Abstract
The pericentric inv(10)(p11.2q21.2) mutation has been frequently identified in cytogenetic laboratories, is phenotypically silent, and is considered to be a polymorphic variant. Cloning and sequencing of the junction fragments on 10p11 and 10q21 revealed that neither inversion breakpoint directly involved any genes or repetitive sequences, although both breakpoint regions contain a number of repeats. All 20 apparently unrelated inv(10) families in our study had identical breakpoints, and detailed haplotype analysis showed that the inversions were identical by descent. Thus, although considered a common variant, inv(10)(p11.2q21.2) has a single ancestral founder among northern Europeans.
A small number of cytogenetically visible human chromosome rearrangements are considered to be polymorphic variants, including several common pericentric inversions.1 These inversions fall into two classes: one in which both breakpoints occur within heterochromatin (chromosomes 1, 3, 9, and 16) and the other in which both breakpoints occur within euchromatin (chromosomes 2, 5, and 10). The heterochromatic variants are the most frequent but may be a consequence of alterations in the amount and distribution of heterochromatin rather than true inversions.
The pericentric inv(10)(p11.2q21.2) mutation is not associated with any phenotypic abnormalities2 and has been frequently identified in cytogenetic laboratories in the United Kingdom,2 France,3 Denmark and Sweden,4 and North America.5 The estimated frequency of inv(10) among prenatal diagnostic referrals to the laboratories taking part in this study is 1 in 3,600 in Germany, 1 in 7,100 in Denmark, and 1 in 12,800 in the United Kingdom. Thus, although the great majority of chromosome inversions appear to be unique rearrangements, the frequency and wide geographical distribution of inv(10)(p11.2q21.2) suggest that it might be a recurrent variation that has arisen independently in different populations.6
Repetitive sequence elements have been implicated in the formation of a range of recurrent structural rearrangements.7 For example, the breakpoints of the most frequently occurring non-Robertsonian translocation, t(11;22), are within palindromic AT-rich repeat sequences,8 and low copy number repeats (LCRs), or duplicons, mediate the formation of microdeletions and microduplications.9
We have studied a series of 20 apparently unrelated families with cytogenetically identical inv(10)s, comprising 9 families from the United Kingdom, 5 from Germany, 3 from Denmark, 2 from Sweden, and 1 from northwestern Russia (table 1). Our study had two specific aims: (1) characterization of the inv(10) breakpoints at the molecular level, to ascertain whether the formation of the inversion is mediated by repetitive sequence elements, and (2) haplotype analysis, to determine the proportion of inv(10)s that arose independently and the proportion that share an ancestral founder and are identical by descent (IBD).
Table 1.
Study Population[Note]
| Family | Patient | Country of Origin |
| 1 | Ger1 | Germany |
| 2 | Sw1 | Sweden |
| 3 | Sw2 | Sweden |
| 4 | Dk1 | Denmark |
| 5 | Dk2 | Denmark |
| 6 | Dk3 | Denmark |
| 7 | UK1 | United Kingdom |
| 8 | UK3 | United Kingdom |
| 9 | UK4 | United Kingdom |
| 10 | UK5 | United Kingdom |
| 11 | UK6 | United Kingdom |
| 12 | UK7 | United Kingdom |
| 13 | UK8 | United Kingdom |
| 14 | UK9 | United Kingdom |
| 15 | UK10 | United Kingdom |
| 16 | Ger2 | Germany |
| 17 | Ger3 | Germany |
| 18 | Ger4 | Germany |
| 19 | Ger5 | Germany |
| 20 | Rus1 | Northwestern Russia |
Note.— Bold italic type = phase known.
