STG | HCG22 |
| Missense, nonsense, or coding indel | 48 | 1 | 0 | 1 | 2 | 8 | 2 | 8 | 3 | 5 | 4 |
| Silent | 16 | 1 | 0 | 4 | 2 | 6 | 0 | 10 | 0 | 3 | 2 |
| Noncoding (UTR) | 24 |
8 |
8 |
2 |
2 |
2 |
6 |
6 |
5 |
2 |
14 |
| Total | 88 | 10 | 8 | 7 | 6 | 16 | 8 | 24 | 8 | 10 | 20 |
| Single mRNA polymorphism of any type unique to risk | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Combination of protein-altering mRNA polymorphisms unique to risk | 68 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| Combination of mRNA polymorphisms, of any type, unique to risk | 6,065 | 0 | 0 | 0 | 0 | 0 | 4 | 47 | 5 | 0 | 0 |
For genes with more than one known splice variant, the maximum number of qualifying polymorphisms or combinations among all variants is reported.
Seven genes in the candidate interval (HCG27, PSORS1C3, OTF3, TCF19, HCR, STG, and HCG22) encode a spliced transcript on one or more psoriasis-associated haplotypes that is identical in sequence to the spliced transcripts of one or more nonrisk haplotypes, which renders each gene a noncandidate for PSORS1 at the level of mRNA and protein. A further two genes in the candidate interval (SEEK1 and SPR1) have no protein alleles specific to risk haplotypes but do have mRNA sequences that are unique to risk. For SPR1, four different pairwise combinations of mRNA alleles are unique to risk and involve five variations: 5′ UTR SNPs −284A→G and −90T→C, missense SNP 248C→T, and 3′ UTR SNPs 506A→G and 673C→G. The four combinations specific to the risk haplotypes all involve the 673G allele with −284G, −90C, 248C, or 506A as the second allele. For the larger of the two known splice variants of SEEK1, one pairwise combination and four four-way combinations of mRNA alleles are unique to risk. Seven polymorphisms are involved in these combinations: 5′ UTR variations −199A→G, −100C→T, −94G→A; missense variations −70C→A and 100A→G; frameshift variation 117delC; and 3′ UTR variation 483T→C. The combinations of polymorphism alleles unique to risk are (1) −100C and −94A; (2) −199A, −100C, 70C, and 100G; (3) −199A, −100C, 70C, and 483C; (4) −199A, −100C, 100G, and 117 undeleted; and (5) −199A, −100C, 117 undeleted, and 483C.
Only two genes in the PSORS1 candidate interval (HLA-C and CDSN) have protein alleles unique to the two risk haplotypes among the 10 sequenced haplotypes. For HLA-C, three polymorphisms—silent SNP 213G→C and missense SNPs 302G→A and 312C→A—each have an allele (C, A, and A, respectively) that is unique to the spliced risk transcripts among the 10 sequenced haplotypes. The two unique missense SNPs code for 77Asn and 80Lys in the HLA-Cw0602 protein of the risk haplotypes, as opposed to 77Ser and 80Asn in the nonrisk haplotypes. However, these variations are not unique to HLA-Cw6 transcripts or proteins, either singly or in combination, among all HLA-C alleles in the most recent release (2.10.0, July 2005) of the IMGT/HLA Sequence Database.20 Two pairs of SNP alleles (213C and 361T and 341A and 361T) are the smallest combinations that distinguish the spliced transcripts of all 12 known HLA-Cw6 alleles from those of all 186 known non–HLA-Cw6 alleles. To specifically distinguish the transcript of HLA-Cw*0602 (the allele found on the four major risk haplotypes of this study) from transcripts of all other HLA-C alleles, including the rare Cw*0603-Cw*0613 alleles, one of nine different nine-way combinations of SNP alleles is required. A pair of amino acids, 90Asp in exon 2 paired with a 97Trp in exon 3 (corresponding to the 341A and 361T allele pair mentioned above), is the smallest combination that distinguishes all HLA-Cw6 protein alleles from all non–HLA-Cw6 protein alleles. Four different nine-way combinations of amino acids uniquely define the Cw*0602 protein among all HLA-C alleles.
Five different combinations of protein-altering variations distinguish the CDSN protein of the two sequenced risk haplotypes from the nonrisk haplotypes. Seven polymorphisms are involved in these combinations—missense SNPs 52A→T, 428G→A, 605C→T, 1222T→G, 1229C→T, and 1579G→A, as well as coding indel 447–449delAAG. The five allele combinations unique to the two risk haplotypes are (1) 605T, 1222T, and 1229C; (2) 428G, 447–449 undeleted, 605T, and 1222T; (3) 52A, 428G, 605T, and 1222T; (4) 447–449 undeleted, 605T, 1222T, and 1579G; and (5) 52A, 605T, 1222T, and 1579G. The smallest combination unique to the CDSN gene carried by the risk haplotypes (605T, 1222T, and 1229C) corresponds to residues 202Phe, 408Ser, and 410Ser in the predicted protein of these haplotypes. These conclusions are unaffected by the presence of two known variations in the CDSN protein among risk haplotypes; the HLA-Cw6-B50 haplotype has a T rather than a C at missense variation 166C→T (which results in a Phe rather than Leu at residue 56), and the HLA-Cw6-B13 haplotype has a G rather than an A at missense variation 1201A→G (which results in a Gly instead of Ser at residue 401).44
Association of Psoriasis with HLA-C and CDSN
After we determined that HLA-C and CDSN were the only genes of the candidate interval encoding protein alleles unique to risk, we proceeded to type our entire sample for the seven coding SNPs that uniquely define HLA-Cw6 and the eight SNPs and one indel that distinguish the known risk alleles of the CDSN gene. Haplotypes were constructed for each gene as described in the “Subjects and Methods” section. As shown in table 10, HLA-C yielded 9 haplotype alleles, and CDSN yielded 12 haplotype alleles, 4 of which are rare and not described elsewhere. The alleles of each gene were then tested for association with psoriasis by use of the TDT and the FBAT.
Table 10.
Association of Psoriasis with Alleles of HLA-C and CDSN
| Gene and Haplotype Number |
Corresponding Allele(s)a | Haplotype Sequenceb | Frequencyc | T:NT (%T)d | TDT Pe | FBAT S*f | FBAT Pe |
| HLA-C: | |||||||
| 1 | *01(0201–04, 06–11), *02(05), *08(12), *12(0301–07, 11–13,15), *14(0202), and *16(01–0401) | GCCTCCG | .1116 | 108:115 (48.4) | .69 | −.020 | .66 |
| 2 | *01(05), *02(0201–04, 06–12), *03(14), *05(01–11), *07(07, 16), *08(0101–11), *12(0201–0203, 08, and 14), *15(0201–14), *16(06–07), and *17(01–03) | GCCACCG | .1814 | 127:190 (40.1) | 4.8 × 10−4 | −.134 | 9.6 × 10−4 |
| 3 | *03(0201–13, 16, and 18–19) | GCCACTG | .1151 | 76:133 (36.4) | 9.8 × 10−5 | −.195 | 1.1 × 10−5 |
| 5 | *03(17), *14(0201, and 0203–07N) | GCCTCTG | .0116 | 10:14 (41.7) | .54 | .115 | .54 |
| 6 | *04(010101–0103, 0401, 05, 08–10, 12–14, and 16–17) | GAAAGTG | .0961 | 84:96 (46.7) | .41 | −.072 | .14 |
| 11 | *06(02–13) | CCATCCG | .2140 | 295:97 (75.3) | 2.5 × 10−24 | .403 | 3.8 × 10−25 |
| 12 | *07(0101–03, 05–06, 08–10, 13–15, and 17–29) | GCAACCG | .2509 | 169:223 (43.1) | .0074 | −.131 | 3.3 × 10−4 |
| 13 | *07(0401–0402 and 11–12) | CCAACCC | .0182 | 17:17 (50.0) | 1.00 | −.007 | .95 |
| 15 | *18(01–02) | CCAAGTG | .0011 | 0:5 (.0) | .50 | … | … |
| CDSN: | |||||||
| 1 | 1.21 | ACG(AAG)TGTCG | .0658 | 88:45 (66.2) | 2.4 × 10−4 | .237 | 8.6 × 10−5 |
| 2 | 2.21–2.25 | ACA(AAG)TATTA | .2216 | 158:201 (44.0) | .027 | −.123 | .0031 |
| 3 | 1.31–1.32 | ATG(AAG)TATCG | .0179 | 23:11 (67.6) | .058 | .281 | .015 |
| 4 | 1.41–1.44 and 1.61 | ACG(AAG)CATCG | .1845 | 131:180 (42.1) | .0064 | −.117 | .0013 |
| 5 | 2.102 | TCG(—)TATTG | .1775 | 141:177 (44.3) | .050 | −.127 | .0049 |
| 6 | 1.51–1.52 | ACG(AAG)TAGCG | .1709 | 131:175 (42.8) | .014 | −.082 | .034 |
| 7 | 1.11–1.14 | ACG(AAG)TATCG | .1485 | 199:77 (72.1) | 1.3 × 10−13 | .336 | 1.6 × 10−15 |
| 8 | 2.101, 2.103–2.104, and 2.107–2.111 | ACG(—)TATTG | .0119 | 10:15 (40.0) | .42 | −.095 | .43 |
| 9 | … | TCG(—)CATTG | .0004 | 0:1 (.00) | 1.00 | … | … |
| 10 | … | ACG(AAG)TGGCG | .0004 | 1:0 (100.0) | 1.00 | … | … |
| 11 | … | TCG(—)TATTA | .0004 | 0:1 (.0) | 1.00 | −.500 | … |
| 12 | … | ATG(AAG)CATCG | .0004 | 1:0 (100.0) | 1.00 | .500 | … |
| 1, 3, and 7 | 1.21, 1.31–1.32, and 1.11–1.14 | AYG(AAG)TRTCG | .2297 | 289:112 (72.1) | 3.9 × 10−19 | .363 | 5.9 × 10−23 |
HLA-C allele designations follow the classification scheme of release 2.10.0 (July 2005) of the IMGT/HLA Sequence Database maintained by the HLA Informatics Group of the Anthony Nolan Research Institute.20 CDSN allele designations follow the classification scheme of Romphruk et al.,71 which is an extension of the system proposed by Guerrin et al.45 and Hui et al.69 Previously unpublished alleles are denoted with ellipses (…).
Haplotype for seven coding SNPs of HLA-C (mRNA positions 213, 218, 341, 361, 387, 459, and 540) and nine missense polymorphisms of CDSN (mRNA positions 52, 166, 428, 447–449, 605, 1201, 1229, and 1579), in 5′→3′ orientation. The TTC signature common to all three CDSN risk haplotypes is shown in bold italics.
Haplotype frequency, based on 2,850 founder chromosomes in 670 pedigrees for HLA-C and 2,856 founder chromosomes in 678 pedigrees for CDSN.
For the biallelic TDT.
