Abstract
Acid sphingomyelinase (ASM) is the lipid hydrolase that is deficient in types A and B Niemann-Pick disease (NPD). Here, we demonstrate that the gene encoding ASM (SMPD1) is paternally imprinted and that differential expression of the mutant alleles in patients with ASM-deficient NPD and in carriers influences the disease phenotype. Comparison of the results of genomic sequencing versus reverse-transcriptase polymerase chain reaction sequencing for several patients with NPD revealed preferential expression of one mutant allele. Further analysis of one family showed that the expressed allele was maternally inherited and that the distinct clinical presentations of the individual patients were correlated with the amount of residual ASM activity expressed from the maternal mutation. Treatment of NPD cell lines with 5-aza-2′-deoxycytidine enhanced the expression of the paternal SMPD1 allele, and bisulfite genomic sequencing identified which CpG dinucleotides within the SMPD1 promoter were methylated. In a related set of studies, we identified a carrier individual who had ∼15% of normal ASM activity and clinical features of ASM-deficient NPD. DNA sequencing confirmed that this individual carried a single SMPD1 mutation and that this mutant allele was preferentially expressed. These data thus demonstrate, for the first time, imprinting at the SMPD1 gene and reveal the influence of this epigenetic modification on the presentation of ASM-deficient NPD.
Acid sphingomyelinase (ASM) is the lipid hydrolase that is deficient in types A and B Niemann-Pick disease (NPD) (MIMs 257200 and 607616, respectively).1 Individuals with this disorder inherit two mutations in the SMPD1 gene (MIM 607608), which is located on the short arm of chromosome 11 within a cluster of imprinted genes (fig. 1).2 The clinical presentation of ASM-deficient NPD is dependent, in large part, on the amount of residual enzymatic activity expressed in various tissues from the individual mutant alleles.1 Severe mutations lead to type A NPD, an infantile form of the disease that follows a uniform and rapid neurodegenerative course, leading to death before age 4 years. Type B NPD, on the other hand, is a very heterogeneous disorder that may present in early childhood or adulthood, affects multiple organ systems, and progresses at variable rates in different patients. The basis of this variability is poorly understood and does not always correlate with the individual SMPD1 mutations. In addition, some carrier individuals for ASM-deficient NPD have features of the disease, which is an unusual finding for an autosomal recessive disorder such as this.3
Figure 1.
Schematic representation of the imprinting cluster on the short arm of human chromosome 11, with the location of the SMPD1 gene shown. Paternally imprinted genes (black boxes), maternally imprinted genes (gray boxes), and genes exhibiting biallelic expression (white boxes) are shown. The two BWS candidate regions (BWSCR1 and BWSCR2) are indicated. Only those human genes for which imprinting has been confirmed are shown. cen = centromere; tel = telomere.
In 2000, a patient with Beckwith-Wiedemann syndrome (BWS) was described who had ∼30% of normal ASM activity in cultured skin fibroblasts.4 This 23-mo-old child had a normal karyotype, which suggested uniparental disomy of paternal chromosome 11p15. Such disomic events are responsible for ∼20% of all BWS cases.5 An accompanying article from the same group suggested that the SMPD1 gene might be imprinted, on the basis of the reduced ASM activity in this patient, the possibility of chromosome 11p15 uniparental disomy, and the fact that the SMPD1 gene had structural features common to other imprinted genes.6
To assess imprinting at the SMPD1 locus, we studied several heteroallelic patients with ASM-deficient NPD. All patients had a confirmed enzymatic diagnosis of ASM-deficient NPD and voluntarily provided written informed consent before participation in this study. Patient 1 was a 22-year-old white man who had presented with NPD at age 18 mo. During childhood, he had recurrent otitis media infections, sinusitis, and pneumonias, and he underwent a partial splenectomy at age 12 years. Neurological and fundoscopic examinations were normal, although pulmonary function testing revealed a diffusion capacity of 54% of the predicted value. Patient 1 also has the typical cholesterol abnormalities associated with ASM-deficient NPD. Patients 2 and 3 are adult siblings who are first cousins of patient 4 and were identified after patient 4 received a diagnosis of ASM-deficient NPD at age 1 year (see pedigree in fig. 2A). Patients 2 and 3 have a mild disease phenotype, with normal neurological and ophthalmologic findings and mild hepatosplenomegaly. Pulmonary function has remained essentially normal in both patients. In contrast, patient 4 has severe ASM-deficient NPD and presented with hepatosplenomegaly in early childhood, which led to bone marrow transplantation at age 2 years.