The inv(10) breakpoints of patients 1 and 2 were located by FISH in the cytogenetic bands 10p11.21 and 10q21.1. For both inv(10) carriers, the BAC clone RP11-92B19 spans the breakpoint on 10p11.21. On 10q21.1, the breakpoints of both carriers were within the overlapping region of BAC clones RP11-22H3 and RP11-806B6. Subsequent analysis showed that the breakpoints of a further seven inv(10) carriers fell in the same spanning BAC clones.10
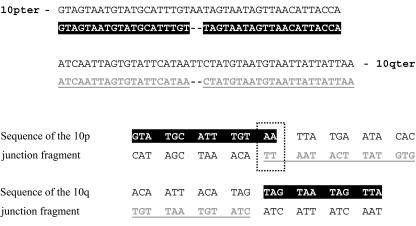
The inversion breakpoints of patient 1 were further refined by Southern blot analysis and were subsequently cloned. Sequence analysis revealed an overall loss of two nucleotides. The break in the 10q junction fragment could be unambiguously assigned, but, in the 10p junction fragment, there was a 2-bp overlap common to both 10p11 and 10q21 sequences (fig. 1). Thus, it is not possible to tell at which breakpoint site the deletion occurred. Apart from the 2-bp identity at the breakpoint, there was no extensive homology between the 10p11 and 10q21 sequences.
Figure 1.
Sequence analysis of junction fragments. A, Genomic sequence encompassing breakpoints. B, Sequence of PCR-amplified junction fragments showing the chromosome 10 genomic sequence (normal text), the sequence flanking the 10p11 breakpoint (white text), the sequence flanking the 10q21 breakpoint (underlined text), and the sequence from either 10p11 or 10q21 (boxed). The genomic sequences shown have the following coordinates in NCBI 35 (November 2005): 10p: 37,148,066–37,148,108/AL390061.9; 4,483–4,525 and 10q: 59,748,173–59,748,217/AC016396.6; 120,582–120,626.
To determine whether other inv(10) carriers in our series contained the same breakpoints, we designed PCR assays to specifically amplify the rearranged chromosome 10 (table 2). PCR fragments of identical length were amplified at both breakpoints in the remaining 19 families, and sequencing revealed that the breakpoints were identical in all the inv(10) carriers.
Table 2.
Amplification of 10p11 and 10q21 Junction Fragments
|
Primer Sequences |
||||
| Breakpoint | Forward | Reverse |
Fragment Size (bp) |
PCR Conditions |
| 10p11.2 | GAGGCCAGGCTTAAAGCAAC | CCCACTATGGTCTGCACCAG | 354 | 39 cycles: 95°C 30 s, 57°C 30 s, 72°C 40 s |
| 10q21.2 | AGCTGCTGTAGCCTTTGCAC | AACTGGTAAAAGAAGATCCTTGG | 513 | 39 cycles: 95°C 30 s, 56°C 30 s, 68°C 40 s |
The 10p11 breakpoint maps to 37,148 kb from 10pter (NCBI 35, November 2005) in a gene desert with no known gene for 300 kb on either side of the breakpoint. The 10q21 breakpoint maps to 59,748 kb within a cluster of four genes (IPMK, CJ070, UBE2D1 [MIM 602961], and TFAM [MIM 600438]). Although a position effect cannot be excluded, no genes are directly disrupted by either breakpoint. This observation is consistent with the benign nature of the inversion.
The breakpoints did not directly involve any repetitive sequences. However, although the breaks occurred within short stretches of unique single-copy sequence, in both cases these were flanked by several repeats. The RepeatMasker program showed that the sequence around both breakpoints was enriched for interspersed repetitive elements. The 10-kb interval on 10p11—5 kb on either side of the breakpoint—contained 34% repetitive sequences (15% short interspersed transposable elements [SINEs] and 14% LTRs), and the 10-kb interval on 10q21 contained 47% repetitive sequences (20% long interspersed transposable elements [LINEs], 10% LTRs, and 9% SINEs). Interspersed repeats may promote instability and the formation of DNA double-strand breaks and/or act as substrates for recombination.7 Therefore, although it seems unlikely that the sequences around each breakpoint are predisposed to the formation of the inversion, we cannot exclude this possibility.