All P values are uncorrected for multiple testing. P values for FBAT were computed only when there are at least 10 families informative for the allele.
Modified S statistic for the FBAT (see the “Subjects and Methods” section for details).
As shown in table 10, only HLA-Cw6 (HLA-C haplotype 11) and three of the CDSN haplotypes (1, 3, and 7) are positively associated with psoriasis. For CDSN, three of the five combinations of polymorphisms that distinguish the two sequenced risk haplotypes also distinguish the three associated CDSN haplotypes from all unassociated haplotypes for the entire pedigree sample. The other two combinations (52A, 428G, 605T, and 1222T and 52A, 605T, 1222T, and 1579G) are also carried by CDSN haplotype 8, which appears to be unassociated with psoriasis. The three CDSN risk haplotypes differed from each other at missense variations 166C→T and 1201A→G and exhibited somewhat different magnitudes for LD, as measured by the percentage transmission values in the TDT (66.2, 67.6, and 72.1 for alleles 1, 3, and 7, respectively; permutation tests for differences in %T were not significant) or the modified S statistic in the FBAT (.237, .281, and 336). Because these three CDSN haplotypes appear to carry similar risk for psoriasis, we combined them into one risk haplotype for subsequent analysis, which we designate “CDSN*TTC,” because the smallest allele combination unique to all three risk haplotypes is T, T, and C at mRNA positions 605, 1222, and 1229.
The association of HLA-Cw6 and CDSN*TTC with psoriasis is highly significant with use of both tests (table 10). HLA-Cw6 seems to be more strongly associated than CDSN*TTC (75.3% vs. 72.1% transmission; 3.8×10-25 vs. 5.9×10-23 FBAT P value). However, the difference in TDT T:NT (transmitted:nontransmitted) ratios for the two risk alleles (295:97 vs. 289:112) is not significant with use of the t test for equality of two proportions (P=.31).
Several of the more common alleles of each gene are negatively associated with psoriasis. However, these negative associations are not unexpected, since neutral alleles will be noticeably undertransmitted whenever one or more risk alleles are common and strongly overtransmitted. Consonant with this expectation, T:NT ratios for the subset of pedigrees lacking the common risk alleles show no significantly undertransmitted alleles (data not shown).
Haplotype-Cluster Analysis
We extended our earlier 34-marker haplotype-clustering analysis14 to include the HLA-C and CDSN genes and 130 additional pedigrees. We also incorporated improved methods for haplotype reconstruction. In general, the results of the new clustering analysis were very similar to those of the original study, but the larger sample size yielded more haplotype clusters (76 vs. 66) and substantially greater power of association tests. Sequencing the candidate interval greatly improved our ability to accurately determine regions that were identical by descent on the basis of microsatellite marker alleles. Consensus alleles and TDT results for all 36-marker haplotype clusters with a frequency of ⩾1% among founder chromosomes are presented in table 11. Data for all 76 haplotype clusters, including correspondence of old and new cluster numbers, can be found on the University of Michigan Psoriasis Genetics Laboratory Web site.
Table 11.
Major 36-Marker Haplotype Clusters: Consensus Alleles and Family-Based Tests of Association with Psoriasis[Note]
|
Alleles at |
||||||||||||||||||||||||||||||||||||||||
|
Category and Clustera |
Frequencyb | D6S273 | M6S124 | TNFB | M6S125 | MICA | M6S166 | HLA-B | M6S101 | M6S151 | HLA-C | M6S167 | M6S105 | M6S168 | M6S178 | M6S102 | M6S172 | M6S176 | M6S145 | M6S143 | M6S111 | M6S169 | M6S200 | M6S198 | M6S190 | CDSN | M6S161 | M6S160 | M6S159 | M6S162 | M6S224 | M6S179 | M6S181 | M6S187 | M6S164 | M6S163 | M6S165 | M6S201 | T:NT (%T)c | Pd |
| Risk: | ||||||||||||||||||||||||||||||||||||||||
| 44 | .0881 | 6 | 13 | 2 | 12 | 5 | 9 | 57 | 15 | 3 | 6 | 13 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 1 | 8 | 2 | 4 | 3 | 1 | 7 | 2 | 2 | 2 | 4 | 20 | 1 | 2 | 4 | 6 | 13 | 8 | 5 | 138:38 (78.4) | 1.6 × 10−14 |
| 47 | .0615 | 3 | 6 | 7 | 7 | 3 | 5 | 13 | 16 | 3 | 6 | 15 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 1 | 9 | 2 | 4 | 3 | 1 | 1 | 2 | 2 | 2 | 4 | 16 | 1 | 2 | 7 | 9 | 9 | 3 | 3 | 84:37 (69.4) | 2.3 × 10−5 |
| 43 | .0204 | 6 | 6 | 9 | 9 | 3 | 10 | 37 | 15 | 3 | 6 | 13 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 1 | 8 | 2 | 4 | 3 | 1 | 7 | 2 | 2 | 2 | 4 | 26 | 1 | 2 | 4 | 6 | 13 | 8 | 5 | 28:12 (70.0) | .017 |
| 41 | .0141 | 2 | 6 | 5 | 3 | 4 | 6 | 50 | 16 |
3 |
6 |
9 |
3 |
2 |
3 |
6 |
1 |
3 |
1 |
1 |
9 |
3 |
4 |
3 |
1 |
3 |
2 |
2 |
2 |
4 |
8 | 2 | 1 | 4 | 3 | 5 | 3 | 9 | 20:8 (71.4) | .036 |
| Nonrisk: | ||||||||||||||||||||||||||||||||||||||||
| 37 | .0926 | 6 | 6 | 11 | 6 | 3 | 5 | 7 | 9 | 3 | 7 | 1 | 7 | … | 5 | 1 | 3 | 3 | 1 | 1 | 4 | 4 | 6 | 3 | 2 | 2 | 2 | 2 | 2 | 4 | 23 | 2 | 2 | 5 | 3 | 5 | 3 | 5 | 66:104 (38.8) | .0044 |
| 49 | .0904 | 8 | 4 | 2 | 16 | 3 | 12 | 8 | 7 | 3 | 7 | 15 | 3 | 5 | 3 | 6 | 1 | 3 | 1 | 1 | 10 | 2 | 4 | 3 | 6 | 5 | 2 | 1 | 2 | 4 | 1 | 1 | 2 | 7 | 3 | 6 | 3 | 16 | 69:80 (46.3) | .41 |
| 11 | .0256 | 5 | 10 | 6 | 3 | 3 | 15 | 44 | 4 | 2 | 5 | 10 | 3 | 13 | 7 | 3 | 3 | 1 | 1 | 1 | 2 | 4 | 5 | 4 | 8 | 5 | 14 | 13 | 4 | 1 | … | 1 | 2 | 4 | 3 | 6 | 1 | … | 18:35 (34.0) | .027 |
| 60 | .0252 | 6 | 10 | 2 | 10 | 2 | 1 | 62 | 14 | 2 | 3 | 15 | 3 | 8 | 6 | 6 | 5 | 2 | 2 | 4 | 2 | 1 | 6 | 3 | 8 | 4 | 2 | 2 | 2 | 4 | 11 | 2 | 1 | 6 | 3 | 6 | 4 | 11 | 20:33 (37.7) | .098 |
| 7 | .0219 | 5 | 10 | 10 | 17 | 5 | 3 | 38 | 14 | 3 | 12 | 2 | 3 | 12 | 7 | 2 | 3 | 1 | 2 | 6 | 9 | 5 | 5 | 3 | 8 | 6 | 21 | 8 | 4 | 16 | 8 | 1 | 2 | 6 | 3 | 6 | 8 | … | 24:25 (49.0) | 1.00 |
| 68 | .0204 | 4 | 5 | 7 | 7 | 4 | 5 | 44 | 3 | 2 | 16 | 15 | 3 | 9 | 6 | 6 | 5 | 2 | 2 | 4 | 2 | 1 | 5 | 3 | 6 | 4 | 2 | 1 | 2 | 3 | 20 | 2 | 1 | 6 | 4 | 3 | 3 | 5 | 21:27 (43.8) | .47 |
| 51 | .0189 | 4 | 6 | 2 | 3 | 4 | 3 | 65 | 5 | 2 | 8 | 13 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 1 | 9 | 2 | 4 | 3 | 8 | 4 | 3 | 10 | 4 | 16 | … | 1 | 2 | 6 | 4 | 6 | 2 | 21 | 26:13 (65.0) | .053 |
| 22 | .0167 | 5 | 6 | 5 | 12 | 5 | 13 | 35 | 19 | 2 | 4 | 10 | 3 | 1 | 3 | 6 | 5 | 2 | 2 | 2 | 2 | 4 | 4 | 3 | 6 | 2 | 15 | 8 | 4 | 1 | 19 | 1 | 1 | 6 | 3 | 6 | 1 | 10 | 11:18 (37.9) | .27 |
| 57 | .0148 | 3 | 5 | 4 | 16 | 3 | 13 | 60 | 12 | 2 | 3 | 15 | 3 | 7 | 6 | 6 | 5 | 2 | 2 | 4 | 2 | 1 | 6 | 3 | 8 | 4 | 2 | 3 | 2 | 4 | 11 | 2 | 1 | 6 | 3 | 8 | 4 | 9 | 6:19 (24.0) | .015 |
| 29 | .0133 | 4 | 5 | 7 | 7 | 4 | 5 | 44 | 11 | 2 | 4 | 8 | 8 | 1 | 3 | 6 | 5 | 2 | 1 | 1 | 4 | 4 | 6 | 4 | 6 | 6 | 7 | 8 | 4 | 16 | 18 | 1 | 2 | 5 | 3 | 6 | 3 | 15 | 10:15 (40.0) | .42 |
| 5 | .0104 | 5 | 9 | 10 | 10 | 1 | 12 | 18 | 14 | 3 | 12 | 2 | 3 | 13 | 7 | 2 | 3 | 1 | 2 | 5 | 9 | 5 | 5 | 3 | 9 | 6 | 18 | 9 | 4 | 14 | 10 | 1 | 2 | 6 | 3 | 8 | 3 | 11 | 8:10 (44.4) | .82 |
| MinorHLA-Cw6: | ||||||||||||||||||||||||||||||||||||||||
| 40 | .0048 | 5 | 10 | 11 | 6 | 4 | 8 | 45 | 16 | 3 | 6 | 17 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 1 | 4 | 4 | 6 | 4 | 3 | 7 | 2 | 2 | 2 | 4 | 20 | 1 | 2 | 6 | 6 | 13 | 3 | … | 5:2 (71.4) | .45 |
| 45 | .0026 | 6 | 13 | 2 | 12 | 5 | 9 | 57 | 15 | 3 | 6 | 13 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 1 | 8 | 2 | 4 | 3 | 1 | 7 | 2 | 1 | 2 | 4 | 1 | 1 | 2 | 7 | 3 | 6 | 3 | 16 | 4:1 (80.0) | .38 |
| 46 | .0022 | 4 | 14 | 10 | 8 | 3 | 13 | ND | 15 | 3 | 6 | 17 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 1 | 9 | 2 | 4 | 3 | 1 | 7 | 2 | 2 | 2 | 4 | 13 | 1 | 2 | 6 | 4 | 7 | 6 | 5 | 3:2 (60.0) | 1.00 |
Note.— The table displays all 36 markers subjected to haplotype clustering, along with HLA-B, in centromeric→telomeric order, from left to right. Ellipses (…) indicate that no allele occurred in at least 50% of the founder haplotypes comprising the cluster. Italicized numbers indicate that the allele occurred in 50%–80% of the founder haplotypes comprising the cluster. Numbers in roman type indicate that the allele occurred in at least 80% of the founder haplotypes comprising the cluster. ND = no data. Alleles at the three gene loci (HLA-B, HLA-C, and CDSN) are shown in bold. Alleles are shaded when they differ among risk haplotypes for a marker but are not indicative of a significant difference in the underlying sequence of the region. The boxed area shows the minimum region of conserved or shaded marker alleles shared in common by all risk haplotypes.
