Abstract
Individuals born of consanguineous union have segments of their genomes that are homozygous as a result of inheriting identical ancestral genomic segments through both parents. One consequence of this is an increased incidence of recessive disease within these sibships. Theoretical calculations predict that 6% (1/16) of the genome of a child of first cousins will be homozygous and that the average homozygous segment will be 20 cM in size. We assessed whether these predictions held true in populations that have preferred consanguineous marriage for many generations. We found that in individuals with a recessive disease whose parents were first cousins, on average, 11% of their genomes were homozygous (n=38; range 5%–20%), with each individual bearing 20 homozygous segments exceeding 3 cM (n=38; range of number of homozygous segments 7–32), and that the size of the homozygous segment associated with recessive disease was 26 cM (n=100; range 5–70 cM). These data imply that prolonged parental inbreeding has led to a background level of homozygosity increased ∼5% over and above that predicted by simple models of consanguinity. This has important clinical and research implications.
Individuals whose parents are related are expected to have an increased proportion of their autosomal genome that is homozygous. The more closely the parents are related, the greater this effect is expected to be.1 Offspring of second cousins are expected to have children with 1/64 of their genome homozygous; offspring of first cousins, 1/16; offspring of double–first cousins, 1/8; and offspring of incestuous union, 1/4.2 Furthermore, in the case of first-cousin offspring, it has been calculated that the average homozygous segment will be 20 cM.3 This degree of homozygosity is far greater than that seen in apparently outbred populations.4 The clinical consequence of this is an increased incidence of autosomal recessive diseases (and, to a lesser extent, homozygous dominant diseases) in consanguineous populations.5–9
There are clear social and economic advantages to consanguinity.10,11 Most populations that have practiced consanguineous marriage have done so for many generations.12 The effect of prolonged consanguinity on the degree of homozygosity seen in such populations has not been estimated directly. We sought to address this question by examining individuals with autosomal recessive disease drawn from two populations with a long history of consanguinity (Pakistani and Arab) and a third population with a short history of consanguinity (Irish traveler, a distinct ethnic group originating from Ireland, with their own culture and language). We also sought to determine the number of homozygous segments exceeding a defined length in both disease-associated and -nonassociated homozygous segments in individuals from these populations. We reexamined linkage data generated from polymorphic microsatellite markers and SNPs.
The coefficient of inbreeding (F) gives the probability that a locus will be identical by descent in an individual (and the proportion of the autosomal genome that will be homozygous). In clinical practice, this can rarely be calculated with confidence, because of incomplete knowledge of a sufficient ancestry.13 Generally, in human genetics, the closest relationship between parents is used to estimate F, under the assumption that all grandparents are only distantly related. Here, we show that, in two populations with a long history of consanguinity, this assumption leads to a significant underestimation of the degree of homozygosity.
Our study population comprised affected individuals from two sets of families with diverse autosomal recessive diseases: Alstrom syndrome (MIM 203800), Fuhrmann syndrome (MIM 228930), Lebers amaurosis (MIM 204000), nonsyndrome mental retardation (MIM 607417), oro-facial-digital syndrome (MIM 258850), retinitis pigmentosa (MIM 268000), primary microcephaly (MIM 251200), Seckel syndrome (MIM 210600), Warburg micro syndrome (MIM 600118), and six different previously uncharacterized neurodegenerative disorders. The first set of families was screened for linkage with SNPs; the second, with microsatellite markers (table 1). We examined the majority of ascertained families and drew pedigrees with careful attention to the parental relationship of affected individuals. Formal calculation of the minimal inbreeding coefficient for each affected offspring was not possible, because pedigrees were known in detail for only 3 or 4 generations.
Table 1.
Details of the Study Individuals and Their Families[Note]
|
Disease Location |
||||||
|
Set, Data Type, and Ethnicity |
No. of Families |
No. of Individuals | No. of First Cousins | Proven by Mutation Analysis |
Proven by Linkage Analysis |
Untested or Unknown |
| 1: | ||||||
| SNP: | ||||||
| Pakistani | 10 | 29 | 22 | 9 | 13 | 7 |
| Arab | 5 | 19 | 16 | 3 | 16 | 0 |
| Irish Traveler | 1 | 10 |
10 |
0 |
10 |
0 |
| Total | 16 | 58 | 48 | 12 | 39 | 7 |
| 2: | ||||||
| Linkage dataa: | ||||||
| Pakistani | 52 | 183 | 149 | 17 | 166 | 0 |
| Arab | 2 |
16 |
12 |
0 |
16 |
0 |
| Total | 54 | 199 | 161 | 17 | 182 | 0 |
Note.— Individuals from set 1 were analyzed using a whole-genome array of SNPs, whereas those from set 2 were analyzed with polymorphic microsatellite markers for the region of linkage only.
