Abstract
Depressive disorders account for a large and increasing global burden of disease. Although the condition of many patients improves with medication, only a minority experience full remission, and patients whose condition responds to one medication may not have a response to others. Individual variation in antidepressant treatment outcome is, at present, unpredictable but may have a partial genetic basis. We searched for genetic predictors of treatment outcome in 1,953 patients with major depressive disorder who were treated with the antidepressant citalopram in the Sequenced Treatment Alternatives for Depression (STAR*D) study and were prospectively assessed. In a split-sample design, a selection of 68 candidate genes was genotyped, with 768 single-nucleotide–polymorphism markers chosen to detect common genetic variation. We detected significant and reproducible association between treatment outcome and a marker in HTR2A (P range 1×10-6 to 3.7×10-5 in the total sample). Other markers in HTR2A also showed evidence of association with treatment outcome in the total sample. HTR2A encodes the serotonin 2A receptor, which is downregulated by citalopram. Participants who were homozygous for the A allele had an 18% reduction in absolute risk of having no response to treatment, compared with those homozygous for the other allele. The A allele was over six times more frequent in white than in black participants, and treatment was less effective among black participants. The A allele may contribute to racial differences in outcomes of antidepressant treatment. Taken together with prior neurobiological findings, these new genetic data make a compelling case for a key role of HTR2A in the mechanism of antidepressant action.
Major depressive disorder (MDD) is a major public health problem and a frequent reason why patients visit internists, family practitioners, psychiatrists, and other physicians.1 MDD constitutes the fourth greatest disease burden worldwide, measured in disability-adjusted life years, which express years of healthy life lost to death and disability.2 MDD is predicted to account for the second greatest global disease burden by 2020.3
Many patients can expect their condition to improve with antidepressant treatment, but only a minority experience full remission, and individual outcomes differ across medications. The largest study to date demonstrated that up to 63% of patients have improvement and that 47% of patients achieve complete remission of symptoms after an adequate trial with a single antidepressant.4 Patients whose treatment is unsuccessful with one antidepressant medication often have a response when treated with an antidepressant of a different chemical class (reviewed by Marangell5).
Little is known about the basis for such marked individual variation in treatment outcome. Indirect evidence suggests that at least some of this variation has a genetic basis.6 Outcome and side-effect patterns vary less between illness episodes than between individuals in some studies7,8 but not all.9 Other studies have shown that outcome of antidepressant treatment runs in families.8,10 It has been suggested that a number of genetic variants influence outcome and/or side effects in comparatively small, naturalistic samples of patients treated for major depression, but these findings have often not been replicated (reviewed by Franchini et al.8 and Malhotra et al.11). Since the effects of individual genes may be small, the definitive identification of alleles involved in antidepressant-treatment outcome may require large, well-characterized samples.
The Sequenced Treatment Alternatives for Depression (STAR*D) study collected DNA from 1,953 subjects with MDD. At the first treatment step, participants received the selective serotonin reuptake inhibitor (SSRI) citalopram, with regular assessment of outcome and side effects.12 A genetic association study of phenotypes measuring outcome of citalopram treatment was undertaken in the STAR*D sample, with use of 768 markers selected to detect common sequence variation within each of 68 candidate genes.
Methods
Sample
The rationale, methods, and design of the STAR*D study have been detailed elsewhere.13 In brief, investigators at 14 regional centers across the United States implemented a standard study protocol at 41 clinical sites.
Subjects provided separate written informed consent for study participation and for the collection of blood samples for genetic studies. Outpatients aged 18–75 years with a baseline Hamilton Depression Rating Scale score14,15 of ⩾14 who met DSM-IV16 criteria for nonpsychotic MDD were eligible. Patients with bipolar, psychotic, or obsessive-compulsive disorders were excluded, as were those with primary eating disorders, general medical conditions that contraindicated study medications, substance dependence requiring inpatient detoxification, and clear nonresponse or intolerance to any protocol antidepressant during current episode or those who were pregnant or breast-feeding.
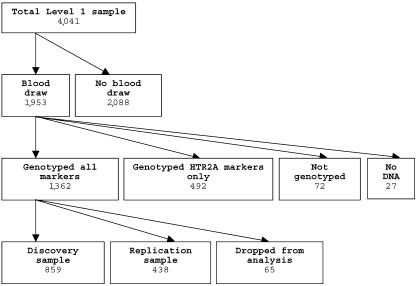
The 16-item Quick Inventory of Depressive Symptomatology–Clinician-rated (QIDS-C16)12,13,17,18 was obtained at baseline and at each treatment visit, to measure symptom severity. The intraclass correlation coefficient for the QIDS-C16, repeated across raters over 4 years, was 0.96 (A.J.R.'s unpublished data). Patients with a baseline QIDS-C16 >10 were eligible if the treating clinician determined that outpatient treatment with an antidepressant medication was indicated and safe. At level 1, the protocol required an adequate dose of citalopram for a sufficient time to maximize the likelihood of treatment success, to ensure that those who did not improve were most likely unresponsive to the medication, not just underdosed.12 No concomitant medications were allowed, aside from benzodiazepines and hypnotics if needed. A CONSORT (Consolidated Standards of Reporting Trials) diagram of the current study sample is shown in figure 1.
Figure 1.
CONSORT chart of genotyping and analysis of STAR*D sample. Samples dropped from analysis comprised 34 subjects who were noncompliant with medications, 19 with initial QIDS-C16 <10 or missing, 5 with missing clinical data, 4 whose molecular and recorded sex did not match, and 3 duplicate samples.
DNA Samples
DNA samples were collected from 1,953 participants. A sample of 20 ml of whole blood was collected in citrate-treated vacuum tubes and was shipped overnight to the Rutgers Cell Repository, where lymphocytes were extracted and cryopreserved using standard methods. DNA was extracted using GenePure chemistry (Qiagen) and was shipped on dry ice to the NIH laboratories. Samples were arrayed using a Tecan Genesis robot (Lipsky Lab), then sex-verified with a set of three X-linked and two Y-linked markers (McMahon Lab). Four sex discrepancies were identified and were excluded before samples were genotyped further.
A summary of the sample characteristics is shown in table 1. Those who consented to have blood drawn were similar to those in the full study sample but showed slight differences in several variables that reached statistical significance because of the large sample size. Subjects who consented to have blood drawn were older and better educated, with higher household income, and were more likely to be married, to be retired, and to describe themselves as white. These subjects were also more likely to come from a primary-care setting and to report more time elapsed since their first major depressive episode (MDE), more episodes, and greater comorbidity. These differences cannot affect the genetic association results, which derive from comparisons among the genotyped subjects. However, these differences may limit the generalizability of our findings, and clinical outcomes in the genotyped sample may differ somewhat from those in the full STAR*D sample.12
Table 1.