All clusters with a frequency of at least 1% among 2,700 founder chromosomes in 620 pedigrees are shown. Clusters are listed by descending frequency within risk and nonrisk categories, where risk is defined as excess transmission with a nominal TDT P<.05. Also shown are minor clusters that carry the HLA-Cw6 allele. The complete table of all clusters is available at the University of Michigan Psoriasis Genetics Laboratory Web site.
Frequency of haplotypes in cluster.
For the biallelic TDT.
Uncorrected exact binomial P value for TDT.
The four identifiable risk haplotypes (defined as “positive association with psoriasis, with nominal TDT P<.05”) share a 300-kb region of homologous marker alleles between HLA-B and M6S224 that is identical to the PSORS1 candidate interval identified by sequencing (fig. 2) and reassessment of our earlier clustering study (fig. 1). It is likely that the risk haplotypes are derived from extended ancestral MHC haplotype 57.1 (HLA-A1-Cw6-B57-DR7-DQ916), represented in its fullest extent by cluster 44, and the 300-kb candidate interval is the minimum fragment of this ancestral haplotype retained by all four.
None of the other 11 common haplotype clusters appear to impart risk for psoriasis, although cluster 51 (designated “cluster 17” in our earlier study14) is overtransmitted (26:13; 65%) and nearly attains nominal significance (TDT P=.053). However, the HLA-Cw8-B65 haplotype of cluster 51 was clearly unassociated with psoriasis in a larger collaborative study17 and in a Sardinian population.46 Haplotype clusters 37 (Cw7-B7), 11 (Cw5-44), and 57 (Cw3-B60) are all significantly undertransmitted. These same haplotypes remain undertransmitted in the subset of pedigrees lacking all four risk haplotypes, but their negative association with psoriasis is no longer significant (data not shown). Whether these haplotypes are truly protective can be determined only by a larger sample with greater power.
The two prime candidates for the PSORS1 risk allele, HLA-Cw6 and CDSN*TTC, are both present on all four risk haplotypes and are both absent from all 11 nonrisk haplotypes. Hence, the common haplotype clusters provide no ability to discriminate between these two candidates. Risk haplotype clusters do vary in constituent CDSN alleles and apparent strength of association with psoriasis. The most common risk haplotype, cluster 44 (HLA-Cw6-B57), which carries the allele 7 variant of CDSN*TTC, shows the strongest association (78.4% transmission). The other three risk haplotypes, cluster 47 (Cw6-B13), 43 (Cw6-B37), and 41 (Cw6-B50)—which carry CDSN*TTC allele variants 1, 7, and 3, respectively—show somewhat weaker associations (69.4%, 70.0%, and 71.4% transmission, respectively), but the difference in T:NT ratios is not statistically significant.
Analysis of Recombinants between HLA-C and CDSN
Determining whether HLA-C or CDSN is the better PSORS1 candidate requires identifying and testing ancestral recombinants that have separated the risk alleles of these two genes. To this end, inspection of all 36-marker haplotypes in 620 families, along with all HLA-C–CDSN haplotypes in 58 additional families, revealed 76 such recombinant haplotypes among the 684 founder chromosomes carrying either risk allele. Recombinants retaining CDSN*TTC (60 in 55 families; 2.1% haplotype frequency) were nearly four times as common as recombinants retaining HLA-Cw6 (16 in 16 families; 0.56% haplotype frequency). Breakpoints for these recombinant haplotypes were conservatively mapped, as described in the “Subjects and Methods” section. Breakpoints are not distributed evenly within the 147-kb interval between the two genes. There appears to be a recombination hotspot just centromeric of CDSN, because nearly half (36 of 76) of the breakpoints mapped to the 23-kb interval between M6S198 and CDSN and nearly 90% (68 of 76) mapped to the 60-kb interval between M6S169 and CDSN. The variety of breakpoint locations meant that most recombinant haplotypes were rare, and only 7 of the 74 recombinant haplotypes were assigned to a cluster representing greater than five founder chromosomes.
As shown in table 12, association testing of the ancestral recombinants excluded CDSN as the PSORS1 gene with high confidence. Recombinant haplotypes retaining HLA-Cw6 but lacking CDSN*TTC were positively associated with psoriasis (T:NT=11:2; PDT Pc=.014 [where Pc is the P value corrected for multiple testing]), whereas recombinants retaining CDSN*TTC but lacking HLA-Cw6 were unassociated (T:NT=15:26; TDT Pc=.33), despite good statistical power (corrected TDT power=98.3%). As depicted in figure 4, grouping recombinants with similar breakpoints allowed us to test whether different overlapping portions of the 300-kb candidate interval carry PSORS1. HLA-Cw6–positive recombinants lacking the M6S190–M6S224 portion of the candidate interval are still positively associated with psoriasis (PDT Pc=.029), and CDSN*TTC–positive recombinants carrying the M6S190–CDSN portion of the interval are still unassociated (TDT Pc=.52; corrected TDT power=98.0%), which allowed us to exclude the 74.8-kb segment from M6S190 to M6S224 from further consideration. Even though we do not have power to confidently exclude more than the telomeric quarter of the candidate interval, note the consistent lack of association for all seven groups of HLA-Cw6–negative recombinants (transmission < 39%; 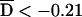 ) and the consistent positive association for all three groups of HLA-Cw6–positive recombinants (transmission > 84%;
) and the consistent positive association for all three groups of HLA-Cw6–positive recombinants (transmission > 84%;  ). If these trends of association are confirmed in a larger sample, we should be able to exclude the entire 129-kb gene-rich region spanning PSORS1C3 and M6S224 from the candidate interval.
). If these trends of association are confirmed in a larger sample, we should be able to exclude the entire 129-kb gene-rich region spanning PSORS1C3 and M6S224 from the candidate interval.
Table 12.
Association of Psoriasis with Haplotypes Carrying a Recombination between the Risk Alleles of HLA-C and CDSN
| TDT |
PDT |
||||||||||
|
P |
Power |
P |
|||||||||
|
Recombinant Haplotype and Test Regiona |
Map Bounds (kb)b | Haplotype Frequencyc | T:NT (%T)e | Uncorrectedf | Correctedg | Uncorrectedh | Correctedi |
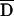 j j
|
Uncorrectedf | Correctedg | Excluded as Risk?d |
| HLA-Cw6+/CDSN*TTC−: | |||||||||||
| CDSN | 590.5–595.4 | .0056 | 11:2 (84.6) | .023 | .076 | … | … | .643 | .0042 | .014 | Yes |
| M6S190–M6S224 | 583.3–658.1 | .0049 | 9:1 (90.0) | .022 | .071 | … | … | .667 | .0074 | .029 | Yes |
| M6S111–M6S224 | 528.9–658.1 | .0028 | 5:0 (100.0) | .033 | .19 | … | … | .750 | .037 | .18 | No |
| HLA-Cw6−/CDSN*TTC+: | |||||||||||
| CDSN | 590.5–595.4 | .0209 | 15:26 (36.6) | .12 | .33 | .995 | .983 | −.248 | .19 | .70 | Yes |
| CDSN-M6S162 | 590.5–655.7 | .0122 | 9:18 (33.3) | .12 | .34 | .938 | .861 | −.295 | .22 | .77 | No |
| CDSN-M6S224 | 590.5–658.1 | .0066 | 5:9 (35.7) | .42 | .86 | .700 | .527 | −.283 | .39 | .95 | No |
| M6S190-CDSN | 583.3–595.4 | .0199 | 15:24 (38.5) | .20 | .52 | .994 | .980 | −.212 | .36 | .93 | Yes |
| M6S198-CDSN | 566.7–595.4 | .0105 | 8:13 (38.1) | .38 | .81 | .899 | .790 | −.261 | .42 | .96 | No |
| M6S200-CDSN | 561.7–595.4 | .0063 | 5:8 (38.5) | .58 | .95 | .677 | .498 | −.214 | .66 | 1.00 | No |
| HCR-CDSN | 551.1–595.4 | .0049 | 4:7 (36.4) | .55 | .94 | .553 | .363 | −.250 | .62 | .97 | No |
Bounds of the test region with use of the markers and genes of figure 2. To be conservative, the smallest possible portion of the extended HLA-Cw6+/CDSN*TTC+ risk haplotype not carried by HLA-Cw6+/CDSN*TTC− recombinant haplotypes and the smallest possible portion of the extended risk haplotype carried by HLA-Cw6−/CDSN*TTC+ recombinant haplotypes were used to determine whether a recombinant haplotype qualified for assessing the exclusion of the test region from the PSORS1 candidate interval.
Map bounds refer to the map coordinate system of figure 2.
Frequency of HLA-Cw6+/CDSN*TTC− recombinant haplotypes not carrying the test region or the frequency of HLA-Cw6−/CDSN*TTC+ recombinants carrying the test region; results are based on 2,867 founder chromosomes in 678 pedigrees.
The test region is considered excluded from the PSORS1 candidate interval if the corrected TDT or PDT P value is <.05 for the positively associated HLA-Cw6+/CDSN*TTC− haplotypes or if the corrected TDT power was at least 95% for the unassociated HLA-Cw6−/CDSN*TTC+ haplotypes.
For the biallelic TDT.
Nominal P value uncorrected for multiple testing.
P value corrected for multiple testing (see the “Subjects and Methods” section for details of correction procedure).
Nominal power of the TDT on the basis of a type I error rate of 0.05, an additive model with a GRR2 of 5, and a disease prevalence of 0.02.
Corrected power of the TDT based on a type I error rate of 0.016, which ensures an experimentwide type I error rate of 0.05 for all tests in the table, an additive model with a GRR2 of 5, and a disease prevalence of 0.02.
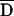 is a standardized measure of disequilibrium for the PDT (see the “Subjects and Methods” section).
is a standardized measure of disequilibrium for the PDT (see the “Subjects and Methods” section).