The presence of the same breakpoints in all inv(10) carriers and the lack of obvious predisposing factors suggest a founder effect—that is, that all 20 families share a common ancestor. To determine whether the inv(10)s were all IBD, we undertook detailed haplotype analysis, using microsatellites and SNPs. DNA was available for more than one inversion carrier from 5 of the 20 families. The five haplotypes for which phase was known were identical or differed at no more than 2 of the 17 microsatellites tested within the inversion (table 3). This suggests that all five inv(10)s are IBD and allowed us to predict the likely ancestral haplotype that was identical to that observed for family 8 (UK3). In contrast to the degree of allele sharing within the inverted region, the flanking haplotypes were completely divergent outside the inversion breakpoints.
Table 3.
Microsatellite Analysis in Phase-Known Families[Note]
|
Allele Sizea |
|||||||||
| Locus |
Locus Size (Mb) |
Heterozygosity | No. of Alleles | Family 2 | Family 3 | Family 8 | Family 9 | Family 15 | Founder |
| D10S600 | 28.7 | .84 | 10 | 178 | 182 | 190 | 186 | 182 | - |
| D10S213 | 29.5 | .83 | 9 | 188 | 180/188 | 188 | 172 | 182 | - |
| D10S204 | 29.7 | .76 | 12 | 291 | 291 | 319 | 329 | 295 | - |
| D10S193 | 30.6 | .82 | 9 | 214 | 220 | 224 | 224 | 220 | - |
| D10S208 | 31.7 | .80 | 9 | 180 | 182 | 182 | 178 | 182 | - |
| D10S199 | 32.4 | .86 | 12 | 173 | 173 | 173 | 179 | 179 | - |
| D10S1666 | 33.7 | .72 | 10 | 266 | 258 | 276 | 274 | 256 | - |
| D10S1175 | 33.9 | 1.00 | Unknown | 320 | 320 | 316 | 310 | 348 | - |
| D10S176 | 36.8 | .70 | 10 | 114 | 94 | 94 | 94 | 94 | - |
| D10S1791 | 37.1 | .73 | 6 | 207 | 201 | 201/207 | 201 | 201 | - |
| 10p11.21 | 37.1 | ||||||||
| D10S508 | 37.8 | .67 | Unknown | 184 | 184 | 184 | 184 | 184 | 184 |
| Centromere | 39–41 | ||||||||
| D10S141 | 42.8 | .85 | 13 | 115 | 115 | 115/131 | 115 | 115 | 115 |
| D10S469 | 42.8 | .87 | 10 | 123 | 123 | 123/137 | 123 | 123 | 123 |
| ZNF22 | 44.8 | .84 | 10 | 151 | 151 | 151 | 151 | 151 | 151 |
| sJRH | 48.0 | .90 | 17 | 303 |
299 | 299 | 299 | 299 | 299 |
| D10S1793 | 49.5 | .86 | 11 | 254 | 254 | 254 | 252 |
254 | 254 |
| D10S1766 | 50.4 | .75 | 5 | 171 | 171 | 171 | 171 | 171 | 171 |
| D10S220 | 51.7 | .84 | 10 | 107 | 107 | 107 | 107 | 109 |
107 |
| D10S196 | 51.8 | .79 | 6 | 100 | 94 |
100 | 100 | 94/100 | 100 |
| D10S1790 | 54.6 | .84 | 11 | 191 |
193 | 193 | 193 | 193 | 193 |
| D10S539 | 54.7 | .76 | 8 | 93 | 93 | 93 | 93 | 93 | 93 |
| D10S1124 | 56.8 | .88 | 14 | 231 | 231 | 231 | 231 | 231 | 231 |
| D10S1788 | 57.3 | .78 | 7 | 249 | 249 | 249 | 249 | 249 | 249 |
| D10S1767 | 58.0 | .71 | 13 | 256 | 256 | 256 | 256 | 256 | 256 |
| D10S1756 | 58.4 | .84 | 9 | 192 | 192/194 | 192 | 192 | 192 | 192 |
| D10S524 | 58.6 | .88 | Unknown | 369 | 365 |
369 | 369 | 369 | 369 |
| D10S1659 | 58.7 | .75 | 8 | 184 | 184 | 184 | 184 | 184/194 | 184 |
| 10q21.1 | 59.8 | ||||||||
| D10S589 | 60.8 | .79 | 8 | 190 | 186 | 184 | 184 | 186 | - |
| D10S464 | 60.9 | .78 | 8 | 140 | 140 | 144 | 134 | 140 | - |
| D10S1652 | 63.8 | .78 | 10 | 167 | 163 | 165 | 161 | 171 | - |
| D10S581 | 65.2 | .80 | 12 | 142 | 134/138 | 136 | 136 | 148 | - |
| D10S1743 | 66.8 | .78 | 9 | 227 | - | 235 | 243 | 241 | - |
| D10S1670 | 68.2 | .76 | 12 | 305 | 301 | 305 | 305/307 | 321 | - |
| D10S210 | 69.4 | .80 | 6 | 135 | - | 133 | 129/133 | 131 | - |