Selected Demographic and Clinical Characteristics of the STAR*D Sample[Note]
|
Patients with Blood Drawn |
Comparison |
|||||
| Characteristic |
Complete Sample (N=4,041) |
Yes (n=1,953) |
No (n=2,088) |
Test Statistica | df | Pb |
| Sociodemographic: | ||||||
| Mean (±SD) age (years) | 40.5 ± 13.3 | 42.7 ± 13.4 | 38.4 ± 12.9 | t = 10.31 | 4,037 | <.0001 |
| Sex: | χ2 = 1.48 | 1 | NS | |||
| Male | 1,509 (37.3) | 748 (38.3) | 761 (36.4) | |||
| Female | 2,532 (62.7) | 1,205 (61.7) | 1,327 (63.6) | |||
| Race: | χ2 = 13.20 | 2 | .0014 | |||
| White | 3,055 (75.7) | 1,526 (78.2) | 1,529 (73.4) | |||
| Black | 709 (17.6) | 313 (16.0) | 396 (19.0) | |||
| Other/mixed | 272 (6.7) | 113 (5.8) | 159 (7.6) | |||
| Mean (±SD) no. of years of education | 13.4 ± 3.2 | 13.6 ± 3.2 | 13.3 ± 3.2 | χ2 = 13.01 | 1 | .0003 |
| Employment: | χ2 = 19.65 | 2 | <.0001 | |||
| Employed | 2,311 (57.3) | 1,092 (55.9) | 1,219 (58.6) | |||
| Unemployed | 1,489 (36.9) | 715 (36.6) | 774 (37.2) | |||
| Retired | 234 (5.8) | 146 (7.5) | 88 (4.2) | |||
| Mean (±SD) monthly household income (U.S. $) | 2,419 ± 3,143 | 2,521 ± 3,202 | 2,318 ± 3,082 | χ2 = 6.23 | 1 | .0125 |
| Medical insurance: | χ2 = .68 | 2 | NS | |||
| Private | 2,022 (51.8) | 998 (52.4) | 1,024 (51.2) | |||
| Public | 553 (14.2) | 270 (14.2) | 283 (14.1) | |||
| None | 1,332 (34.1) | 638 (33.5) | 694 (34.7) | |||
| Marital status: | χ2 = 15.12 | 3 | .0017 | |||
| Single | 1,207 (29.9) | 545 (27.9) | 662 (31.8) | |||
| Married/cohabiting | 1,663 (41.2) | 838 (42.9) | 825 (39.6) | |||
| Divorced/separated | 1,037 (25.7) | 493 (25.2) | 544 (26.1) | |||
| Widowed | 128 (3.2) | 77 (3.9) | 51 (2.4) | |||
| Clinical: | ||||||
| Mean (±SD) age at first MDE (years) | 25.5 ± 14.4 | 26.1 ± 14.9 | 24.9 ± 13.9 | χ2 = 3.40 | 1 | NS |
| Mean (±SD) time since first MDE (years) | 15.0 ± 13.1 | 16.6 ± 13.9 | 13.5 ± 12.1 | χ2 = 42.07 | 1 | <.0001 |
| Mean (±SD) no. of MDEs | 5.9 ± 11.4 | 6.4 ± 12.5 | 5.4 ± 10.2 | χ2 = 11.09 | 1 | .0009 |
| Suicide ever attempted: | χ2 = 6.26 | 1 | .0123 | |||
| Yes | 667 (16.5) | 293 (15.0) | 374 (17.9) | |||
| No | 3,370 (83.5) | 1,659 (85.0) | 1,711 (82.1) | |||
| No. of psychiatric comorbidities: | χ2 = 24.39 | 4 | <.0001 | |||
| 0 | 1,510 (38.2) | 781 (40.9) | 729 (35.7) | |||
| 1 | 1,028 (26.0) | 510 (26.7) | 518 (25.4) | |||
| 2 | 607 (15.4) | 282 (14.8) | 325 (15.9) | |||
| 3 | 342 (8.7) | 133 (7.0) | 209 (10.2) | |||
| ⩾4 | 465 (11.8) | 204 (10.7) | 261 (12.8) | |||
| Current episode: | ||||||
| Clinical setting: | χ2 = 29.97 | 1 | <.0001 | |||
| Primary | 1,575 (39) | 846 (43.3) | 729 (34.9) | |||
| Specialty | 2,466 (61) | 1,107 (56.7) | 1,359 (65.1) | |||
| Mean (±SD) duration of current episode (mo) | 24.5 ± 52.0 | 24.8 ± 53.1 | 24.3 ± 51.0 | χ2 = .78 | 1 | .3764 |
| HDRS-17c | 18.8 ± 6.5 | 18.4 ± 6.2 | 19.6 ± 6.9 | t = 1.65 | 330 | .099 |
| QID-S16 | 13.8 ± 4.2 | 13.4 ± 4.1 | 14.5 ± 4.5 | t = 2.20 | 373 | .0283 |
Phenotypes
All phenotype definitions and assignments were settled in advance and were assigned before genotyping. Patients were scored for treatment outcome in two ways: designated remission and response (fig. 2). In the absence of external validators, our choice of categorical phenotypes was guided (1) by careful work with the STAR*D clinicians—in advance of the genotyping—to develop distinctions that had face validity and took advantage of the large body of data available from the STAR*D trial; (2) by ensuring maximal contrast between the outcome groups, to improve power, and creating “probable” groups that approximated the more narrowly defined categories, to test their robustness; (3) and by paying special attention to full remission of symptoms, since this was the primary target outcome of treatment.
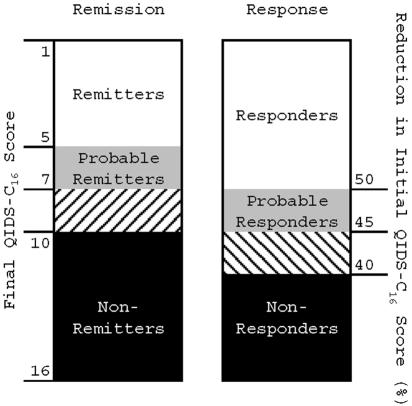
Figure 2.
Treatment-outcome phenotypes. Subjects who completed at least 6 wk of treatment with citalopram were assigned a remission and response phenotype that was based on the QIDS-C16 score at the last treatment visit. Those who met score criteria for remission or response after 3–6 wk of treatment were grouped with probable remitters or probable responders, respectively.
Remitters achieved a QIDS-C16 score of ⩽5 at the last treatment visit; probable remitters achieved a score of 6 or 7. Nonremitters had a QIDS-C16 score of ⩾10 at the last visit. Those with a final QIDS-C16 score in the borderline range of 8 and 9 were excluded from analysis.
Responders achieved at least a 50% reduction in baseline QIDS-C16 at the last treatment visit; probable responders achieved a 45%–50% reduction. Nonresponders did not achieve even a 40% reduction in baseline QIDS-C16 score at the last treatment visit. Those with a reduction in QIDS-C16 in the borderline range of 40%–45% were excluded from analysis.
Only patients who completed at least 6 wk of treatment were included in the primary analysis. Patients who achieved the required QIDS-C16 scores after <6 wk of treatment but who received at least 3 wk of treatment were assigned to the appropriate outcome group but were classified as “probable.” Those who did not complete at least 3 wk of treatment were excluded from analysis. Similarly, subjects who were classified as “intolerant” or “probably intolerant” were removed from the nonremitter and nonresponder groups but were retained in the remitter and responder groups, since intolerant subjects were probably not able to take the full effective dose of citalopram but might have responded if they had. Assessment of tolerability is discussed below. Subjects who did not adhere to the treatment regimen were excluded from analysis.