Figure 4.
Recombinant haplotypes and test regions used to map the PSORS1 locus. Key genes and their direction of transcription, along with microsatellite markers used in this study, are depicted immediately above the coordinate axis. All founder haplotypes bearing an ancestral recombination between the HLA-Cw6 and CDSN*TTC psoriasis risk alleles are shown above the axis, and those regions tested for exclusion of the PSORS1 locus are shown below the axis. Haplotypes and test regions of the unassociated (see table 12) HLA-Cw6−/CDSN*TTC+ recombinants are shown in red; those of the associated HLA-Cw6+/CDSN*TTC− recombinants are shown in blue. Line segments for the recombinant haplotypes indicate that portion of the extended HLA-Cw6+/CDSN*TTC+ risk haplotype within the 298-kb PSORS1 candidate region that is retained by the recombinant; to be conservative, the largest possible portion of the extended risk haplotype carried by HLA-Cw6+ recombinants and the smallest possible portion of the extended haplotype not carried by HLA-Cw6− recombinants are plotted. The test regions and their bounds are shown in the same order as in table 12. The numbers of founder chromosomes in the pedigree sample are shown for each recombinant haplotype, as are the numbers of qualifying recombinant founder chromosomes for each of the tests for exclusion.
One group of recombinants, represented by the last line of table 12, is equivalent to the HLA-Cw7-B58 recombinant found to be positively associated with psoriasis in both a family and case-control sample of Sardinian psoriatics.46 Although this haplotype appears to be unassociated in our families (T:NT=4:7, TDT Pc=.94), our sample has only 36% power to detect an association. We fine mapped the recombination breakpoints of this haplotype with 15 additional markers. As shown in figure 5, if the results of both studies are correct, then PSORS1 would lie within the 38 kb of overlap between that portion of the HLA-Cw7-B58 haplotype derived from the ancestral 57.1 haplotype and our reduced 224-kb candidate interval. Only two genes in this overlap region (SPR1 and SEEK1) have spliced transcript alleles unique to the sequenced risk haplotypes; however, both genes lie in the segment from M6S198 to CDSN that can be excluded with good confidence, by the TDT, from the candidate interval (79% corrected power) (table 12).
Figure 5.
Comparison of the reduced PSORS1 candidate region, established by this study, with two rare haplotypes that are possibly associated with psoriasis. Above the coordinate axis, the I-shaped line segment depicts the boundaries of the 224-kb reduced PSORS1 candidate interval, and unblackened rectangles depict the maximum possible regions of the “double recombinant” HLA-Cw6-B45 (cluster 40) and HLA-Cw7-B58 putative risk haplotypes that are homologous to the ancestral HLA-Cw6-B57 risk haplotype. Known genes and their direction of transcription are depicted below the axis. A summary of the psoriasis-risk status of these two haplotypes, as determined by this study and other studies, is also shown.
Analysis of Rare HLA-Cw6 Haplotypes
On the basis of the preceding analyses, HLA-Cw6 is the best candidate for PSORS1. If HLA-Cw6 is indeed the PSORS1 risk allele, then any haplotype carrying it should bear risk. As already seen, this certainly appears true for the four most common HLA-Cw6 haplotype clusters (table 11) and for the rare HLA-Cw6+/CDSN*TTC− recombinants (table 12). Three relatively uncommon 36-marker haplotype clusters (40, 45, and 46) also carry HLA-Cw6 (table 11). All three are overtransmitted to affected children, but power is too low to achieve a statistically significant association, because these clusters have haplotype frequencies <0.5% (table 11). Combining all HLA-Cw6–positive haplotypes outside of the four common risk clusters into a single group (frequency 2.9%) increases power and yields a significant association (T:NT=36:19; TDT P=.030; PDT P=.018). Although the four common risk clusters combined yield a higher percentage transmission than do other HLA-Cw6 haplotypes (76.0% vs. 65.5%), this difference is not significant (P=.34), and the disparity in strength of LD is not much greater than what is predicted (74.7% vs. 70.8%) by simulation under the null hypothesis that they have identical effects and differ only in frequency.
Another test of the hypothesis that HLA-Cw6 is the disease allele is the comparison of the strength of association for two different kinds of HLA-Cw6 haplotypes—those that perfectly match the consensus risk alleles of microsatellite markers in the reduced 224-kb candidate interval and those that have at least one allele mismatch for these markers. For this analysis, only those 12 markers for which the risk allele is found on at least 99% of the haplotypes in the four common risk clusters were used (i.e., M6S101, M6S167, M6S111, and M6S169 were excluded). Perfect-match haplotypes were abundant (19.4% haplotype frequency) and strongly associated with psoriasis (264:87; TDT P=7.3×10-22), whereas mismatch haplotypes were much less frequent (1.8%) but still positively associated (22:10; TDT P=.050; PDT P=.014). The difference in transmission ratios (75.2% vs. 68.8%) is not significant (P=.81). To reduce the possibility that mismatches with the consensus risk-haplotype signature were simply the result of typing or phasing errors, we also tested the association of all HLA-Cw6 haplotypes with at least two mismatches for the 12 conserved markers of the candidate interval. This group of haplotypes shows strong LD with psoriasis (10:3; 76.9% 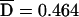 ), but the association is no longer significant (TDT P=.092; PDT P=.083) because of the reduction in sample size. In summary, the study of our families suggests that the PSORS1 risk allele is present on all HLA-Cw6 haplotypes.
), but the association is no longer significant (TDT P=.092; PDT P=.083) because of the reduction in sample size. In summary, the study of our families suggests that the PSORS1 risk allele is present on all HLA-Cw6 haplotypes.
The most common of the minor HLA-Cw6 haplotype clusters, cluster 40, is an HLA-Cw6-B45 haplotype that appears to have originated as a double recombinant of the 57.1 ancestral haplotype. Fine mapping of its breakpoints shows that it retains the centromeric and telomeric ends of the candidate interval, including the HLA-Cw6 and CDSN*TTC risk alleles, but it lacks the risk haplotype sequence for the intervening segment that includes PSORS1C3, OTF3, TCF19, HCR, SPR1, and part of SEEK1 (fig. 5). In our families, this haplotype appears to be associated with psoriasis (T:NT=7:2 and 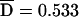 when cluster 40 is extended to include all haplotypes matching its consensus alleles within the 300-kb candidate interval), but we have no power to demonstrate significant association. This same haplotype, also known as “cluster D,” has been demonstrated to be overtransmitted to affected children in a study of European families15 and to be significantly associated with psoriasis in a case-control study of Gujarati Indians.47 If the HLA-Cw6-B45 haplotype does indeed bear PSORS1, then, in conjunction with our results, the candidate interval is further reduced to the 158-kb segment between HLA-B and PSORS1C3, which excludes all known genes except HLA-C and HCG27.
when cluster 40 is extended to include all haplotypes matching its consensus alleles within the 300-kb candidate interval), but we have no power to demonstrate significant association. This same haplotype, also known as “cluster D,” has been demonstrated to be overtransmitted to affected children in a study of European families15 and to be significantly associated with psoriasis in a case-control study of Gujarati Indians.47 If the HLA-Cw6-B45 haplotype does indeed bear PSORS1, then, in conjunction with our results, the candidate interval is further reduced to the 158-kb segment between HLA-B and PSORS1C3, which excludes all known genes except HLA-C and HCG27.
If HLA-C is indeed the PSORS1 gene, then, in the absence of allelic heterogeneity, no haplotype lacking HLA-Cw6 should be associated with psoriasis. This is exactly what is observed for haplotypes common enough to provide adequate power for association testing (table 11), even when excluding all families bearing any HLA-Cw6 haplotypes (data not shown).
Discussion
We used a combination of genomic DNA sequencing and haplotype analysis in an effort to complete the genetic dissection of PSORS1. Our principal findings are: (1) haplotypes that belong to the same cluster are extremely similar; (2) the PSORS1 gene must reside in a 297.8-kb interval defined by a region of unbroken sequence homology between the HLA-B57-Cw6 and HLA-B50-Cw6 haplotypes; (3) only 98 of 5,545 polymorphisms in this interval have alleles specific to risk haplotypes, but many combinations of alleles at the remaining polymorphisms can specifically tag the risk haplotype; (4) HLA-C and CDSN are the only genes in the candidate interval that encode protein variations unique to risk among the 10 sequenced haplotypes; (5) analysis of recombinants mapping between HLA-C and CDSN implicates HLA-Cw6 rather than CDSN*TTC as the likely disease allele, and further analysis of these recombinants allows us to exclude the telomeric quarter of the 297.8-kb risk interval with high confidence; and (6) in our families, all HLA-Cw6 haplotypes and only HLA-Cw6 haplotypes appear to carry risk for psoriasis.
To our knowledge, our study is the most extensive resequencing of the MHC class I region reported to date. We uncovered remarkable variation in nucleotide diversity among different haplotypes across the proximal class I region, particularly in the vicinity of HLA-B and HLA-C (fig. 2). Our results confirm and extend previous findings of high nucleotide diversity in this region.24,48 In contrast to sequences drawn from different haplotype clusters, sequences drawn from the same haplotype cluster are extremely similar (table 8), which increases confidence that a single sequence can adequately represent the entire cluster.
We defined a 297.8-kb risk interval on the basis of extended sequence similarity between the HLA-Cw6-B57 and HLA-Cw6-B50 risk haplotypes (fig. 2). That these haplotypes should resemble each other over such a long interval is not surprising, since they are clearly derived from a common ancestor who carried the risk determinant on extended MHC haplotype 57.1. However, the divergence between them in the risk interval (0.25 variations per kb) is much greater than the divergence of any two sequences drawn from the same haplotype cluster (0.0088–0.034 variations per kb) (table 8), which suggests that the PSORS1 determinant is relatively old.
We identified extensive stretches of low nucleotide divergence between the risk haplotypes and the nonrisk haplotypes HLA-Cw7-B8, HLA-Cw8-B65, and HLA-Cw12-B38 (fig. 2), in agreement with analysis of microsatellite haplotype clusters14 (table 11). Indeed, the similarity between HLA-Cw8-B65 and HLA-Cw6 risk haplotypes falsely reinforced our confidence that this haplotype was risk conferring.14 In retrospect, it is apparent that recurrence of long haplotype stretches is not a rare event. The long stretches of homologous haplotypes among sequenced chromosomes probably reflect the combined effects of natural selection on the MHC and a relatively low meiotic recombination rate across this region.12 Our haplotype analysis found a recombination fraction of θ=0.004 for a 1.2-Mb interval encompassing the proximal half of the MHC class I region, which is one-third the predicted genomewide rate.
Regions of sequence similarity between risk and nonrisk haplotypes allowed us to eliminate most individual sequence variants as PSORS1 candidates. Sequencing of other common nonrisk haplotypes (e.g., clusters 11, 68, 22, 57, 29, and 5 in table 11) could allow further reduction of the number of variants unique to risk haplotypes. However, the number of unique combinations of polymorphisms would still be enormous, which is why we restricted our combinatorial analysis to spliced transcripts of known genes as the most likely candidates.