| D10S1647 | 70.3 | .82 | 9 | 204 | 208 | 212 | 208 | 206 | - |
| D10S1665 | 70.6 | .87 | 12 | 240 | - | 238 | 218 | 234/240 | - |
| D10S537 | 71.7 | .83 | 9 | - | - | 298 | 290 | 292 | - |
| D10S1650 | 72.6 | .85 | 12 | 136 | - | 132 | 124 | 138 | - |
Note.— Breakpoints and the centromere are shaded in gray. All microsatellite details are available from the Genome Database, and distances were taken from Ensembl. Alleles outside the inversion are in italics. Shared alleles and the common haplotype are shown in bold italics, and allele differences are underlined.
Allele sizes are taken from the total size of the PCR product and are given in base pairs, rounded to the nearest whole number.
We also typed the same microsatellites in the 15 families where DNA was available from only a single carrier (table 4). This demonstrated that all 20 families are IBD. The alleles in 8 of the 20 families were consistent with the common haplotype, whereas in 12 families there was at least one difference. In total, there were nine allele differences: five were private mutations, whereas four were seen in more than one family. The most common allele change observed was at the microsatellite D10S220, from a PCR product length of 107 bp in the ancestral haplotype to 109 bp in five families.
Table 4.
Microsatellite Results for All 20 Inv(10) Families[Note]
|
Family Allele Size |
|||||||||||||||||||||
| Locus |
Ancestral Haplotype |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| 10p11.21 | |||||||||||||||||||||
| D10S508 | 184 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Centromere | |||||||||||||||||||||
| D10S141 | 115 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D10S469 | 123 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ZNF22 | 151 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| sJRH | 299 | - | 303 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 303 | 303 | - | - |
| D10S1793 | 254 | - | - | - | - | - | - | - | - | 252 | - | - | - | - | - | - | - | - | - | - | - |
| D10S1766 | 171 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D10S220 | 107 | 109 | - | - | - | - | - | - | - | - | 109 | 109 | - | - | - | 109 | - | - | - | - | 109 |
| D10S196 | 100 | - | - | 94 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D10S1790 | 193 | - | 191 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D10S539 | 93 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D10S1124 | 231 | - | - | - | - | - | - | - | - | - | - | 213/223 | - | - | - | - | - | - | - | - | - |
| D10S1788 | 249 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D10S1767 | 256 | - | - | - | - | - | - | - | - | - | - | - | 254 | 254 | - | - | - | - | - | - | - |
| D10S1756 | 192 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 190 | 190 | - | - |
| D10S524 | 369 | - | - | 365 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D10S1659 | 184 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 10q21.1 | |||||||||||||||||||||
Note.— Breakpoints and the centromere are shaded in gray. A hyphen indicates that the same allele is present as that in the ancestral haplotype. Where no allele is shared with the ancestral haplotype (i.e., a microsatellite mutation), the size of the divergent allele is shown. Allele sizes are taken from the total size of the PCR product and are given in base pairs, rounded to the nearest whole number. Only microsatellites within the inverted region are shown.