Figure 2.
Expression pattern of SMPD1 mutant alleles in ASM-deficient NPD. A, Pedigree of the family with three affected members. Patients 2 and 3 are adult siblings with mild ASM-deficient NPD. Patient 4, a paternally related cousin, is a child with severe ASM-deficient NPD. Genotype analysis identified the shared paternal (C92W) and unique maternal (S248R and P184L) mutations. Symbols with a dot indicate obligate carriers. Blackened symbols indicate affected individuals. B, Normal and mutant C92W alleles, distinguishable by restriction-enzyme digestion with MwoI after RT-PCR amplification. The enzyme digested the amplified 239-bp fragment containing the wild-type (C92) maternal sequence into a smaller 129-bp fragment. After digestion of radioactive RT-PCR products from each of the three patients and quantification of the signal from the paternal (W92 mutant) and maternal (C92 wild-type) fragments, expression from the maternal allele was found to account for ⩾∼80% of the products. An unrelated, unaffected individual was included as a control. In this individual, both the maternal and the paternal alleles had the normal C92 sequence; thus, only the lower band is evident. Lanes show PCR reactions with (“+”) and without (“−”) the cDNA template. The numbers below each lane marked with a plus sign (+) indicate the percentages of paternal (top number) and maternal (bottom number) alleles, as determined by densitometry and IMAGEQUANT software.
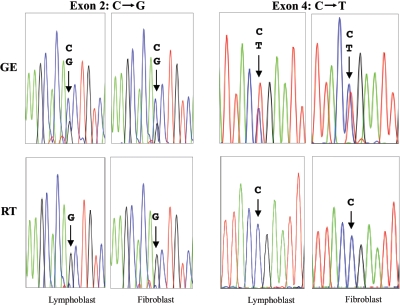
As illustrated in figure 3, two distinct SMPD1 mutations were readily identified in these patients after direct sequencing of genomic DNA. However, sequencing of RT-PCR products from the same individuals revealed that one of the mutant alleles (in this case, the allele expressing the mutant G in exon 2 and the wild-type C in exon 4) was preferentially expressed.
Figure 3.
Comparison of genomic sequencing (GE) versus RT-PCR sequencing (RT) in ASM-deficient NPD. Results of sequencing two different cell lines (lymphoblasts and fibroblasts) from patient 1 are shown. This individual had two distinct genomic mutations caused by nucleotide substitutions in exons 2 and 4 (C→G and C→T, respectively). When genomic sequencing was performed on either cell line, both mutations were evident (arrows). Note that the signal intensities (peak heights) for the wild-type and mutant nucleotides were approximately equivalent, which is consistent with heterozygosity of each mutation. However, when RNA was prepared from the same cell lines and the reverse-transcribed cDNA products were sequenced, only one of the alleles was preferentially expressed (arrows). This preferentially expressed allele carried the mutant nucleotide (G) in exon 2 and the wild-type nucleotide (C) in exon 4.
To further evaluate preferential expression of the maternal or paternal SMPD1 alleles, we investigated the family with the three NPD-affected individuals. As mentioned above, two of the members of this family (patients 2 and 3) were adult siblings with clinically mild NPD, whereas the third (patient 4) was a paternally related first cousin with severe NPD (fig. 2A). The shared paternal mutation in these patients was at codon 92 (W92) and could be distinguished from the wild-type (maternal) allele (C92) by restriction-enzyme digestion with MwoI, which detects the C→G nucleotide change at nucleotide position 276 of the full-length cDNA sequence.7 Figure 2B illustrates that, after semiquantitative RT-PCR and restriction-enzyme analysis of each patient and an unrelated normal control individual, the maternal SMPD1 allele was found to be preferentially expressed, which is consistent with paternal imprinting at this locus.