As summarized in table 9, our analysis of PSORS1 candidate genes is the most comprehensive to date. We found that 7 of the 11 known genes in the risk interval—HCG27, PSORS1C3, OTF3, TCF19, HCR, STG, and HCG22—are unlikely candidates because, for each gene, the spliced transcript of at least one common nonrisk haplotype is identical in sequence to that of the risk haplotypes. We are the first to study the association of HCG22 and HCG27 with psoriasis. Others have found TCF19 and STG to be unassociated,49,50 and PSORS1C3 was found to be more weakly associated with psoriasis than was HLA-Cw*0602.51 The WWCC protein allele of HCR has been reported to be strongly associated with psoriasis,52–54 and HCR is expressed differently in psoriatic skin55 and in transgenic mice carrying the HCR*WWCC risk allele.56 However, none of these studies could differentiate the genetic association of HCR*WWCC with disease from that of HLA-Cw6, which are carried together on nearly all risk haplotypes in the sampled populations. Indeed, we57 and others58 found the association of HCR to be weaker than that of HLA-C, as would be predicted by our current sequence analysis. Although one study found OTF3 to be unassociated with psoriasis in a Chinese population,59 another study found that an allele of a HindIII polymorphism in OTF3 was more strongly associated with psoriasis in a Spanish population than was HLA-Cw*0602 itself.60 Our data definitely do not support this OTF3 allele as a PSORS1 candidate, because the HindIII risk allele (1321T) is found on both splice variants of at least three common nonrisk haplotypes. Furthermore, its location in the 3′ UTR of the gene lessens the likelihood that it has functional consequences.
SPR1 and SEEK1 have mRNA alleles unique to risk among the 10 haplotypes we sequenced, but these do not affect protein sequence, so there are no protein alleles for these genes unique to risk haplotypes. SPR1 was found to be associated with psoriasis in two other studies.61,62 The first study reported significant associations for two mRNA haplotypes carrying the same four pairwise combinations of alleles that we find to be unique to risk; the second study detected association even though analysis was restricted to single SNPs, none of which included the critical 3′ UTR 673G→C SNP common to all combinations that uniquely tag the risk haplotype in our analysis. Neither study could demonstrate an SPR1 association independent of association with HLA-Cw6. Conflicting results have been reported for SEEK1. A study of Chinese psoriatics63 could find no association with SEEK1 polymorphisms, considered either individually or as haplotypes, but a study of Swedish psoriatics62 did find a HLA-Cw*0602–independent association with two SNP alleles (−94A and −100C) that constitute one of the combinations we found to be unique among our sequenced haplotypes. However, the P values for HLA-Cw*0602–independent association of SEEK1 obtained in that study would not have been significant if corrected for multiple testing, and the positive association reported for SNP −100C in that study is actually for the allele not carried on HLA-Cw6 haplotypes and hence does not comprise the combination that we found to be unique to risk. Among our sequenced haplotypes, this allele is found only on those carrying HLA-Cw*1203, which are definitely nonrisk in our families. Moreover, these two SEEK1 polymorphisms lie outside the 224-kb reduced risk interval determined by recombinant haplotype mapping (fig. 5). On the basis of these considerations, we conclude that variation at SEEK1 does not adequately explain the risk of psoriasis conferred by PSORS1.
HLA-C and CDSN are the only candidates with predicted protein alleles unique to risk among the 10 sequenced haplotypes. We genotyped our full family sample for the risk alleles of these two genes that were identified by our sequence analysis and other studies. Of the 15 groups of HLA-C alleles distinguishable by our typing assay (table 10), 9 occur in our families, and only HLA-Cw6 is positively associated with psoriasis. Of the 12 alleles typed for CDSN, 3 are positively associated. Because we could not demonstrate significant differences in the degree of risk conferred by the three alleles, we lumped them into a single risk allele, CDSN*TTC, named for the smallest combination of polymorphism alleles distinguishing it from nonrisk variants. Association for both HLA-Cw6 and CDSN*TTC is very strong (>72% transmission; PDT 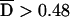 ) and highly significant (FBAT P<6×10-23) (table 10).
) and highly significant (FBAT P<6×10-23) (table 10).
CDSN is an excellent functional candidate for PSORS1, since it is a secreted protein that mediates homophilic adhesion between terminally differentiated keratinocytes. Its degradation by serine proteases results in their desquamation from the stratum corneum.64 Psoriasis is characterized by a markedly defective stratum corneum, with immature corneocytes and large increases in transepidermal water loss.65 Moreover, CDSN is expressed and secreted into the extracellular space in the upper spinous layers of lesional psoriatic skin, whereas this does not occur until keratinocytes reach the granular layer in normal and nonlesional psoriatic skin.66 Many groups have studied the association of CDSN with psoriasis. Several groups—three from Japan67–69 and one each from Spain,60 China,59 and Finland52—could find no association between CDSN and psoriasis. These negative studies, however, all suffer from one or more factors that reduced their power to detect association, including small sample size, lack of haplotyping, and incomplete description of allelic variation. In contrast, many other studies have detected positive association of psoriasis and CDSN. Many of these studies offer no compelling evidence that differentiates the association of psoriasis with CDSN from its association with risk alleles of other genes in strong LD with CDSN, especially HLA-Cw6 and HCR*WWCC.44,53,68–71 Others present evidence that the observed association between CDSN and psoriasis is at least partially independent of that seen for HLA-C.46,72–76 Yet others argue that both HLA-C and CDSN are necessary for disease.15,47 Finally, a recently published study77 localized PSORS1 to a haplotype block containing HLA-C but not HCR or CDSN.
In light of the conflicting evidence for the role of CDSN versus HLA-C in psoriasis, we designed a study that addresses many of the shortcomings of previous work. We recruited a sample larger than any published to date and used families rather than cases and controls, to guard against spurious associations and to allow for more accurate haplotype reconstruction. We required that the proband of each family have early-onset (type I) psoriasis, which increases the probability of MHC involvement.10 We rigorously typed HLA-C, using a novel approach that distinguishes HLA-Cw6 alleles from all other 186 known alleles of the gene, even in the absence of external phase information from a family or population. In addition to the three SNPs constituting the CDSN*TTC variant, we typed CDSN for four other protein-altering polymorphisms involved in combinations distinguishing the CDSN of risk haplotypes among our 10 sequenced chromosomes, as well as for two variants causing CDSN protein changes among different HLA-Cw6–bearing haplotypes. Finally, we haplotyped CDSN and HLA-C with 14 intervening and 20 flanking microsatellite markers, greatly reducing the chances of identifying false recombinants that are simply the result of typing or phasing errors.
Our single-gene association analysis favors HLA-Cw6 over CDSN*TTC because of its somewhat higher levels of LD with psoriasis (table 10), but the difference is not statistically significant. Among haplotype clusters common enough to have power for association testing, the four that are positively associated with psoriasis carry the risk alleles for both genes (table 11). We then identified recombinants between the two risk alleles, which are infrequent (2.6% haplotype frequency) in our families and hence require a large sample for meaningful analysis. Using simulation methods to establish an appropriate correction for multiple testing, we are able to exclude CDSN*TTC as the PSORS1 risk allele with high confidence (table 12). By grouping haplotypes on the basis of their breakpoints, we are also able to exclude the telomeric 25% of the risk interval, extending from M6S190 to M6S224 (fig. 5).
We restricted our analysis of candidate genes to those known to be translated or transcribed. The original 300-kb candidate interval also contains four pseudogenes (WASF5P, RPL32P, USP8P, and POLR2LP) and one computer-predicted locus (LOC442199). Of these, only WASF5P and USP8P have sequences unique to the risk among the 10 sequenced haplotypes (data not shown). Although both of these pseudogenes map within 20 kb of HLA-C, their lack of expression makes them poor candidates for PSORS1. We can also exclude a transcribed fragment of an endogenous retroviral dUTPase advanced elsewhere as a candidate for PSORS178 (data not shown).
All demonstrable risk haplotypes in our families carry HLA-Cw6, and all HLA-Cw6 haplotypes, even the rare ones when considered in aggregate, appear to impart risk for psoriasis. Conversely, all demonstrable nonrisk haplotypes lack HLA-Cw6. The same cannot be said for any protein allele of any other gene in the candidate region or for any spliced mRNA allele of any gene other than SPR1 and SEEK1. Because PSORS1 probably lies within the 158-kb interval of conserved risk sequence between HLA-B and PSORS1C3 that is common to our reduced candidate interval and the HLA-Cw6-B45 risk haplotype (fig. 5), we can also exclude the risk-associated mRNA alleles of SPR1 and SEEK1 as candidates. Given these results, HLA-Cw6 is clearly the best candidate for PSORS1. Nevertheless, we cannot rule out intronic variations in known genes of the reduced candidate interval that influence transcription, mRNA processing, or long-range chromatin structure. We also cannot rule out regulatory variations flanking known genes or undiscovered genes or regulatory RNAs in this remaining interval. Ongoing international collaborative studies involving thousands of additional subjects should allow these remaining questions to be addressed through further detailed analysis of informative recombinants.
Although Orrù et al. identified the HLA-Cw7-B58 haplotype as risk-conferring,46 and this haplotype does carry a segment homologous to the ancestral HLA-Cw6-B57 risk haplotype (fig. 5), the few individuals carrying this haplotype in our sample provide no support for this conclusion (T:NT=4:7). Moreover, figure 5 demonstrates that the region of overlap between this haplotype and the reduced risk interval established by the present study is not carried by the HLA-Cw6-B45–bearing “double recombinant” putative risk haplotype also known as “cluster D.”15,47 Interestingly, an allele of the MICA gene that is in strong LD with the HLA-Cw7-B58 haplotype79 has recently been associated with psoriatic arthritis.80 Consistent with this finding, 60% (6 of 10) of the evaluable psoriatics in our sample who carried HLA-Cw7-B58 had joint complaints, as opposed to 28.5% (285 of 1,001) of evaluable psoriatics not carrying this haplotype. Orrù et al.46 did not mention how many of the psoriatics in their sample were affected with arthritis. Clearly, it will be important to stratify any future analysis of this haplotype for the presence or absence of joint disease.
Our studies do not exclude the possibility that allelic heterogeneity in psoriasis involves the closely related HLA-B and HLA-C genes. Indeed, the HLA-Cw1-B46 haplotype, which is essentially specific to East Asian populations81 and is not present in our sample, has been associated with psoriasis in several studies.82–86 Also, in addition to HLA-Cw6, HLA-B38 and HLA-B39 have been associated with psoriatic arthritis in multiple studies.87–91 However, further studies will be required to determine whether these additional disease-associated HLA-B and/or HLA-C alleles play causal roles or reflect LD with a nearby causal gene.