For SNP analysis (table 5), we compared three families for which phase was known and four families for which phase was unknown. In contrast to the microsatellites, which spanned the whole inversion, SNPs were chosen over a few kilobases in the immediate vicinity of the breakpoints. All seven inv(10) families tested had exactly the same haplotype (table 6), providing further evidence that the inversions are IBD. Families 2 (Sw1) and 3 (Sw2) had identical haplotypes even though they differed at 4 of the 17 microsatellites. This is likely to be due to the higher mutation rates in microsatellites compared with SNPs. Thirty-six control SNP haplotypes were generated from 18 normal individuals (from 9 trios) to assess the frequency of the inversion haplotype. There were 19 different haplotypes and, in total, 6 of the 36 control chromosomes carried the inversion haplotype (one homozygous and four heterozygous individuals). Thus, it is unlikely that the SNP haplotype shared by the inv(10) carriers is coincidental.
Table 5.
dbSNP Accession Numbers and Details for the Analyzed SNPs
|
Primers |
||||||||||
| Chromosome and dbSNP Accession Number | Alleles | Allele Frequencya | Heterozygositya | Locationb | Method | Forward | Reverse |
PCR Length (bp) |
Enzyme |
Restriction Fragment Lengths (bp) |
| 10p: | ||||||||||
| rs3898062 | A/G | NK | NK | 37143715 | Enzyme digestion | AGGAGAATGGCGTGAATC | GATATTATCATGGAGGCTTTAGG | 344 | MseI | A: 29, 42, 117, 156; G: 42, 146, 156 |
| rs2488748 | C/G | G: .951; C: .049 | .095 | 37145779 | Enzyme digestion | GTGGCAAGAAAGCTAGTAAGT | TCAAATAGCAGAGTCGCA | 320 | Fnu4HI | C: 81, 239; G: 58, 81, 181 |
| rs12257945 | A/G | NK | NK | 37147006 | Sequencing | TTGTATTAGAGGAACCCGAAGCA | GGAGCCAGTGTAACACGGTAGAT | 324 | … | … |
| rs12572077 | A/C | A: .472; C: .528 | .498/.166 | 37147261/37147272 | Sequencing | CATCTTTTCCCCACCATAGTGTT | CTCAGCCACAGATTCAAGTTCC | 272 | … | … |
| rs2695081 | G/C | G: .092; C: .908 | ||||||||
| rs4934835 | G/C | NK | NK | 37147530 | Sequencing | AGGCTGCTCAAATAAACACGTGAA | GTGGTGGTGTACCCGTGGTC | 224 | … | … |
| rs11010897 | A/G |
NK |
NK |
37147709 |
Enzyme digestion |
GTACACCACCACAGCTAATG |
GTTGGCTATAATACGGAGTTCTA |
354 |
AluI |
A: 14, 108, 232; G: 14, 340 |
| Breakpoint |
||||||||||
| rs2463226 | A/T | NK | NK | 37148241 | Sequencing | CAAGGATCTTCTTTTACCAGTTA | AATGAAACTAATTTTATCATAGGCT | 155 | … | … |
| rs10827731 | C/T | T: .127; C: .873 | .222 | 37148704 | Sequencing | GCACCAATATAAAAATGACTCAAA | CTCAAGAGCAGCCAGAGTG | 170 | … | … |
| rs11817755 | G/T | NK | NK | 37148915 | Enzyme digestion | GAGGTGTGGAAGGAGAGGCGCAGGT | AGCTGAGGCCCGGCAAGAATTTGAGT | 239 | AluI | G: 2, 20, 74, 143; T: 2, 20, 60, 74, 83 |
| rs3867222 | C/T | C: .444; A: .556 | .494 | 37151520 | Enzyme digestion | GTTCCATCACAGGTCATCTT | CAATAAAATGTTGGGCTATTAAA | 332 | Tsp45I | C: 332; T: 83, 249 |
| rs2490841 | G/T | G: .134; T: .866 | .232 | 37152375 | Enzyme digestion | TGGCTGTGTTTTCAGATTGG | GAGAGGGAAAGAGCAATGACA | 272 | AluI | G: 78, 194; T: 272 |
| rs16851 | C/T | C: .022; T: .978 | .042 | 59743596 | Enzyme digestion | TATCTACACATTCCATTATTCCC | CATAATATATGTCAGGCGTTTG | 195 | TaqI | C:44, 151; T:195 |
| rs11818916 | A/C | NK | NK | 59744939 | Enzyme digestion | TGGATGTTTGATGGAGTTGGTAGTTTTG | GTAGTGTCTGCTGGGTTCACCGAA | 199 | Tsp45I | A: 61, 138; C: 199 |