We next introduced the maternal (expressed) SMPD1 mutations found in these three patients (i.e., P184L for patients 2 and 3, and S248R for patient 4) into the full-length ASM cDNA and transiently expressed the mutant constructs in COS-1 cells. The P184L mutation found in the two adult patients with mild disease expressed ∼8% of normal ASM activity in COS-1 cells, whereas the S248R mutation found in the child with severe disease expressed no residual ASM activity. Moreover, the P184L mutation had been previously identified in several unrelated patients with type B NPD and often has been associated with a less severe form of the disorder.8 Thus, the enzymatic activity produced from the expressed maternal allele in these patients correlated with the clinical presentation of the disease.
To extend these observations, cultured lymphoblasts from patients 2, 3, and 4 and a normal individual were grown with and without the demethylating agent 5-aza-2′-deoxycytidine (5Aza-dC) for 5–6 d. After 5Aza-dC treatment of the lymphoblasts, the expression of the paternal allele was enhanced and became equivalent to that of the maternal allele, as illustrated in figure 4 for patient 4. Before 5Aza-dC treatment, expression of the paternal allele was ∼20% that of the maternal allele. These results suggest that methylation was responsible for the reduced expression of the SMPD1 paternal allele.
Figure 4.
Enhanced expression of the paternal SMPD1 gene after 5Aza-dC treatment. Cultured fibroblasts from a representative ASM-deficient individual (patient 4) were grown for 5 d with (20 μM) and without (0 μM) 5Aza-dC. In the presence of 5Aza-dC, expression of the mutant (paternal) allele was enhanced, as shown. Lanes represent amplification reactions with (“+”) and without (“−” [negative control]) the cDNA template. The numbers below the lanes marked with a plus sign (+) indicate the percentages of mutant paternal (top number) and wild-type maternal (bottom number) alleles.
We next performed bisulfite genomic sequencing of two PCR fragments derived from the wild-type SMPD1 promoter region before and after 5Aza-dC treatment. Fragment 1 included nt −340 to +279, and fragment 2 included nt −889 to −316 (numbered in accordance with Schuchman et al.9). Although none of the 55 CpG dinucleotides in fragment 1 were methylated under any conditions, fragment 2 contained six methylated CpG sites, some of which were demethylated in the presence of 5Aza-dC. Sixteen nonmethylated CpG sites were also present in fragment 2. Figure 5 summarizes the methylation patterns of the relevant six CpG sites in patient 3. Each circle represents one of the six sites, and methylation is indicated by a blackened circle. For these studies, a PCR fragment containing the six sites was amplified from cells with and without 5Aza-dC treatment, and the amplified fragments were subsequently subcloned into a plasmid vector for bisulfite sequencing. A total of 26 clones from cells without 5Aza-dC treatment and 23 clones from cells with treatment were subjected to bisulfite sequencing. The methylation pattern of individual clones are schematically depicted in figure 5A, and the data are summarized in figure 5B.
Figure 5.
Methylated CpG dinucleotides in the SMPD1 promoter, identified by bisulfite sequencing. Two fragments within the putative SMPD1 promoter region were PCR amplified from cells grown with and without 5Aza-dC, and the amplified fragments were subjected to bisulfite sequencing. No methylated sites were found in fragment 1, whereas six were found in fragment 2. A, Schematic depiction of the six CpG sites that were differentially methylated in fragment 2. A total of 26 clones were sequenced from cells grown without 5Aza-dC, and 23 clones from cells grown with 5Aza-dC. Methylation patterns of the individual clones are shown, and the number of clones found with each pattern is indicated (1×, 5×, etc.). Blackened circles represent methylated sites; unblackened circles represent unmethylated sites. B, Summary of bisulfite sequencing data for the six methylated CpG sites in fragment 2. The number of clones containing each of the methylated sites is shown for the two groups of clones (26 from untreated cells [0 μM] and 23 from treated cells [20 μM]). Note that 5Aza-dC treatment led to substantial demethylation of sites 1–4.