As a secondary test, relative change in QIDS-C16 score at the last visit (expressed as percentage change from initial score) was tested as a quantitative trait, after removal of intolerant and nonadherent subjects.
Tolerability
Medication tolerability comprises an individual’s objective and perceived side-effect burden and typically increases over time and with response to treatment.19 Since failure to consider tolerability could lead to misclassification of intolerant patients as nonresponders, we scored all subjects as tolerant, probably tolerant, intolerant, or probably intolerant on the basis of an algorithm that considered study exit data and the Global Rating of Side Effect Burden (GRSEB).13 In brief, all subjects who elected to continue citalopram at the end of the level 1 treatment period were considered tolerant, whereas subjects who refused to continue citalopram or who left the study because of side effects were considered intolerant. The remaining subjects were classified on the basis of GRSEB score into probably tolerant (no more than moderate side effects) or probably intolerant (more than moderate side effects). A small number of subjects with missing GRSEB scores were classified according to whether they took citalopram for <4 wk (probably intolerant) or ⩾4 wk (probably tolerant).
Candidate Genes
Sixty-eight genes were chosen for study from among a larger list of plausible candidates. Genes primarily involved in drug metabolism were excluded, by prior agreement, since these will be studied by another group using the same set of DNA samples. Genes were scored by an expert panel (D.C., W. Drevets, H.M., and F.J.M.) on the basis of (1) prior evidence of association with antidepressant outcome (1–3 points), (2) prior evidence of association with major mood disorders (1–3 points), and (3) known functional variant(s) (0–1 points). Under this scoring system, candidate genes could receive 0–7 points; higher scores conferred higher priority for study. Genes with a score of ⩾4 were used to seed sets of related genes broadly encompassing five main pathways: serotonin related (n=20), glutamate related (n=16), dopamine related (n=3), adrenergic (n=4), and neurotrophic (n=4), along with selected genes in other pathways (n=21). The complete list of genes studied is shown in table 2.
Table 2.
List of Genes Screened
| Hypothesis and Gene Symbol | Gene Name |
| Dopamine hypothesis: | |
| TH | Tyrosine hydroxylase |
| COMT | Catechol-O-methyltransferase |
| MAOA | Monoamine-oxidase A |
| Adrenergic hypothesis: | |
| ADRA2A | alpha-2a adrenergic receptor |
| ADRA2C | alpha-2c adrenergic receptor |
| DBH | Dopamine beta-hydroxylase |
| SLC6A2 | Norepinephrin transporter |
| Serotonin hypothesis: | |
| SLC6A4 | Serotonin transporter |
| TPH1 | Tryptophane hydroxylase-1 |
| TPH2 | Tryptophane hydroxylase-2 |
| HTR1A | Serotonin receptors |
| HTR1B | Serotonin receptors |
| HTR1D | Serotonin receptors |
| HTR1E | Serotonin receptors |
| HTR1F | Serotonin receptors |
| HTR2A | Serotonin receptors |
| HTR2B/PSMD1 | Serotonin receptors |
| HTR2C | Serotonin receptors |
| HTR3A | Serotonin receptors |
| HTR3B | Serotonin receptors |
| HTR3C | Serotonin receptors |
| HTR3D | Serotonin receptors |
| HTR3E | Serotonin receptors |
| HTR4 | Serotonin receptors |
| HTR5A | Serotonin receptors |
| HTR6 | Serotonin receptors |
| HTR7 | Serotonin receptors |
| Glutamate hypothesis: | |
| GRIA1 | AMPA receptors |
| GRIA2 | AMPA receptors |
| GRIA3 | AMPA receptors |
| GRIA4 | AMPA receptors |
| GRIN1 | NMDA receptors |
| GRIN2A | NMDA receptors |
| GRIN2B | NMDA receptors |
| GRIN2C | NMDA receptors |
| GRIN2D | NMDA receptors |
| GRIN3A | NMDA receptors |
| GRIK1 | Kainate receptors |
| GRIK2 | Kainate receptors |
| GRIK3 | Kainate receptors |
| GRIK4 | Kainate receptors |
| GRIK5 | Kainate receptors |
| SLC1A1 | Glutamate/aspartate transporter |
| Neurotrophin hypothesis: | |
| BDNF | BDNF |
| NTRK2 | Trk-B |
| BCL2 | B-cell CLL/lymphoma 2 |
| BAG1 | BCL2-associated athanogene |
| Other signaling pathways: | |
| PPP1R1B | DARP-32 |
| NR3C2 | Mineralocorticoid receptor |
| CREB1 | CREB |
| MAPK1 | Mitogen-activated protein kinase 1 |
| GSK3B | Glycogen synthase kinase 3 beta |
| CAMK1 | Calcium/calmodulin-dependent protein kinase I |
| PPP3R2 | Calcineurin B (located within GRIN3A) |
| Other genes: | |
| FKBP5 | FK506 binding protein 5 |
| LAMA4 | Laminin alpha-4 |
| GNB3 | Guanine nucleotide binding protein |
| OGG1 | 8-Oxoguanine DNA glycosylase |
| NET-5 | Tetraspan NET-5 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 |
| GRWD1 | Glutamate-rich WD repeat containing 1 |
| RPP30 | Ribonuclease P (30 kDa) |
| RNF20 | Hypothetical protein FLJ20690 |
| FBXO38 | F-box only protein 38 |
| ARHGAP10 | rho GTPase activating protein 21 |
| NR1I2 | Orphan nuclear receptor PAR2 |
| KDELR1 | KDELR1 protein |
| ATP1A3 | ATPase, Na+/K+ transporting, alpha-3 polypeptide |
Selection of SNP Markers
For each candidate gene, genotype data spanning the coding region and up to 2 kb of flanking sequence were downloaded from the International HapMap Project, accessed November 2004.20 Since the STAR*D sample is mostly white, data from the CEPH sample (Utah residents with northern and western European ancestry) were used. The program LDSelect21 was used to select an optimal set of available SNPs to genotype, at an r2 threshold of ⩾0.8. From the remaining SNPs, we further excluded those with a minor-allele frequency <7.5%, since we expected the alleles that contribute to treatment outcome in this data set to be common. Six nonsynonymous SNPs and four SNPs reported elsewhere22,23 to be associated with treatment outcome were added to the set, which brought the total to 768. Illumina then performed a bioinformatic screen that identified 12 markers that would likely fail in their BeadArray assay. Predicted failures were replaced by a nearby marker that was in strong linkage disequilibrium (LD) with the excluded SNP, if available. Absent this, a nearby marker with an allele frequency similar to that of the excluded marker was selected. The complete list of SNPs genotyped, along with flanking sequence and expected alleles, is available on request.