| rs2486489 | G/T | G: .023; T: .977 | .046 | 59746059 | Sequencing | TTCCCCCACAAAACATCTCAACTG | TGGTTTCCAGCTAGTAGATTTGAATCCA | 221 | … | … |
| rs12248484 | A/G |
A: .002; G: .998 |
.005 |
59747654 |
Sequencing |
AGTCTGATTGTGGCTATTTGC |
TATACTGTTAGCCTCTGACCCAT |
252 |
… |
… |
| Breakpoint |
||||||||||
| rs7072568 | A/G | NK | NK | 59748801 | Sequencing | AGGTGGGCGGGATGTTAATGT | TGACCGGGAGAAAAGGCTTAAGA | 272 | … | … |
| rs12241885 | C/T | NK | NK | 59756553 | Enzyme digestion | CATAAATTGCCCGATTGCCGACT | AGGGATCTTGCAGCCGTCAGAA | 307 | Fnu4HI | C: 12, 50, 61, 184; T: 12, 50, 245 |
| rs1007915 | A/G | G: .009; A: .991 | .018 | 59760590 | Enzyme digestion | TACACTTCCTTCCTCCTGCGTAG | AGATGTGGGCACCAGGATATG | 334 | Mse I | G: 81, 116, 137; A: 12, 81, 116, 125 |
NK = Not known.
Location from UCSC Genome Browser, May 2004.
Table 6.
Conserved SNP Haplotype[Note]
| Chromosome and dbSNP Accession Number |
Alleles | Inv(10) |
| 10p: | ||
| rs3898062 | A/G | G |
| rs2488748 | C/G | G |
| rs12257945 | A/G | A |
| rs12572077 | A/C | A |
| rs2695081 | C/G | G |
| rs4934835 | C/G | C |
| rs11010897 | A/G |
A |
| Breakpoint |
||
| rs2463226 | A/T | A |
| rs10827731 | C/T | C |
| rs11817755 | G/T | T |
| rs3867222 | C/T | T |
| rs2490841 | G/T | T |
| 10q: | ||
| rs16851 | C/T | T |
| rs11818916 | A/C | A |
| rs2486489 | G/T | T |
| rs12248484 | A/G |
G |
| Breakpoint |
||
| rs7072568 | A/G | G |
| rs12241885 | C/T | C |
| rs1007915 | C/T | T |
Note.— Twelve SNPs around the 10p breakpoint and seven SNPs around the 10q breakpoint were selected for SNP analysis by enzyme digestion or sequencing. All details are given in table 5.
The haplotype analysis demonstrated complete suppression of recombination within the inverted segment. Our data cannot distinguish between a direct effect—namely, that crossing over does not occur—and indirect selection against unbalanced recombinant products. The inversion breakpoints are close to the centromeric areas of low recombination. No recombinants were seen in two studies of 33 and 15 inv(10) families.2,4
It is difficult to make an accurate estimation of the age of the inversion. The geographical distribution of the 20 inversion carriers, the accumulation of microsatellite mutations within the inversion—estimates for which range from 10−2 to 10−4 per locus per generation—and the occurrence of crossovers very close to both the 10p11 and 10q21 breakpoints in most, if not all, families suggest that the rearrangement is not a recent event. This is consistent with the calculation of average reproductive fitness for inversions of 0.926 ± 0.085.11
The breakpoints of a small number of other pericentric inversions have also been determined. In contrast to inv(10), these inversions were studied because they were associated with specific abnormal phenotypes, and, consequently, the majority of breakpoints were identified within the introns of genes.12–16 Graw et al.17 cloned the breakpoints of the inv(8)(p23.1q22.1), which is associated with various clinical manifestations, including mental retardation and heart defects in unbalanced carriers (Rec 8 syndrome [MIM 179613]). The results were similar to inv(10) in a number of ways: No genes were directly disrupted by the inversion, the breakpoint sequences showed little homology, the breakpoints lay in unique sequences flanked by repetitive elements, and the inversion has spread widely from a single founder.