As can be seen, all CpG sites were methylated in only 1 (“1×”) of the 26 clones analyzed in the absence of 5Aza-dC treatment. In five clones (“5×”), sites 1–4 were methylated; in another clone, sites 1, 2, 3, and 6 were methylated; and so forth. The most common pattern observed for these clones was methylation at sites 1, 2, and 4 (in six clones). Of the six sites in this region that were methylated, sites 1–4 were differentially modified in the 23 clones from cells treated with 5Aza-dC. These results suggest that methylation at one or all of these four sites is likely to be responsible for the imprinting observed at the SMPD1 locus.
As noted above, there have been previous reports describing individuals who carry a single SMPD1 mutation and exhibit features of ASM-deficient NPD (e.g., the report by Lee et al.3). This is an unusual finding for a recessive disorder, and we therefore hypothesized that some individuals who are carriers for ASM-deficient NPD might exhibit features of the disease because of inheritance of the mutant allele on the maternal (i.e., preferentially expressed) chromosome. To investigate this hypothesis, we studied an individual (patient 5) who had received the diagnosis of ASM-deficient NPD as a child (on the basis of splenomegaly, bone marrow foamy cells, and recurrent pulmonary infections) but who was found to have ∼15% of normal ASM activity in cultured cells, which is more consistent with carrier status than with patient status. As an adult, most of his NPD-related symptoms had resolved, but, on reevaluation at age 42 years, he was found to have abnormal low-density lipoprotein and high-density lipoprotein cholesterol, splenomegaly, and several other features associated with ASM deficiency.
Genomic DNA sequencing revealed the presence of a single fsP330 mutation in this individual (fig. 6), which is consistent with his Ashkenazi Jewish ancestry.10 Notably, RT-PCR sequencing demonstrated preferential expression of this mutant allele. Although the parents of patient 5 were not available for study, we predict, on the basis of our previous data, that the preferentially expressed mutant allele was maternally inherited. Notably, this carrier individual has two children, one of whom inherited the fsP330 mutation. However, no NPD-related symptoms in this child have been described, which is consistent with the fact that he inherited the fsP330 mutation on his paternal (imprinted) chromosome. Also, ASM activity in cultured skin fibroblasts from this child was ∼54% of normal, which was significantly greater than that seen in the father and thus consistent with preferential expression of the maternal wild-type allele.
Figure 6.
Comparison of genomic sequencing (GE) versus RT-PCR sequencing (RT) in an ASM-deficient NPD heterozygote. Cultured skin fibroblasts from a carrier individual (patient 5) were studied. This individual had one genomic mutation due to deletion of a cytosine (Del C), leading to a frameshift mutation (fsP330). As can be seen, when direct genomic sequencing was performed, both the wild-type and the mutant alleles were evident. Note the overlap of the sequencing signals at the site of the deletion (arrows), consistent with a frameshift. Sequencing reactions from both directions (sense and antisense) are shown. In contrast to the genomic sequencing results, when the reverse-transcribed cDNA products were sequenced from the same individual, preferential expression of the mutant allele (containing five cytosines instead of six) was found.
Thus, we have demonstrated paternal imprinting at the SMPD1 gene and have revealed that the maternal mutation has a predominant influence on the presentation of ASM-deficient NPD. These findings may explain some of the clinical heterogeneity in this disorder. For example, two heteroallelic individuals with the same genotype may have distinct phenotypes, depending on whether the severe mutation is inherited on the expressed (maternal) chromosome or the imprinted (paternal) chromosome. This observation may also explain why some carrier individuals exhibit some features of NPD.3 For example, NPD carriers who inherit the wild-type allele on their paternal chromosome (and thus preferentially express the maternal mutant allele) may have reduced ASM activity (i.e., less than the expected 50%) and perhaps may develop clinical features of NPD. We confirmed this hypothesis in one such carrier individual (patient 5), who preferentially expressed a mutant allele carrying a severe frameshift mutation. As mentioned above, patient 5 had ∼15% of normal ASM activity and, as a child, was diagnosed with NPD on the basis of splenomegaly, bone marrow foamy cells, and recurrent pulmonary infections.