Genotyping Methods
Since all of the collected samples were not available at the start of the experiment, only the first 1,380 samples were shipped to Illumina, where they were genotyped on the Illumina BeadArray platform, a highly accurate, high-throughput assay.24 At Illumina, 99.78% of samples were successfully genotyped, and 97.92% of SNPs produced usable data, so that a total of 1,034,602 of a possible 1,035,504 genotypes were returned, including 11,280 blind duplicate genotypes, all of which matched exactly.
On the basis of the results of the first 1,380 samples, five SNPs in the remaining samples were genotyped using Taqman chemistry, were scored on a fluorescent plate reader (Molecular Dynamics) without regard to phenotype. By design, 394 samples were genotyped at rs7997012 and rs1928040 both by Illumina and in-house (McMahon Lab). No discrepancies were detected.
Analysis Plan
The primary experiment was based on comparison of allele and genotype frequencies between subjects who benefited or did not benefit from citalopram therapy. Because the number of tests in this experiment was large, a split sample design was employed. The 1,380 samples genotyped for all SNPs were divided, a priori, into a discovery sample and a replication sample. The discovery sample consisted of two-thirds of the total sample genotyped at Illumina; the replication sample consisted of the remaining one-third. The choice of asymmetric sample sizes for the discovery and test samples was based on the large overall sample size. Splitting the sample in this fashion allowed for the detection of genetic effects of equal size but with a more conservative nominal significance level in the discovery sample than in the test sample. The split samples were matched on sex and reported race (collapsed into “white,” “black,” and “other/mixed”). Any SNPs meeting the a priori significance levels in both the discovery and test samples (see below) were tested for robustness in the total data set.
Power Analysis
The power to detect association was estimated in two ways. In the first, standard statistical methods25 were used to determine the effect size for approximate sample sizes for the discovery and replication samples and for a third sample that included the discovery, replication, and all remaining samples. The discovery sample was screened with a nominal significance level of .01, which had a power of 0.9 to detect an allelic effect size of 0.16. The power to detect a similar effect size in the smaller replication sample was ∼0.8 at the P=.05 level.
In addition, the power to detect allelic and genotypic (additive) effects was estimated for logistic regression analysis for a variety of genetic models (Quanto v. 1.0 beta [Gene×Environment, Gene×Gene Interaction home page]).26 There are a large number of possible underlying genetic models. Estimates of power under a specified model are appropriate only when the specified model is, in fact, the true underlying model. For example, in an unmatched case-control design (case:control ratio of 1:0.5), for a log-additive model with allele frequency of .3, a significance level of .01, and a power of .90, the detectable genetic relative risks for sample sizes of 600 and 300 were ∼1.5 and ∼1.7, respectively. At a power of .80 and a significance level of .05, corresponding genetic relative risks were ∼1.4 and ∼1.6, respectively.
Race and Ethnicity
Subjects were enrolled into the STAR*D study without regard to race or ethnicity. Detailed data on ethnicity were not collected, but most subjects identified themselves as “white,” “black,” or “other.” We attempted to verify this self-reported race using STRUCTURE,27 which assigns a probability of group membership to individuals on the basis of marker-allele frequencies. Since the number of subjects of “other” ethnicity in the sample was small, they were excluded from the analysis. We prepared a test data set of 1,284 individuals containing 57 unlinked loci selected from among those genotyped at Illumina, and we ran this in STRUCTURE under an admixture model with correlated allele frequencies, performing 20,000 burn-ins and 20,000 repetitions. This set of markers robustly distinguished between black and white subjects in this sample, with median posterior probabilities of group membership of 0.8 for the black subjects and 0.6 for the white subjects. We then divided the sample into self-identified whites and self-identified blacks and ran STRUCTURE again with the same markers. In each subject, STRUCTURE identified only one major cluster, and a one-population model gave the best fit to the data (data not shown).
Statistical Methods
All pairs of individuals were compared using RELCHECK,28 which verified that individuals were unrelated. Because the sample consisted entirely of patients with major depression, a marker that increased risk for major depression in many of these individuals would not necessarily be in Hardy-Weinberg equilibrium.29 Thus, SNPs were not removed from analysis because of deviation from Hardy-Weinberg expectations.
Tests of association included the Pearson χ2 test (2×2 for allelic and 2×3 for genotypic associations), likelihood-ratio χ2 test (2×2 for allelic and 2×3 for genotypic associations), and Fisher’s exact test (2×2 for allelic associations only). Since each test has complementary advantages, all tests were used to make decisions about replication. This strategy might increase false-positive results due to multiple testing, but the high correlation between the tests makes this unlikely. Some of the SNPs tested had relatively rare alleles, which led to contingency tables with small cell sizes. The Pearson χ2 test is only an approximation for these SNPs but makes no assumptions about the underlying model. Fisher’s exact test is robust to small cells but assumes that the marginal distributions of the contingency table are fixed, which may not hold in this sample. When small cell size warnings were ignored, correlations among P values from the Pearson χ2, likelihood-ratio χ2, and Fisher’s exact test were high (> 0.9) for the main SNP of interest. Fisher’s exact test was the most conservative of the tests considered, so those are the findings presented in the “Results” section. For the quantitative outcome phenotype, genotypic means were compared with the F test. All P values were from two-tailed tests. LD between adjacent SNPs was estimated using Haploview 3.2,30 which generates estimates of D′ and r2 that are based on the input genotypes.
Results
Marker Coverage
The genotyped SNPs sampled the common variation within the genes selected for study at a median pairwise D′ value of 0.81. Overall, 75% of adjacent marker pairs were in LD at a D′>0.73. Only 5% of SNP pairs were associated with each other at an r2 value of >0.8, demonstrating that our SNP-selection strategy effectively excluded redundant markers.
Allelic Association Results
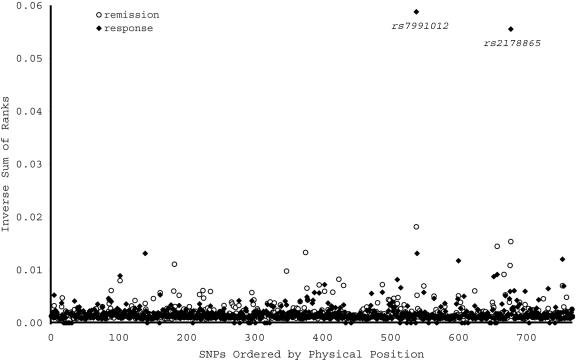
Each SNP was tested for association with treatment response and remission in the discovery sample (n=1,380). A total of 12 SNPs met or exceeded the nominal significance level of .01 for one or both phenotypes. Of these, only one SNP met or exceeded the nominal significance level of .05 in the replication sample for the same allele and phenotype (fig. 3). This SNP, rs7997012, resides in the second intron of the gene HTR2A, which encodes the serotonin 2A receptor. None of the other 767 SNPs met these strict criteria for association and replication in this experiment.
Figure 3.
Allelic association between treatment outcome and each of 768 SNPs representing 68 candidate genes. Allelic association P values were ranked in the discovery and replication samples, then the ranks were summed across samples. For visual clarity, the inverse of the sum of ranks is shown on the Y-axis. The remission phenotype is indicated by the unblackened circles, the response phenotype by the blackened circles. Only rs7997012 met the a priori P value thresholds in both samples. Another SNP, rs2178865, ranked highly in both samples but fell short of the a priori significance level in the replication sample.