Our genetic findings in support of HLA-Cw6 as a major disease allele at PSORS1 are congruent with current concepts of psoriasis immunopathogenesis. Whereas the most prominent clinical features of psoriasis are epidermal hyperplasia and increased cutaneous blood flow, multiple lines of evidence indicate that these changes are triggered and maintained by infiltrating immunocytes. Psoriasis has been triggered or cured by bone-marrow transplantation, depending on whether the donor or the host was psoriatic, and T cell–specific immunosuppressants such as cyclosporine A and FK506 exert dramatic therapeutic effects.4,92 Studies of T cell receptor (TCR) rearrangements have documented the presence of oligoclonal T cell expansions in multiple psoriatic lesions of the same subject, which persist after improvement followed by relapse (references in the work of Lin et al.93). These oligoclonal proliferations involve CD8+ as well as CD4+ T cells.94 HLA-Cw6 seems well suited for a role in this process, since HLA-C presents peptide antigens to CD8+ T cells, and CD8+ T cells comprise at least 80% of the T cells in lesional psoriatic epidermis.5 HLA-C also regulates the activity of natural killer cells by interacting with killer immunoglobulinlike receptors (KIR).95 Interestingly, variation in KIR genes has been associated with psoriasis96,97 and psoriatic arthritis.98,99
In aggregate, these data strongly suggest that HLA-Cw6 is the major disease allele at the PSORS1 locus that confers early-onset disease in white populations. Future genetic studies of this locus should focus on large collaborative collections of subjects so that more critical recombinants can be identified to further reduce the PSORS1 candidate interval. The polymorphisms we have identified here will be very useful for this task. Also, additional sequenced haplotypes can be applied to candidate-locus analysis, further decreasing the number of polymorphisms and combinations of polymorphisms unique to risk. Now that the extreme similarity of unrelated examples of the same haplotype has been demonstrated (table 8) and the risk status of most common HLA-C-B haplotypes is known (table 11), these sequences no longer need to be cloned in an allele-specific fashion from psoriasis-affected families as we have done but can be derived from homozygous HLA typing cell lines identified on the basis of their HLA-C-B haplotypes.24 Future functional studies of PSORS1 should be directed toward elucidating the role of HLA-Cw6 in the immunopathogenesis of psoriasis.
Acknowledgments
The authors thank all the patients with psoriasis and family members who volunteered to participate in this study. This work was supported by the National Institute of Arthritis, Musculoskeletal, and Skin Diseases, National Institutes of Health grants R01 AR042742 and R01 AR050511; by the Ann Arbor Veterans Affairs Hospital; by the Dudley and Dawn Holmes Fund; by the Babcock Memorial Trust; by the National Psoriasis Foundation, by grant M01 RR00042 from the National Center for Research Resources, National Institutes of Health, to the University of Michigan General Clinical Research Center; and by German National Genome Research Network grant BMFT 01GS 0171/BMBF NUW-S23T10.
Web Resources
The URLs for data presented herein are as follows:
- Anthony Nolan Trust, http://www.anthonynolan.org.uk/HIG/ (for the IMGT/HLA sequence database)
- BLAST 2 Sequences, http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- GenBank, http://www.ncbi.nih.gov/Genbank/ (see tables and for accession numbers)
- Entrez Gene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
- Harvard School of Public Health, http://www.biostat.harvard.edu/~fbat/ (for FBAT software)
- HGNC, http://www.gene.ucl.ac.uk/nomenclature/
- Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/
- Merlin software, http://www.sph.umich.edu/csg/abecasis/Merlin/
- MHC Haplotype Project, http://www.sanger.ac.uk/HGP/Chr6/MHC/ (for Sanger Institute sequences of HLA-homozygous MHC haplotypes)
- NCBI Human Genome Assembly Release Notes, http://www.ncbi.nlm.nih.gov/genome/guide/human/release_notes.html#b34
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PSORS1) [PubMed]
- Pedmanager, http://www.broad.mit.edu/ftp/distribution/software/pedmanager/
- Stanford Center for Tuberculosis Research, http://www.stanford.edu/group/molepi/free_software.html (for MINCOV software)
- Unigene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene
- University of Michigan Psoriasis Genetics Laboratory, http://www.psoriasis.umich.edu/
References
- 1.Stern RS, Nijsten T, Feldman SR, Margolis DJ, Rolstad T (2004) Psoriasis is common, carries a substantial burden even when not extensive, and is associated with widespread treatment dissatisfaction. J Investig Dermatol Symp Proc 9:136–139 10.1046/j.1087-0024.2003.09102.x [DOI] [PubMed] [Google Scholar]
- 2.Gupta MA, Schork NJ, Gupta AK, Kirkby S, Ellis CN (1993) Suicidal ideation in psoriasis. Int J Dermatol 32:188–190 [DOI] [PubMed] [Google Scholar]
- 3.Gladman DD (1994) Natural history of psoriatic arthritis. Baillieres Clin Rheumatol 8:379–394 10.1016/S0950-3579(94)80024-3 [DOI] [PubMed] [Google Scholar]
- 4.Schon MP, Boehncke WH (2005) Psoriasis. N Engl J Med 352:1899–1912 10.1056/NEJMra041320 [DOI] [PubMed] [Google Scholar]
- 5.Gudjonsson JE, Johnston A, Sigmundsdottir H, Valdimarsson H (2004) Immunopathogenic mechanisms in psoriasis. Clin Exp Immunol 135:1–8 10.1111/j.1365-2249.2004.02310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elder JT, Nair RP, Guo SW, Henseler T, Christophers E, Voorhees JJ (1994) The genetics of psoriasis. Arch Dermatol 130:216–224 10.1001/archderm.130.2.216 [DOI] [PubMed] [Google Scholar]
- 7.Capon F, Trembath RC, Barker JN (2004) An update on the genetics of psoriasis. Dermatol Clin 22:339–347 10.1016/S0733-8635(03)00125-6 [DOI] [PubMed] [Google Scholar]
- 8.Sagoo GS, Tazi-Ahnini R, Barker JW, Elder JT, Nair RP, Samuelsson L, Traupe H, Trembath RC, Robinson DA, Iles MM (2004) Meta-analysis of genome-wide studies of psoriasis susceptibility reveals linkage to chromosomes 6p21 and 4q28-q31 in Caucasian and Chinese Hans population. J Invest Dermatol 122:1401–1405 10.1111/j.0022-202X.2004.22607.x [DOI] [PubMed] [Google Scholar]
- 9.Russell TJ, Schultes LM, Kuban DJ (1972) Histocompatibility (HL-A) antigens associated with psoriasis. N Engl J Med 287:738–740 [DOI] [PubMed] [Google Scholar]
- 10.Henseler T, Christophers E (1985) Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol 13:450–456 [DOI] [PubMed] [Google Scholar]
- 11.Mallon E, Newson R, Bunker CB (1999) HLA-Cw6 and the genetic predisposition to psoriasis: a meta-analysis of published serologic studies. J Invest Dermatol 113:693–695 10.1046/j.1523-1747.1999.00724.x [DOI] [PubMed] [Google Scholar]
- 12.Walsh EC, Mather KA, Schaffner SF, Farwell L, Daly MJ, Patterson N, Cullen M, Carrington M, Bugawan TL, Erlich H, Campbell J, Barrett J, Miller K, Thomson G, Lander ES, Rioux JD (2003) An integrated haplotype map of the human major histocompatibility complex. Am J Hum Genet 73:580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S (2004) Gene map of the extended human MHC. Nat Rev Genet 5:889–899 10.1038/nrg1489 [DOI] [PubMed] [Google Scholar]
- 14.Nair RP, Stuart P, Henseler T, Jenisch S, Chia NVC, Westphal E, Schork NJ, Kim J, Lim HW, Christophers E, Voorhees JJ, Elder JT (2000) Localization of psoriasis-susceptibility locus PSORS1 to a 60-kb interval telomeric to HLA-C. Am J Hum Genet 66:1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veal CD, Capon F, Allen MH, Heath EK, Evans JC, Jones A, Patel S, Burden D, Tillman D, Barker JNWN, Trembath RC (2002) Family-based analysis using a dense single-nucleotide polymorphism–based map defines genetic variation at PSORS1, the major psoriasis-susceptibility locus. Am J Hum Genet 71:554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, Cattley S, Martinez P, Kulski J (1999) Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev 167:275–304 [DOI] [PubMed] [Google Scholar]
- 17.Cluster 17 Collaboration (2005) Fine mapping of the psoriasis susceptibility gene PSORS1: a reassessment of risk associated with a putative risk haplotype lacking HLA-Cw6. J Invest Dermatol 124:921–930 10.1111/j.0022-202X.2005.23729.x [DOI] [PubMed] [Google Scholar]
- 18.Nair RP, Henseler T, Jenisch S, Stuart P, Bichakjian CK, Lenk W, Westphal E, Guo SW, Christophers E, Voorhees JJ, Elder JT (1997) Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet 6:1349–1356 10.1093/hmg/6.8.1349 [DOI] [PubMed] [Google Scholar]
- 19.Jenisch S, Henseler T, Nair RP, Guo S-W, Westphal E, Stuart P, Krönke M, Voorhees JJ, Christophers E, Elder JT (1998) Linkage analysis of Human Leukocyte Antigen (HLA) markers in familial psoriasis: strong disequilibrium effects provide evidence for a major determinant in the HLA-B/ -C region. Am J Hum Genet 63:191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 31:311–314 10.1093/nar/gkg070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair R, Guo S, Jenisch S, Henseler T, Lange E, Terhune M, Westphal E, Christophers E, Voorhees J, Elder J (1995) Scanning chromosome 17 for psoriasis susceptibility: lack of evidence for a distal 17q locus. Hum Hered 45:219–230 [DOI] [PubMed] [Google Scholar]