Our findings may also provide insights into the pathogenesis of BWS. As noted above, the basis for this work was the identification of an individual with BWS who had reduced ASM activity. Presumably, this reduced activity was due to uniparental disomy of paternal chromosome 11. In the future, it will be informative to study other individuals with BWS who have confirmed uniparental disomy of paternal chromosome 11p15 (∼20% of all cases) and to examine their ASM levels. In addition, the status of the SMPD1 gene should be examined in individuals with BWS who have other types of rearrangements involving the short arm of chromosome 11. Indeed, because ASM is involved in regulating cell growth through the ceramide-mediated signaling pathway,11,12 reduced activity of this enzyme may contribute to the overgrowth and embryonic tumors associated with this disorder.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant R01 HD28607-15 (to E.H.S.). M.M.M. is the recipient of an NIH Mid-Career Patient-Oriented Research Career Development Award (K24 RR021991-01).
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for types A and B NPD and SMPD1)
References
- 1.Schuchman EH, Desnick RJ (2001) Types A and B Niemann-Pick disease: acid sphingomyelinase deficiencies. In: Scriver C, Beaudet A, Sly W, Valle D (eds) The metabolic and molecular basis of inherited disease. 8th ed. McGraw-Hill, New York, pp 3589–3610 [Google Scholar]
- 2.da Veiga Pereira L, Desnick RJ, Adler DA, Disteche CM, Schuchman EH (1991) Regional assignment of the human acid sphingomyelinase gene (SMPD1) by PCR analysis of somatic cell hybrids and in situ hybridization to 11p15.1-p15.4. Genomics 9:229–234 10.1016/0888-7543(91)90246-B [DOI] [PubMed] [Google Scholar]
- 3.Lee CY, Krimbou L, Vincent J, Bernard C, Larramee P, Genest J Jr, Marcil M (2003) Compound heterozygosity at the sphingomyelin phosphodiesterase-1 (SMPD1) gene is associated with low HDL cholesterol. Hum Genet 112:552–562 [DOI] [PubMed] [Google Scholar]
- 4.Réthy LA, Kálmánchey R, Klubjer V, Koós R, Fekete G (2000) Acid sphingomyelinase deficiency in Beckwith-Weidemann syndrome. Pathol Oncol Res 6:295–297 [DOI] [PubMed] [Google Scholar]
- 5.Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, Bowdin SC, Riccio A, Sebastio G, Bliek J, Schofield PN, Reik W, Macdonald F, Maher ER (2005) Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur J Hum Genet 13:1025–1032 10.1038/sj.ejhg.5201463 [DOI] [PubMed] [Google Scholar]
- 6.Réthy LA (2000) Growth regulation, acid sphingomyelinase gene and genomic imprinting: lessons from an experiment of nature. Pathol Oncol Res 6:298–300 [PubMed] [Google Scholar]
- 7.Schuchman EH, Suchi M, Takahashi T, Sandhoff K, Desnick RJ (1991) Human acid sphingomyelinase: isolation, nucleotide sequence, and expression of the full-length and alternatively spliced cDNAs. J Biol Chem 266:8531–8539 [PubMed] [Google Scholar]
- 8.Simonaro CM, Desnick RJ, McGovern MM, Wasserstein MP, Schuchman EH (2002) The demographics and distribution of type B Niemann-Pick disease: novel mutations lead to new genotype/phenotype correlations. Am J Hum Genet 71:1413–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuchman EH, Levran O, Pereira LV, Desnick RJ (1992) Structural organization and complete nucleotide sequence of the gene encoding human acid sphingomyelinase (SMPD1). Genomics 12:197–205 10.1016/0888-7543(92)90366-Z [DOI] [PubMed] [Google Scholar]
- 10.Levran O, Desnick RJ, Schuchman EH (1993) Type A Niemann-Pick disease: a frame-shift mutation in the acid sphingomyelinase gene (fsP330) occurs in Asheknazi Jewish patients. Hum Mut 2:317–319 10.1002/humu.1380020414 [DOI] [PubMed] [Google Scholar]
- 11.Gulbins E, Kolesnick R (2003) Raft ceramide in molecular medicine. Oncogene 22:7070–7077 10.1038/sj.onc.1207146 [DOI] [PubMed] [Google Scholar]
- 12.Jaffrezou JP, Laurent G (2004) Ceramide: a new target in anticancer research? Bull Cancer 91:E133–E161 [PubMed] [Google Scholar]