In HTR2A, significant association was detected, in both samples, between the same (A) allele of rs7997012 and treatment response (table 3). (Evidence of association was also detected between this allele and the remission phenotype, but this did not meet our a priori significance levels in both samples.) An additional SNP in HTR2A (rs1928040) showed evidence of association with response and remission in the discovery sample but not in the replication sample. LD analysis demonstrated that these two SNPs were not in strong LD with each other (fig. 4).
Table 3.
Results of Association Analysis of Genotyped HTR2A SNPs, Stratified by Race[Note]
|
All |
White |
Black |
|||||||
|
P |
P |
P |
|||||||
| Phenotype and SNP | N | Allelewise | Genotypewise | n | Allelewise | Genotypewise | n | Allelewise | Genotypewise |
| Remission: | |||||||||
| rs7997012 | 1,149 | .000024 | .000035 | 911 | .00107 | .00183 | 170 | NS | NS |
| rs1928040 | 1,148 | .0446 | .0701 | 910 | .0626 | NS | 170 | NS | NS |
| rs6313 | 1,183 | NS | NS | 942 | NS | NS | 172 | NS | NS |
| rs6311 | 1,180 | NS | NS | 939 | NS | NS | 172 | .0431 | .0874 |
| Response: | |||||||||
| rs7997012 | 1,329 | .000037 | .000002 | 1,049 | .00183 | .000157 | 199 | NS | NS |
| rs1928040 | 1,327 | .0709 | NS | 1,048 | NS | NS | 199 | NS | NS |
| rs6313 | 1,372 | NS | NS | 1,086 | NS | NS | 202 | NS | NS |
| rs6311 | 1,371 | NS | NS | 1,084 | NS | NS | 203 | .0918 | .0149 |
| Change in QIDS-C16: | |||||||||
| rs7997012 | 1,749 | .000007 | .00000146 | 1,380 | .00123 | .000516 | 261 | NS | NS |
| rs1928040 | 1,747 | .0214 | .0072 | 1,378 | .0738 | .0887 | 261 | NS | NS |
| rs6313 | 1,802 | NS | .0878 | 1,425 | NS | NS | 264 | NS | .0353 |
| rs6311 | 1,804 | .0599 | .0494 | 1,426 | NS | NS | 265 | .0094 | .0261 |
Note.— SNPs are shown in physical order (by row) within each phenotype. Results were similar when subjects with probable phenotypes were included (data not shown).
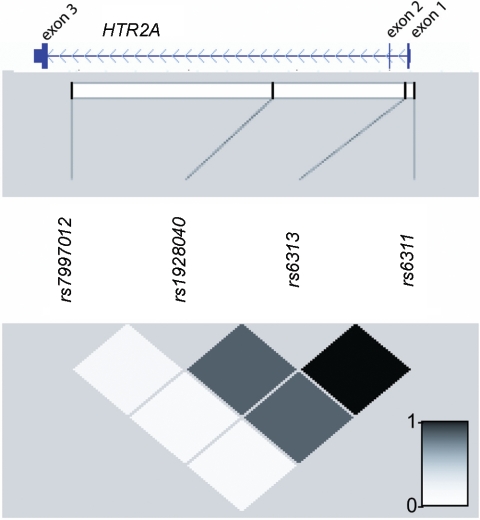
Figure 4.
Physical positions and LD relationships among SNP markers genotyped in the STAR*D sample. This figure shows the Refseq gene model for HTR2A taken from the UCSC Genome Browser (build 43), juxtaposed on a graphical representation of intermarker r2 values produced by Haploview 3.2.30
On the basis of these results, we genotyped rs7997012 and rs1928040 in the remaining subjects and tested association with treatment outcome in the total sample. Significant evidence of association was again observed between rs7997012 and treatment outcome (table 3), with P values on the order of 10-6 for the treatment response phenotype. Evidence of association between rs1928040 and treatment outcome was not substantially stronger in the total sample.
Since rs7997012 and rs1928040 are both intronic SNPs with no known function, we also genotyped two SNPs in HTR2A (rs6313 and rs6311) that may have functional importance.31–33 Both SNPs were in tight LD with each other and with rs1928040, but they were not in significant LD with rs7997012. Neither rs6316 nor rs6311 showed significant association with treatment response or remission in the total sample (table 3).
Further support for association with rs7997012 was obtained in the quantitative-trait analysis (table 3). The marker rs7997012 was significantly associated with relative change in initial QIDS-C16 score, with a P value of 7.0×10-5 for allelic and 1.0×10-6 for genotypic association. Less-significant evidence of association was also observed at other HTR2A SNPs (table 3).
Stratified Analyses
Having established an association between treatment outcome and HTR2A in this sample, we next sought to characterize the association with respect to medication tolerability and race, using data sets stratified by the variable of interest. We used rs7997012 as the test SNP and remission as the outcome variable, since they had yielded the strongest evidence of association in the earlier analyses.
In the primary analysis, medication-intolerant subjects were dropped from the nonremitter and nonresponder groups. However, this clinically justified decision created an imbalance with the remitter and responder groups, which included medication-intolerant subjects. To rule out any role of this imbalance in the primary association finding, we dropped the medication-intolerant subjects from the remitter and responder groups, then reanalyzed the data. Significant association between rs7997012 and treatment outcome remained in the sample of medication-tolerant subjects (P=2.3×10-5, n=1,140 for the remission; P=2.3×10-5, n=1,303 for the response phenotypes).
It has been suggested that black patients respond less well than white patients to antidepressant medications, particularly SSRIs, but firm data are lacking. When we divided this sample by race into “white” and “black” strata, two important differences emerged. First, we noted that the A allele of rs7997012 that was associated with better treatment outcome in the mixed-race sample was more than six times more frequent in the white than in the black participants (allele frequency in whites=0.42, in blacks, 0.06). Second, it was apparent that the association between HTR2A and treatment outcome was largely confined to the white participants. No significant association between rs7997012 and either treatment response or remission was detected in the black participants, although nominally significant results appeared for rs6313 and rs6311 in some analyses (table 3).
Potential Confounders
Carriers of the A allele of rs7997012 were compared with the other cases by means of several variables that could themselves be associated with treatment outcome: maximum citalopram dose (in mg), tolerability, sex, and initial QIDS-C16 score. Carriers of the A allele did not differ from the other cases for any of these variables (data not shown).
Strength of Association
For treatment-outcome phenotypes, the usual measures of strength of association, on the basis of odds ratios, are perhaps less informative than are measures that directly capture differences in clinical outcome.34 Thus, we compared the rates of treatment response among participants grouped by genotype at rs7997012. Overall, 79.9% of those homozygous for the A allele were classified as “responders,” compared with 62.4% of those homozygous for the G allele. Among white participants, the values were 79.7% and 63.5%, respectively. Thus, the AA genotype at rs7997013 confers a 16%–18% reduction in absolute risk of being a nonresponder in this sample.