The 20 inv(10) families studied were all from northern Europe. It would be interesting to establish whether all cases worldwide are also derived from the same founder. Of the inv(10) cases in the literature, only one has been reported as de novo.18 Breakpoint sequencing and haplotype analysis should be applied to any potentially unrelated or non-European inv(10) carriers. We have contacted several cytogenetic laboratories worldwide whose populations are unlikely to be of European origin. To date, we have had replies from three laboratories (in Egypt, Mexico, and Singapore), none of which have identified a single inv(10). The only non-European cases in the literature are from the United States and Canada,3 and these individuals could conceivably be of European origin.
Thus, the overall evidence suggests that, although it is considered a common variant, inv(10) may well be a unique rather than a recurrent rearrangement, with a single European founder. It would be interesting to apply the approaches used in this study to other common inversions, such as the variant inv(2)(p11q13), to establish whether they are also IBD.
Acknowledgments
We thank the cytogenetic staff of the Wessex Regional Genetics Laboratory, especially Morag Collinson, and of the Institute of Human Genetics in Kiel. We are grateful to Minna Becker, Susanne Freier, and Hannelore Madle, for cell culture and chromosome preparations; to Dr. Panos Deloukas (Sanger Centre, U.K.), for providing BAC clones; and to Drs. Merete Bugge and Ulf Kristofferson, for collecting patient DNA. We also thank Drs. Ashraf Ibrahim, Hanaa Adib, Horacio Rivera, and Leena Gole, for information on inv(10) identification in their laboratories, and Jan Hansen from the Danish Cytogenetic Central Registry and Professor Patricia Jacobs, for critical reading of this manuscript. The Institute of Human Genetics and the Max Planck Institute for Molecular Genetics are supported by grants from the Bundeministerium für Bildung und Forschung (BMBF) within the National Genome Project. The Wilhelm Johannsen Center for Functional Genome Research is supported by the Danish National Research Foundation.
Web Resources
URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Ensembl, http://www.ensembl.org/
- Genome Database, http://www.gdb.org/
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for UBE2D1, TFAM, and Rec 8 syndrome)
- RepeatMasker, http://www.repeatmasker.org/
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Gardner RJM, Sutherland GT (1996) Chromosome abnormalities and genetic counselling, 2nd ed. Oxford University Press, Oxford, pp 115–182 [Google Scholar]
- 2.Collinson MN, Fisher AM, Walker J, Currie J, Williams L, Roberts P (1997) Inv(10)(p11.2q21.2), a variant chromosome. Hum Genet 101:175–180 10.1007/s004390050609 [DOI] [PubMed] [Google Scholar]
- 3.Goupe de Cytogénéticiens Français (1986) Pericentric inversions in man: a French collaborative study. Ann Genet 29:129–168 [PubMed] [Google Scholar]
- 4.Sherman SL, Iselius L, Gallano P, Buckton K, Collyer S, De Mey R, Kristoffersson U, Lindsten J, Mikkelsen M, Morton NE, Newton M, Nordensson I, Petersen MB, Wahlstrom J (1986) Segregation analysis of balanced pericentric inversions in pedigree data. Clin Genet 30:87–94 [DOI] [PubMed] [Google Scholar]
- 5.Daniel A, Hook EB, Wulf G (1989) Risks of unbalanced progeny at amniocentesis to carriers of chromosome rearrangements: data from United States and Canadian laboratories. Am J Med Genet 33:14–53 10.1002/ajmg.1320330105 [DOI] [PubMed] [Google Scholar]