- 22.Wigginton JE, Abecasis GR (2005) PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics 21:3445–3447 10.1093/bioinformatics/bti529 [DOI] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 24.Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, et al (2004) Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 14:1176–1187 10.1101/gr.2188104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431:931–945 10.1038/nature03001 [DOI] [PubMed] [Google Scholar]
- 26.Salamon H, Tarhio J, Ronningen K, Thomson G (1996) On distinguishing unique combinations in biological sequences. J Comput Biol 3:407–423 [DOI] [PubMed] [Google Scholar]
- 27.Abecasis GR, Wigginton JE (2005) Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet 77:754–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuart P, Nair RP, Abecasis GR, Nistor I, Hiremagalore R, Chia NV, Qin ZS, Thompson RA, Jenisch S, Weichenthal M, Janiga J, Lim HW, Christophers E, Voorhees JJ, Elder JT (2006) Analysis of RUNX1 binding site and RAPTOR polymorphisms in psoriasis: no evidence for association despite adequate power and evidence for linkage. J Med Genet 43:12–17 10.1136/jmg.2005.032193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabinowitz D, Laird N (2000) A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered 50:211–223 10.1159/000022918 [DOI] [PubMed] [Google Scholar]
- 32.Horvath S, Xu X, Laird NM (2001) The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet 9:301–306 10.1038/sj.ejhg.5200625 [DOI] [PubMed] [Google Scholar]
- 33.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM (2004) Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol 26:61–69 10.1002/gepi.10295 [DOI] [PubMed] [Google Scholar]
- 34.Spielman RS, Ewens WJ (1999) TDT clarification. Am J Hum Genet 64:668–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knapp M (1999) The transmission/disequilibrium test and parental-genotype reconstruction: the reconstruction-combined transmission/disequilibrium test. Am J Hum Genet 64:861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dudbridge F, Koeleman BPC, Todd JA, Clayton DG (2000) Unbiased application of the transmission/disequilibrium test to multilocus haplotypes. Am J Hum Genet 66:2009–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- 38.Martin ER, Bass MP, Kaplan NL (2001) Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet 68:1065–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis D, Sham PC (1995) A note on the application of the transmission disequilibrium test when a parent is missing. Am J Hum Genet 56:811–812 [PMC free article] [PubMed] [Google Scholar]
- 41.Laird NM, Horvath S, Xu X (2000) Implementing a unified approach to family-based tests of association. Genet Epidemiol Suppl 19:S36–S42 [DOI] [PubMed] [Google Scholar]
- 42.Manly BFJ, McAlevey L, Stevens D (1986) A randomization procedure for comparing group means on multiple measurements. Br J Math Stat Psychol 39:183–189 [Google Scholar]
- 43.Antonarakis SE, Nomenclature Working Group (1998) Recommendations for a nomenclature system for human gene mutations. Hum Mutat 11:1–3 [DOI] [PubMed] [Google Scholar]
- 44.Jenisch S, Koch S, Henseler T, Nair RP, Elder JT, Watts CE, Westphal E, Voorhees JJ, Christophers E, Kronke M (1999) Corneodesmosin gene polymorphism demonstrates strong linkage disequilibrium with HLA and association with psoriasis vulgaris. Tissue Antigens 54:439–449 10.1034/j.1399-0039.1999.540501.x [DOI] [PubMed] [Google Scholar]
- 45.Guerrin M, Vincent C, Simon M, Tazi Ahnini R, Fort M, Serre G (2001) Identification of six novel polymorphisms in the human corneodesmosin gene. Tissue Antigens 57:32–38 10.1034/j.1399-0039.2001.057001032.x [DOI] [PubMed] [Google Scholar]
- 46.Orrù S, Giuressi E, Carcassi C, Casula M, Contu L (2005) Mapping of the major psoriasis-susceptibility locus (PSORS1) in a 70-Kb interval around the corneodesmosin gene (CDSN). Am J Hum Genet 76:164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capon F, Toal IK, Evans JC, Allen MH, Patel S, Tillman D, Burden D, Barker JN, Trembath RC (2003) Haplotype analysis of distantly related populations implicates corneodesmosin in psoriasis susceptibility. J Med Genet 40:447–452 10.1136/jmg.40.6.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaudieri S, Dawkins RL, Habara K, Kulski JK, Gojobori T (2000) SNP profile within the human major histocompatibility complex reveals an extreme and interrupted level of nucleotide diversity. Genome Res 10:1579–1586 10.1101/gr.127200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teraoka Y, Naruse TK, Oka A, Matsuzawa Y, Shiina T, Iizuka M, Iwashita K, Ozawa A, Inoko H (2000) Genetic polymorphisms in the cell growth regulated gene, SC1 telomeric of the HLA-C gene and lack of association of psoriasis vulgaris. Tissue Antigens 55:206–211 10.1034/j.1399-0039.2000.550303.x [DOI] [PubMed] [Google Scholar]
- 50.Sanchez F, Holm SJ, Mallbris L, O’Brien KP, Stahle M (2004) STG does not associate with psoriasis in the Swedish population. Exp Dermatol 13:413–418 10.1111/j.0906-6705.2004.00170.x [DOI] [PubMed] [Google Scholar]
- 51.Holm SJ, Sanchez F, Carlen LM, Mallbris L, Stahle M, O’Brien KP (2005) HLA-Cw*0602 associates more strongly to psoriasis in the Swedish population than variants of the novel 6p21.3 gene PSORS1C3. Acta Derm Venereol 85:2–8 10.1080/00015550410023527 [DOI] [PubMed] [Google Scholar]
- 52.Asumalahti K, Laitinen T, Itkonen-Vatjus R, Lokki M, Suomela S, Snellman E, Saarialho-Kere U, Kere J (2000) A candidate gene for psoriasis near HLA-C, HCR (Pg8) is highly polymorphic with a disease-associated susceptibility allele. Hum Mol Genet 9:1533–1542 10.1093/hmg/9.10.1533 [DOI] [PubMed] [Google Scholar]
- 53.Asumalahti K, Veal C, Laitinen T, Suomela S, Allen M, Elomaa O, Moser M, de Cid R, Ripatti S, Vorechovsky I, Marcusson JA, Nakagawa H, Lazaro C, Estivill X, Capon F, Novelli G, Saarialho-Kere U, Barker J, Trembath R, Kere J (2002) Coding haplotype analysis supports HCR as the putative susceptibility gene for psoriasis at the MHC PSORS1 locus. Hum Mol Genet 11:589–597 10.1093/hmg/11.5.589 [DOI] [PubMed] [Google Scholar]
- 54.Chang YT, Shiao YM, Chin PJ, Liu YL, Chou FC, Wu S, Lin YF, Li LH, Lin MW, Liu HN, Tsai SF (2004) Genetic polymorphisms of the HCR gene and a genomic segment in close proximity to HLA-C are associated with patients with psoriasis in Taiwan. Br J Dermatol 150:1104–1111 10.1111/j.1365-2133.2004.05972.x [DOI] [PubMed] [Google Scholar]
- 55.Suomela S, Elomaa O, Asumalahti K, Kariniemi AL, Karvonen SL, Peltonen J, Kere J, Saarialho-Kere U (2003) HCR, a candidate gene for psoriasis, is expressed differently in psoriasis and other hyperproliferative skin disorders and is downregulated by interferon-γ in keratinocytes. J Invest Dermatol 121:1360–1364 10.1046/j.1523-1747.2003.12642.x [DOI] [PubMed] [Google Scholar]
- 56.Elomaa O, Majuri I, Suomela S, Asumalahti K, Jiao H, Mirzaei Z, Rozell B, Dahlman-Wright K, Pispa J, Kere J, Saarialho-Kere U (2004) Transgenic mouse models support HCR as an effector gene in the PSORS1 locus. Hum Mol Genet 13:1551–1561 10.1093/hmg/ddh178 [DOI] [PubMed] [Google Scholar]
- 57.Chia NV, Stuart P, Nair RP, Henseler T, Jenisch S, Lim HW, Christophers E, Voorhees JJ, Elder JT (2001) Variations in the HCR (Pg8) gene are unlikely to be causal for familial psoriasis. J Invest Dermatol 116:823–824 10.1046/j.1523-1747.2001.01245-2.x [DOI] [PubMed] [Google Scholar]
- 58.O’Brien KP, Holm SJ, Nilsson S, Carlen L, Rosenmuller T, Enerback C, Inerot A, Stahle-Backdahl M (2001) The HCR gene on 6p21 is unlikely to be a psoriasis susceptibility gene. J Invest Dermatol 116:750–754 10.1046/j.0022-202x.2001.01323.x [DOI] [PubMed] [Google Scholar]
- 59.Chang YT, Tsai SF, Lee DD, Shiao YM, Huang CY, Liu HN, Wang WJ, Wong CK (2003) A study of candidate genes for psoriasis near HLA-C in Chinese patients with psoriasis. Br J Dermatol 148:418–423 10.1046/j.1365-2133.2003.05166.x [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez S, Martinez-Borra J, Del Rio JS, Santos-Juanes J, Lopez-Vazquez A, Blanco-Gelaz M, Lopez-Larrea C (2000) The OTF3 gene polymorphism confers susceptibility to psoriasis independent of the association of HLA-Cw*0602. J Invest Dermatol 115:824–828 10.1046/j.1523-1747.2000.00133.x [DOI] [PubMed] [Google Scholar]
- 61.Chang YT, Tsai SF, Lin MW, Liu HN, Lee DD, Shiao YM, Chin PJ, Wang WJ (2003) SPR1 gene near HLA-C is unlikely to be a psoriasis susceptibility gene. Exp Dermatol 12:307–314 10.1034/j.1600-0625.2003.00039.x [DOI] [PubMed] [Google Scholar]
- 62.Holm SJ, Carlen LM, Mallbris L, Stahle-Backdahl M, O’Brien KP (2003) Polymorphisms in the SEEK1 and SPR1 genes on 6p21.3 associate with psoriasis in the Swedish population. Exp Dermatol 12:435–444 10.1034/j.1600-0625.2003.00048.x [DOI] [PubMed] [Google Scholar]
- 63.Chang YT, Liu HN, Shiao YM, Lin MW, Lee DD, Liu MT, Wang WJ, Wu S, Lai CY, Tsai SF (2005) A study of PSORS1C1 gene polymorphisms in Chinese patients with psoriasis. Br J Dermatol 153:90–96 10.1111/j.1365-2133.2005.06570.x [DOI] [PubMed] [Google Scholar]
- 64.Caubet C, Jonca N, Brattsand M, Guerrin M, Bernard D, Schmidt R, Egelrud T, Simon M, Serre G (2004) Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J Invest Dermatol 122:1235–1244 10.1111/j.0022-202X.2004.22512.x [DOI] [PubMed] [Google Scholar]
- 65.Goon AT, Yosipovitch G, Chan YH, Goh CL (2004) Barrier repair in chronic plaque-type psoriasis. Skin Res Technol 10:10–13 10.1111/j.1600-0846.2004.00046.x [DOI] [PubMed] [Google Scholar]