Discussion
To our knowledge, this is the first demonstration of significant, reproducible association between genetic variation and outcome of antidepressant treatment. The association signal reproduces in both our discovery and our replication samples and increases in significance with the inclusion of additional subjects. We evaluated three related definitions of treatment effectiveness: two categorical outcomes (response and remission) and a quantitative outcome based on relative change in the final symptom score. The categorical outcomes were defined in a way that created gaps between responders/remitters and nonresponders/nonremitters, thus increasing the contrast between these groups. The quantitative-trait analysis (which included all patients) supported the results of the categorical phenotypes and demonstrated that the observed association was not a consequence of the arbitrary outcome categories. Furthermore, the HTR2A allele that was associated with better treatment outcome was more than six times more frequent in white than in black participants, who had an overall less favorable treatment outcome in this sample.12
This study has several limitations and strengths. We did not perform a complete genomewide association study, so we cannot conclude that we have found all the genes that may be relevant to antidepressant-treatment outcome. Instead, we screened 68 genes selected by an expert panel as the best candidates. Given the available marker coverage, we did not sample all common variation in these genes, but pairwise LD analysis indicated that much of the common variation was sampled. The large sample size allowed detection of weak effects with high statistical confidence, along with replication testing. The association results, however, do not necessarily point to functional variants in HTR2A. Our strongest results are not attributable to either of the suspected functional variants in HTR2A. Instead, these results point to intronic variants whose functional relevance is unknown. (Indeed, given the local patterns of LD, we cannot fully exclude the possibility that the association signal actually reflects another, nearby gene.) However, our results set the stage for a focused search for functional variants in the 3′ end of HTR2A in future studies. We employed phenotypes on the basis of a uniform rating of clinical symptoms that allowed for comparatively simple scoring of treatment outcome. Alternative methods for scoring treatment outcome exist and could point to different genes. The STAR*D study employed a naturalistic ascertainment and treatment protocol. Thus, these results should have general relevance in the clinic population but may differ from those obtained in more narrowly ascertained study groups.
As expected for a single gene, the clinical impact of HTR2A on treatment outcome is modest. In the total sample, white subjects homozygous for the allele associated with better treatment outcome had an 18% decrease in absolute risk of nonresponse to citalopram, compared with subjects homozygous for the other allele. Additional alleles predictive of treatment outcome will need to be discovered before clinically more-relevant effect sizes are obtainable. In this regard, one pertinent negative finding should be highlighted. Outcome after treatment with SSRIs has been most often associated with variation in the gene encoding the serotonin transporter, SLC6A4, the primary target of SSRI action (reviewed by Anguelova et al.35). We found no evidence of association among any of the four genotyped SLC6A4 markers and treatment outcome in these data. Our split sample design, with a requirement that the same allele show association with the same phenotype by the same test in both samples, was implemented as a way to control for multiple testing but may have reduced the power to detect modest association signals.36
We found an interesting two-way relationship between race, HTR2A variation, and treatment outcomes in this sample. First, significantly fewer black patients had favorable treatment outcomes in this study, partly because of more early dropouts.12 This is consistent with the few published reports that black patients respond less often than do whites to antidepressant medications,37 although the differences have generally been attributed to psychosocial factors. Second, the association found between HTR2A and antidepressant outcome in this study was largely confined to the white subjects. Although the sample size of the black population was smaller, no trend of association with rs7997012 was detected for any treatment-outcome phenotype. Third, the allele that was associated with better treatment outcome was more than six times more frequent in whites than blacks in this sample. These results suggest that genetic variation in HTR2A should be considered along with psychosocial factors in attempts to explain racial differences in antidepressant-treatment outcomes.
Genetic variation in HTR2A has been widely implicated in a variety of neuropsychiatric disorders (reviewed by Norton and Owen38), but it has not been convincingly demonstrated that it affects antidepressant-treatment outcome. One previous study found suggestive evidence that HTR2A was associated with a delayed and sustained pattern of treatment outcome in a small sample.39 Another small study found suggestive evidence that a different SNP in HTR2A was associated with short-term treatment outcome.40 One study of 443 depressed inpatients detected a marginally significant association between HTR2A variants and outcome.41 Finally, one small study observed differential treatment response in patients with one or two C alleles of the T102C polymorphism rs6311.42
Although the precise molecular mechanisms by which antidepressants exert their beneficial effects remain to be fully elucidated, considerable data implicate the serotonergic system.43,44 On the basis of radioligand binding, signal transduction, and amino acid sequences, 5-HT effectors currently comprise seven distinct receptors (5HT1–7). The 5-HT2A, B, and C subtypes are positively coupled with the enzyme phospholipase C (PLC).45 The 5-HT2A receptors are postsynaptic receptors that are highly enriched in neocortex and regulate the function of prefrontal-subcortical circuits implicated in the pathophysiology of depression.46,47 The 5-HT2A receptors interact with Gq/G11 guanine nucleotide binding proteins (G proteins) and thereby stimulate PLC to produce the intracellular second messengers sn-1,2-DAG (an endogenous activator of protein kinase C) and inositol-1,4,5-trisphosphate (IP3), which stimulates the release of Ca++ from intracellular stores.
Considerable neurobiological data suggest that the 5-HT2A receptor plays an important role in antidepressant-drug action. Different classes of antidepressants, including citalopram, downregulate 5-HT2A receptors in rodent and primate forebrain within a time frame paralleling therapeutic effects in humans.44,46,48,49 Selective 5-HT2A antagonists are effective in animal models of depression, and antisense oligonucleotides directed against 5-HT2A receptors regulate the development of depressivelike behavior in the learned-helplessness model.50 Although citalopram does not directly bind to 5-HT2A receptors, several antidepressants do bind 5-HT2A receptors as antagonists, which likely plays an important role in their therapeutic action.43 Finally, a growing body of work suggests that antidepressants bring about their delayed therapeutic effects by the induction of neuronal plasticity mediated by increased expression of brain-derived neurotrophic factor (BDNF).47,51,52 In this context, it is noteworthy that 5-HT2A antagonists block stress-induced downregulation of BDNF mRNA in rodents.53 Together, these results suggest that many of the neuroplastic events believed to underlie the efficacy of SSRIs are mediated, in part, via 5HT2A receptors.53 Although future studies are needed to delineate the precise cellular mechanisms by which the HTR2A SNP described herein affects response to the therapeutic effects of citalopram, the new genetic data presented here, taken together with the existing neurobiologic findings, make a compelling case for a key role of HTR2A in the mechanism of antidepressant action.
In conclusion, this study demonstrates that genetic variation in HTR2A is reproducibly associated with outcome of citalopram treatment in a large sample of outpatients with MDD. This same variation may contribute to racial differences in outcomes with SSRI treatment. Further studies are needed to define the functional changes in HTR2A that account for the association signal in this sample.