- 6.Youings S, Ellis K, Ennis S, Barber J, Jacobs P (2004) A study of reciprocal translocations and inversions detected by light microscopy with special reference to origin, segregation and recurrent abnormalities. Am J Med Genet A 126:46–60 10.1002/ajmg.a.20553 [DOI] [PubMed] [Google Scholar]
- 7.Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13:R57–R64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- 8.Kurahashi H, Shaikh TH, Zackai EH, Celle L, Driscoll DA, Budarf ML, Emanuel BS (2001) Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet 10:2605–2517 10.1093/hmg/10.23.2605 [DOI] [PubMed] [Google Scholar]
- 9.Emanuel BS, Shaikh TH (2001) Segmental duplications: an “expanding” role in genomic instability. Nat Rev Genet 2:791–800 10.1038/35093500 [DOI] [PubMed] [Google Scholar]
- 10.Metzke-Heidemann S, Gesk S, Martin-Subero JI, Harder L, Caliebe A, Kautza M, Duba C, Erdal M, Jenderny J, French L, Earthrowl ME, Grote W, Deloukas P, Seibert R (2004) Molecular cytogenetic mapping of the breakpoints of the constitutional pericentric inversion inv(10)(p11.2q21.2). Eur J Hum Genet 13(S1):P0255 [Google Scholar]
- 11.Jacobs PA, Frackiewicz A, Law P, Hilditch J, Morton NE (1975) The effect of structural aberrations of the chromosomes on reproductive fitness in man. II. Results. Clin Genet 8:169–178 [DOI] [PubMed] [Google Scholar]
- 12.Saito-Ohara H, Fukuda Y, Ito M, Agarwala KL, Hayashi M, Matsuo M, Imoto I, Yamakawa K, Nakamura Y, Inazawa J (2002) The Xq22 inversion breakpoint interrupted a novel Ras-Like GTPase gene in a patient with Duchenne muscular dystrophy and profound mental retardation. Am J Hum Genet 71:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beiraghi S, Zhou M, Talmadge CB, Went-Sumegi CB, Davis JR, Huang D, Saal H, Seemayer TA, Sumegi J (2003) Identification and characterisation of a novel gene disrupted by a pericentric inversion inv(4)(p13.1q21.1) in a family with cleft lip. Gene 309:11–21 10.1016/S0378-1119(03)00461-X [DOI] [PubMed] [Google Scholar]
- 14.Iida A, Emi M, Matsuoka R, Hiratsuka R, Okui K, Ohashi H, Inazawa J, Fukushima Y, Imai T, Nakamura Y (2000) Identification of a gene disrupted by inv(11)(q13.5q25) in a patient with left-right axis malformation. Hum Genet 106:277–287 10.1007/s004390051038 [DOI] [PubMed] [Google Scholar]
- 15.Sood R, Bader PI, Speer MC, Edwards YH, Eddings EM, Blair RT, Hu P, Faruque MU, Robbins CM, Zhang H, Leuders J, Morrison K, Thompson D, Schartzberg PL, Meltzer PS, Trent JM (2004) Cloning and characterisation of an inversion breakpoint at 6q23.3 suggests a role for Map7 in sacral dysgenesis. Cytogenet Genome Res 106:61–67 10.1159/000078563 [DOI] [PubMed] [Google Scholar]
- 16.Tadin-Strapps M, Warburton D, Baumeister FA, Fischer SG, Yonan J, Gilliam TC, Christiano AM (2004) Cloning of the breakpoints of a de novo inversion of chromosome 8, inv(8)(p11.2q23.1) in a patient with Ambras syndrome. Cytogenet Genome Res 107:68–76 10.1159/000079573 [DOI] [PubMed] [Google Scholar]
- 17.Graw SL, Sample T, Bleskan J, Sujansky E, Patterson D (2000) Cloning, sequencing and analysis of Inv8 chromosome breakpoints associated with recombinant 8 syndrome. Am J Hum Genet 66:1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warburton D (1991) De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet 49:995–1013 [PMC free article] [PubMed] [Google Scholar]