- 66.Allen M, Ishida-Yamamoto A, McGrath J, Davison S, Iizuka H, Simon M, Guerrin M, Hayday A, Vaughan R, Serre G, Trembath R, Barker J (2001) Corneodesmosin expression in psoriasis vulgaris differs from normal skin and other inflammatory skin disorders. Lab Invest 81:969–976 [DOI] [PubMed] [Google Scholar]
- 67.Ishihara M, Yamagata N, Ohno S, Naruse T, Ando A, Kawata H, Ozawa A, Ohkido M, Mizuki N, Shiina T, Ando H, Inoko H (1996) Genetic polymorphisms in the keratin-like S gene within the human major histocompatibility complex and association analysis on the susceptibility to psoriasis vulgaris. Tissue Antigens 48:182–186 [DOI] [PubMed] [Google Scholar]
- 68.Ameen M, Allen MH, Fisher SA, Lewis CM, Cuthbert A, Kondeatis E, Vaughan RW, Murakami H, Nakagawa H, Barker JN (2005) Corneodesmosin (CDSN) gene association with psoriasis vulgaris in Caucasian but not in Japanese populations. Clin Exp Dermatol 30:414–418 10.1111/j.1365-2230.2005.01789.x [DOI] [PubMed] [Google Scholar]
- 69.Hui J, Oka A, Tamiya G, Tomizawa M, Kulski JK, Penhale WJ, Tay GK, Iizuka M, Ozawa A, Inoko H (2002) Corneodesmosin DNA polymorphisms in MHC haplotypes and Japanese patients with psoriasis. Tissue Antigens 60:77–83 10.1034/j.1399-0039.2002.600110.x [DOI] [PubMed] [Google Scholar]
- 70.Enerback C, Enlund F, Inerot A, Samuelsson L, Wahlstrom J, Swanbeck G, Martinsson T (2000) S gene (corneodesmosin) diversity and its relationship to psoriasis; high content of cSNP in the HLA-linked S gene. J Invest Dermatol 114:1158–1163 10.1046/j.1523-1747.2000.00002.x [DOI] [PubMed] [Google Scholar]
- 71.Romphruk AV, Oka A, Romphruk A, Tomizawa M, Choonhakarn C, Naruse TK, Puapairoj C, Tamiya G, Leelayuwat C, Inoko H (2003) Corneodesmosin gene: no evidence for PSORS 1 gene in North-eastern Thai psoriasis patients. Tissue Antigens 62:217–224 10.1034/j.1399-0039.2003.00056.x [DOI] [PubMed] [Google Scholar]
- 72.Tazi-Ahnini R, Camp NJ, Cork MJ, Mee JB, Keohane SG, Duff GW, di Giovine FS (1999) Novel genetic association between the corneodesmosin (MHC S) gene and susceptibility to psoriasis. Hum Mol Genet 8:1135–1140 10.1093/hmg/8.6.1135 [DOI] [PubMed] [Google Scholar]
- 73.Tazi-Ahnini R, di Giovine FS, Cox A, Keohane SG, Cork MJ (1999) Corneodesmosin (MHC S) gene in guttate psoriasis. Lancet 354:597 10.1016/S0140-6736(05)77950-2 [DOI] [PubMed] [Google Scholar]
- 74.Allen MH, Veal C, Faassen A, Powis SH, Vaughan RW, Trembath RC, Barker JN (1999) A non-HLA gene within the MHC in psoriasis. Lancet 353:1589–1590 10.1016/S0140-6736(99)01618-9 [DOI] [PubMed] [Google Scholar]
- 75.Schmitt-Egenolf M, Windemuth C, Hennies HC, Albis-Camps M, von Engelhardt B, Wienker T, Reis A, Traupe H, Blasczyk R (2001) Comparative association analysis reveals that corneodesmosin is more closely associated with psoriasis than HLA-Cw*0602-B*5701 in German families. Tissue Antigens 57:440–446 10.1034/j.1399-0039.2001.057005440.x [DOI] [PubMed] [Google Scholar]
- 76.Orrù S, Giuressi E, Casula M, Loizedda A, Murru R, Mulargia M, Masala MV, Cerimele D, Zucca M, Aste N, Biggio P, Carcassi C, Contu L (2002) Psoriasis is associated with a SNP haplotype of the corneodesmosin gene (CDSN). Tissue Antigens 60:292–298 10.1034/j.1399-0039.2002.600403.x [DOI] [PubMed] [Google Scholar]
- 77.Helms C, Saccone NL, Cao L, Daw JA, Cao K, Hsu TM, Taillon-Miller P, Duan S, Gordon D, Pierce B, Ott J, Rice J, Fernandez-Vina MA, Kwok PY, Menter A, Bowcock AM (2005) Localization of PSORS1 to a haplotype block harboring HLA-C and distinct from corneodesmosin and HCR. Hum Genet 118:466–476 10.1007/s00439-005-0048-2 [DOI] [PubMed] [Google Scholar]
- 78.Foerster J, Nolte I, Junge J, Bruinenberg M, Schweiger S, Spaar K, van der Steege G, Ehlert C, Mulder M, Kalscheuer V, Blumenthal-Barby E, Winter J, Seeman P, Stander M, Sterry W, Te Meerman G (2005) Haplotype sharing analysis identifies a retroviral dUTPase as candidate susceptibility gene for psoriasis. J Invest Dermatol 124:99–102 10.1111/j.0022-202X.2004.23504.x [DOI] [PubMed] [Google Scholar]
- 79.Fischer G, Arguello JR, Perez-Rodriguez M, McWhinnie A, Marsh SGE, Travers PJ, Madrigal JA (2000) Sequence-specific oligonucleotide probing for MICB alleles reveals associations with MICA and HLA-B. Immunogenetics 51:591–599 10.1007/s002510000179 [DOI] [PubMed] [Google Scholar]
- 80.Korendowych E, Ravindran J, Owen PA, Carmichael CR, McHugh NJ, Dawkins RL (2006) Disease-specific alleles of the MHC class I related gene, MICA, are associated with type I psoriasis and psoriatic arthritis. Br J Dermatol 154:154 10.1111/j.1365-2133.2005.06957.x [DOI] [PubMed] [Google Scholar]
- 81.Barber LD, Percival L, Valiante NM, Chen L, Lee C, Gumperz JE, Phillips JH, Lanier LL, Bigge JC, Parekh RB, Parham P (1996) The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med 184:735–740 10.1084/jem.184.2.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vejbaesya S, Eiermann TH, Suthipinititharm P, Bancha C, Stephens HA, Luangtrakool K, Chandanayingyong D, Choonhakarn C, Romphruk A, Puapairoj C, Jirarattanapochai K, Leelayuwat C (1998) Serological and molecular analysis of HLA class I and II alleles in Thai patients with psoriasis vulgaris. Tissue Antigens 52:389–392 [DOI] [PubMed] [Google Scholar]
- 83.Choonhakarn C, Romphruk A, Puapairoj C, Jirarattanapochai K, Leelayuwat C (2002) Haplotype associations of the major histocompatibility complex with psoriasis in Northeastern Thais. Int J Dermatol 41:330–334 10.1046/j.1365-4362.2002.01496.x [DOI] [PubMed] [Google Scholar]
- 84.Nakagawa H, Asahina A, Akazaki S, Tokunaga K, Matsuki K, Ishibashi Y, Juji T (1990) Association of Cw11 in Japanese patients with psoriasis vulgaris. Tissue Antigens 36:241–242 [DOI] [PubMed] [Google Scholar]
- 85.Koizumi H, Fukaya T, Tsukinaga I, Ohkawara A, Wakisaka A, Aizawa M (1988) HLA antigens in psoriasis vulgaris. Acta Dermatol Kyoto 83:483–488 [Google Scholar]
- 86.Ozawa A, Miyahara M, Sugai J, Iizuka M, Kawakubo Y, Matsuo I, Ohkido M, Naruse T, Ando H, Inoko H, Kobayashi H, Ohkawara A, Takahashi H, Iizuka H, Morita E, Yamamoto S, Hide M, Taniguchi Y, Shimizu M (1998) HLA class I and II alleles and susceptibility to generalized pustular psoriasis: significant associations with HLA-Cw1 and HLA-DQB1*0303. J Dermatol 25:573–581 [DOI] [PubMed] [Google Scholar]
- 87.Gladman DD, Anhorn KA, Schachter RK, Mervart H (1986) HLA antigens in psoriatic arthritis. J Rheumatol 13:586–592 [PubMed] [Google Scholar]
- 88.Murray C, Mann DL, Gerber LN, Barth W, Perlmann S, Decker JL, Nigra TP (1980) Histocompatibility alloantigens in psoriasis and psoriatic arthritis: evidence for the influence of multiple genes in the major histocompatibility complex. J Clin Invest 66:670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crivellato E, Zacchi T (1987) HLA-B39 and the axial type of psoriatic arthritis. Acta Derm Venereol 67:249–250 [PubMed] [Google Scholar]
- 90.Espinoza LR, Vasey FB, Gaylord SW, Dietz C, Bergen L, Bridgeford P, Germain BF (1982) Histocompatibility typing in the seronegative spondyloarthropathies: a survey. Semin Arthritis Rheum 11:375–381 10.1016/0049-0172(82)90058-0 [DOI] [PubMed] [Google Scholar]
- 91.Trabace S, Cappellacci S, Ciccarone P, Liaskos S, Polito R, Zorzin L (1994) Psoriatic arthritis: a clinical, radiological and genetic study of 58 Italian patients. Acta Derm Venereol Suppl (Stockh) 186:69–70 [PubMed] [Google Scholar]
- 92.Krueger JG, Bowcock A (2005) Psoriasis pathophysiology: current concepts of pathogenesis. Ann Rheum Dis Suppl 2 64:ii30–ii36 10.1136/ard.2004.031120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin WJ, Norris DA, Achziger M, Kotzin BL, Tomkinson B (2001) Oligoclonal expansion of intraepidermal T cells in psoriasis skin lesions. J Invest Dermatol 117:1546–1553 10.1046/j.0022-202x.2001.01548.x [DOI] [PubMed] [Google Scholar]
- 94.Chang JC, Smith LR, Froning KJ, Schwabe BJ, Laxer JA, Caralli LL, Kurland HH, Karasek MA, Wilkinson DI, Carlo DJ, Brostoff SW (1994) CD8+ T cells in psoriatic lesions preferentially use T-cell receptor Vβ3 and/or Vβ13.1 genes. Proc Natl Acad Sci USA 91:9282–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parham P (2005) MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5:201–214 10.1038/nri1570 [DOI] [PubMed] [Google Scholar]
- 96.Suzuki Y, Hamamoto Y, Ogasawara Y, Ishikawa K, Yoshikawa Y, Sasazuki T, Muto M (2004) Genetic polymorphisms of killer cell immunoglobulin-like receptors are associated with susceptibility to psoriasis vulgaris. J Invest Dermatol 122:1133–1136 10.1111/j.0022-202X.2004.22517.x [DOI] [PubMed] [Google Scholar]
- 97.Luszczek W, Manczak M, Cislo M, Nockowski P, Wisniewski A, Jasek M, Kusnierczyk P (2004) Gene for the activating natural killer cell receptor, KIR2DS1, is associated with susceptibility to psoriasis vulgaris. Hum Immunol 65:758–766 10.1016/j.humimm.2004.05.008 [DOI] [PubMed] [Google Scholar]
- 98.Martin MP, Nelson G, Lee JH, Pellett F, Gao X, Wade J, Wilson MJ, Trowsdale J, Gladman D, Carrington M (2002) Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol 169:2818–2822 [DOI] [PubMed] [Google Scholar]
- 99.Williams F, Meenagh A, Sleator C, Cook D, Fernandez-Vina M, Bowcock AM, Middleton D (2005) Activating killer cell immunoglobulin-like receptor gene KIR2DS1 is associated with psoriatic arthritis. Hum Immunol 66:836–841 10.1016/j.humimm.2005.04.005 [DOI] [PubMed] [Google Scholar]