Acknowledgments
This research was supported in part by the Intramural Research Programs of the NIMH, the National Institute on Alcohol Abuse and Alcoholism, and the National Human Genome Research Institute, NIH. The authors appreciate the efforts of the STAR*D research team in performing the clinical study and gathering the DNA samples. Data and sample collection was funded with federal funds from the NIMH, NIH, under contract N01MH90003 to University of Texas Southwestern Medical Center at Dallas (principal investigator, A. J. Rush). We thank Nirmala Akula and Jo Steele, for technical advice, and the Rutgers Cell and DNA Repository, for extracting DNA and providing samples to our laboratories. The content of this publication does not necessarily reflect the views or policies of the DHHS, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We appreciate the support of Forest Laboratories for providing citalopram at no cost for the STAR*D study. Most important, we thank the study participants, without whom this study would not be possible.
Web Resources
The URLs for data presented herein are as follows:
- Gene×Environment, Gene×Gene Interaction home page, http://hydra.usc.edu/gxe/ (for Quanto, v. 1.0 beta)
- International HapMap Project, http://www.hapmap.org/
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Cassano P, Fava M (2002) Depression and public health: an overview. J Psychosom Res 53:849–857 10.1016/S0022-3999(02)00304-5 [DOI] [PubMed] [Google Scholar]
- 2.Ustun TB, Ayuso-Mateos JL, Chatterji S, Mathers C, Murray CJ (2004) Global burden of depressive disorders in the year 2000. Br J Psychiatry 184:386–392 10.1192/bjp.184.5.386 [DOI] [PubMed] [Google Scholar]
- 3.Murray CJ, Lopez AD (1996) Evidence-based health policy—lessons from the Global Burden of Disease Study. Science 274:740–743 10.1126/science.274.5288.740 [DOI] [PubMed] [Google Scholar]
- 4.Thase ME, Haight BR, Richard N, Rockett CB, Mitton M, Modell JG, VanMeter S, Harriett AE, Wang Y (2005) Remission rates following antidepressant therapy with bupropion or selective serotonin reuptake inhibitors: a meta-analysis of original data from 7 randomized controlled trials. J Clin Psychiatry 66:974–981 [DOI] [PubMed] [Google Scholar]
- 5.Marangell LB (2001) Switching antidepressants for treatment-resistant major depression. J Clin Psychiatry Suppl 18 62:12–17 [PubMed] [Google Scholar]
- 6.Serretti A, Artioli P, Quartesan R (2005) Pharmacogenetics in the treatment of depression: pharmacodynamic studies. Pharmacogenet Genomics 15:61–67 [DOI] [PubMed] [Google Scholar]
- 7.Fava M, Schmidt ME, Zhang S, Gonzales J, Raute NJ, Judge R (2002) Treatment approaches to major depressive disorder relapse. Part 2. Reinitiation of antidepressant treatment. Psychother Psychosom 71:195–199 10.1159/000063644 [DOI] [PubMed] [Google Scholar]
- 8.Franchini L, Serretti A, Gasperini M, Smeraldi E (1998) Familial concordance of fluvoxamine response as a tool for differentiating mood disorder pedigrees. J Psychiatr Res 32:255–259 10.1016/S0022-3956(98)00004-1 [DOI] [PubMed] [Google Scholar]
- 9.Remillard AJ, Blackshaw SL, Dangor A (1994) Differential responses to a single antidepressant in recurrent episodes of major depression. Hosp Community Psychiatry 45:359–361 [DOI] [PubMed] [Google Scholar]
- 10.Stern SL, Rush AJ, Mendels J (1980) Toward a rational pharmacotherapy of depression. Am J Psychiatry 137:545–552 [DOI] [PubMed] [Google Scholar]
- 11.Malhotra AK, Murphy GM Jr, Kennedy JL (2004) Pharmacogenetics of psychotropic drug response. Am J Psychiatry 161:780–796 10.1176/appi.ajp.161.5.780 [DOI] [PubMed] [Google Scholar]
- 12.Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, Norquist G, Howland RH, Lebowitz B, McGrath PJ, Shores-Wilson K, Biggs MM, Balasubramani GK, Fava M, STAR*D Study Team (2006) Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 163:28–40 10.1176/appi.ajp.163.1.28 [DOI] [PubMed] [Google Scholar]
- 13.Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, Thase ME, Nierenberg AA, Quitkin FM, Kashner TM, Kupfer DJ, Rosenbaum JF, Alpert J, Stewart JW, McGrath PJ, Biggs MM, Shores-Wilson K, Lebowitz BD, Ritz L, Niederehe G (2004) Sequenced treatment alternatives to relieve depression (STAR*D): rationale and design. Control Clin Trials 25:119–142 10.1016/S0197-2456(03)00112-0 [DOI] [PubMed] [Google Scholar]
- 14.Hamilton M (1960) A rating scale for depression. J Neurol Neurosurg Psychiatry 23:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton M (1967) Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol 6:278–296 [DOI] [PubMed] [Google Scholar]
- 16.American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association, Washington, DC [Google Scholar]
- 17.Rush AJ, Trivedi MH, Ibrahim HM, Carmody TJ, Arnow B, Klein DN, Markowitz JC, Ninan PT, Kornstein S, Manber R, Thase ME, Kocsis JH, Keller MB (2003) The 16-item Quick Inventory of Depressive Symptomatology (QIDS), Clinician Rating (QIDS-C), and Self-Report (QIDS-SR): a psychometric evaluation in patients with chronic major depression. Biol Psychiatry 54:573–583 10.1016/S0006-3223(02)01866-8 [DOI] [PubMed] [Google Scholar]
- 18.Trivedi MH, Rush AJ, Ibrahim HM, Carmody TJ, Biggs MM, Suppes T, Crismon ML, Shores-Wilson K, Toprac MG, Dennehy EB, Witte B, Kashner TM (2004) The Inventory of Depressive Symptomatology, Clinician Rating (IDS-C) and Self-Report (IDS-SR), and the Quick Inventory of Depressive Symptomatology, Clinician Rating (QIDS-C) and Self-Report (QIDS-SR) in public sector patients with mood disorders: a psychometric evaluation. Psychol Med 34:73–82 10.1017/S0033291703001107 [DOI] [PubMed] [Google Scholar]
- 19.Cassano P, Fava M (2004) Tolerability issues during long-term treatment with antidepressants. Ann Clin Psychiatry 16:15–25 [DOI] [PubMed] [Google Scholar]
- 20.The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- 21.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, Papiol S, et al (2004) Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet 36 :1319–1325 [DOI] [PubMed] [Google Scholar]
- 23.Hahn MK, Mazei-Robison MS, Blakely RD (2005) Single nucleotide polymorphisms in the human norepinephrine transporter gene affect expression, trafficking, antidepressant interaction, and protein kinase C regulation. Mol Pharmacol 68:457–466 10.1124/mol.105.011270 [DOI] [PubMed] [Google Scholar]
- 24.Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, Bierle J, Doucet D, Milewski M, Yang R, Siegmund C, Haas J, Zhou L, Oliphant A, Fan JB, Barnard S, Chee MS (2004) Decoding randomly ordered DNA arrays. Genome Res 14:870–877 10.1101/gr.2255804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraemer HC, Thiemann S (1987) How many subjects? Statistical power analysis in research. Sage Publications, London [Google Scholar]
- 26.Gauderman WJ (2002) Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol 155:478–484 10.1093/aje/155.5.478 [DOI] [PubMed] [Google Scholar]
- 27.Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittke-Thompson JK, Pluzhnikov A, Cox NJ (2005) Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet 76:967–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 31.Bray NJ, Buckland PR, Hall H, Owen MJ, O’Donovan MC (2004) The serotonin-2A receptor gene locus does not contain common polymorphism affecting mRNA levels in adult brain. Mol Psychiatry 9:109–114 10.1038/sj.mp.4001366 [DOI] [PubMed] [Google Scholar]
- 32.Erdmann J, Shimron-Abarbanell D, Rietschel M, Albus M, Maier W, Korner J, Bondy B, Chen K, Shih JC, Knapp M, Propping P, Nothen MM (1996) Systematic screening for mutations in the human serotonin-2A (5-HT2A) receptor gene: identification of two naturally occurring receptor variants and association analysis in schizophrenia. Hum Genet 97:614–619 [DOI] [PubMed] [Google Scholar]
- 33.Parsons MJ, D’Souza UM, Arranz MJ, Kerwin RW, Makoff AJ (2004) The -1438A/G polymorphism in the 5-hydroxytryptamine type 2A receptor gene affects promoter activity. Biol Psychiatry 56:406–410 10.1016/j.biopsych.2004.06.020 [DOI] [PubMed] [Google Scholar]
- 34.Jaeschke R, Guyatt G, Barratt A, Walter S, Cook D, McAlister F, Attia J (2002) Measures of association. In: Gyuatt G, Rennie D (eds) User’s guides to the medical literature: a manual of evidence-based clinical practice. AMA Publications, Chicago, pp 351–368 [Google Scholar]
- 35.Anguelova M, Benkelfat C, Turecki G (2003) A systematic review of association studies investigating genes coding for serotonin receptors and the serotonin transporter. I. Affective disorders. Mol Psychiatry 8:574–591 10.1038/sj.mp.4001328 [DOI] [PubMed] [Google Scholar]
- 36.Skol AD, Scott LJ, Abecasis GR, Boehnke M (2006) Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38:209–213 10.1038/ng1706 [DOI] [PubMed] [Google Scholar]
- 37.Brown C, Schulberg HC, Sacco D, Perel JM, Houck PR (1999) Effectiveness of treatments for major depression in primary medical care practice: a post hoc analysis of outcomes for African American and white patients. J Affect Disord 53:185–192 10.1016/S0165-0327(98)00120-7 [DOI] [PubMed] [Google Scholar]
- 38.Norton N, Owen MJ (2005) HTR2A: association and expression studies in neuropsychiatric genetics. Ann Med 37:121–129 10.1080/07853890510037347 [DOI] [PubMed] [Google Scholar]
- 39.Peters EJ, Slager SL, McGrath PJ, Knowles JA, Hamilton SP (2004) Investigation of serotonin-related genes in antidepressant response. Mol Psychiatry 9:879–889 10.1038/sj.mp.4001502 [DOI] [PubMed] [Google Scholar]
- 40.Choi MJ, Kang RH, Ham BJ, Jeong HY, Lee MS (2005) Serotonin receptor 2A gene polymorphism (-1438A/G) and short-term treatment response to citalopram. Neuropsychobiology 52:155–162 10.1159/000087847 [DOI] [PubMed] [Google Scholar]
- 41.Cusin C, Serretti A, Zanardi R, Lattuada E, Rossini D, Lilli R, Lorenzi C, Smeraldi E (2002) Influence of monoamine oxidase A and serotonin receptor 2A polymorphisms in SSRI antidepressant activity. Int J Neuropsychopharmacol 5:27–35 10.1017/S1461145701002711 [DOI] [PubMed] [Google Scholar]
- 42.Minov C, Baghai TC, Schule C, Zwanzger P, Schwarz MJ, Zill P, Rupprecht R, Bondy B (2001) Serotonin-2A-receptor and -transporter polymorphisms: lack of association in patients with major depression. Neurosci Lett 303:119–122 10.1016/S0304-3940(01)01704-9 [DOI] [PubMed] [Google Scholar]
- 43.Leysen JE (2004) 5-HT2 receptors. Curr Drug Targets CNS Neurol Disord 3:11–26 10.2174/1568007043482598 [DOI] [PubMed] [Google Scholar]
- 44.Mann JJ, Curreier D, Quiroz JA, Manji HK (2005) Neurobiology of severe mood and anxiety disorders. In: Siegel GJ, Agranoff BW, Albers RW, Fisher SK, Uhler MD (eds) Basic Neurochemistry. Lippincott-Raven, New York, pp 887–911 [Google Scholar]
- 45.Humphrey PP, Hartig P, Hoyer D (1993) A proposed new nomenclature for 5-HT receptors. Trends Pharmacol Sci 14:233–236 10.1016/0165-6147(93)90016-D [DOI] [PubMed] [Google Scholar]
- 46.Drevets WC (2000) Neuroimaging studies of mood disorders. Biol Psychiatry 48:813–829 10.1016/S0006-3223(00)01020-9 [DOI] [PubMed] [Google Scholar]
- 47.Manji HK, Drevets WC, Charney DS (2001) The cellular neurobiology of depression. Nat Med 7:541–547 10.1038/87865 [DOI] [PubMed] [Google Scholar]
- 48.Peremans K, Audenaert K, Hoybergs Y, Otte A, Goethals I, Gielen I, Blankaert P, Vervaet M, van Heeringen C, Dierckx R (2005) The effect of citalopram hydrobromide on 5-HT2A receptors in the impulsive-aggressive dog, as measured with 123I-5-I-R91150 SPECT. Eur J Nucl Med Mol Imaging 32:708–716 10.1007/s00259-005-1772-5 [DOI] [PubMed] [Google Scholar]
- 49.Strome EM, Clark CM, Zis AP, Doudet DJ (2005) Electroconvulsive shock decreases binding to 5-HT2 receptors in nonhuman primates: an in vivo positron emission tomography study with [18F]setoperone. Biol Psychiatry 57:1004–1010 10.1016/j.biopsych.2005.01.025 [DOI] [PubMed] [Google Scholar]
- 50.Papolos DF, Yu YM, Rosenbaum E, Lachman HM (1996) Modulation of learned helplessness by 5-hydroxytryptamine2A receptor antisense oligodeoxynucleotides. Psychiatry Res 63:197–203 10.1016/0165-1781(96)02935-6 [DOI] [PubMed] [Google Scholar]
- 51.Coyle JT, Duman RS (2003) Finding the intracellular signaling pathways affected by mood disorder treatments. Neuron 38:157–160 10.1016/S0896-6273(03)00195-8 [DOI] [PubMed] [Google Scholar]
- 52.Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM (2002) Neurobiology of depression. Neuron 34:13–25 10.1016/S0896-6273(02)00653-0 [DOI] [PubMed] [Google Scholar]
- 53.Vaidya VA, Marek GJ, Aghajanian GK, Duman RS (1997) 5-HT2A receptor-mediated regulation of brain-derived neurotrophic factor mRNA in the hippocampus and the neocortex. J Neurosci 17:2785–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]