Data were complete for 97 individuals, 65 of whom were first cousins. Disease location was proven by mutation analysis for 48 individuals, and, by linkage analysis, for 17 individuals.
The first set comprised 58 affected individuals from 16 families; 48 of the 58 were born of first-cousin marriages—22 Pakistani and 16 Arab—and the remaining 10 were Irish travelers. These 10 individuals were born of double–first-, second-, or third-cousin marriages. In 12 of the 58 individuals, the pathogenic disease-causing mutation has been found. In a further 39 of 58, a linkage to a homozygous segment containing the causative gene has been found, with a LOD score >3.3 at recombination (θ)=0. For 7 of 58, linkage studies either have not yet been performed or await analysis.
In this first set of 58 individuals, genomic DNA was extracted, amplified, and hybridized to Affymetrix 10K-SNP chips, per the manufacturer's instructions. The resultant SNP allele output was analyzed using the ExcludeAR program.14 The data generated were used to determine the number of homozygous segments >3 cM, their size, the total amount of homozygosity, and the proportion of excess of homozygosity over that expected from random mating. The size of the homozygous segment including the gene locus was also determined in the 51 individuals whose mutation or linkage was known. Chromosomal segments were accepted to be homozygous if they contained ⩾20 homozygous SNPs, for an average detectable DNA segment length of 7 cM (for rationale, see appendix A [online only]). The selection of the number of homozygous consecutive SNPs was inevitably a compromise: a greater number of SNPs, say 30, would have led to rejection of 5%–20% of homozygous segments and a reduction in total homozygosity of 5%–10%; fewer SNPs, say 15, would have led to the inclusion of 10%–20% more segments, the larger of which would probably have been mostly false-positive results, with subsequent inflation of the number of segments and amount of homozygosity by 5%–15%.
The families in the second set had all been studied prior to routine availability of SNP analysis, and the disease loci had been placed using polymorphic microsatellite markers. In the course of linkage being determined, a marker density of <2 cM was used to surround each disease locus (table 1). The length of the homozygous region was taken to be from the most proximal to the most distal homozygous marker, with use of genetic distances from the deCode genetic map (Marshfield genetic distance data gave very similar results [Center for Medical Genetics]) (UCSC Genome Bioinformatics).15 This second set included 199 individuals from 54 families for which either a homozygous pathogenic mutation was found or the family LOD score was >3.3 at θ=0. Of these 199 individuals, 65 were born of first cousins, and the size of the homozygous region encompassing their disease-causing mutation could be determined. A further 96 individuals were also born of first cousins, but only a minimum size of the homozygous region encompassing the disease/mutation–causing region could be determined, since one or both heterozygous boundaries of the homozygous segment has not been defined
Individuals with autosomal recessive disease whose parents were first cousins constituted the majority of our data, from both family sets. This is a direct consequence of the fact that first-cousin union is the most common form of consanguinity in the Pakistani and Arab populations, which we study for autosomal recessive diseases.6,12,16,17 We restricted our detailed analysis to first-cousin offspring and describe below the data for other degrees of consanguinity.
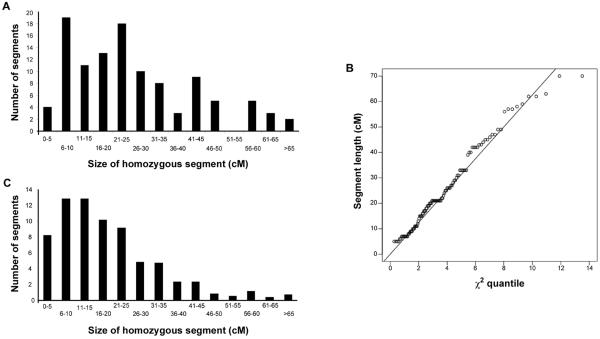
We first determined the size of the homozygous segment that contains a recessive disease–causing mutation in the offspring of first cousins from whom we had complete information (a homozygous pathogenic mutation or a LOD score >3.3) (fig. 1 and table 2). From family set 1, we had data for 35 offspring of first-cousin marriages. From the complete data set of 58 individuals of set 1, we excluded non–first-cousin offspring (10 of the 58); the 10 Irish travelers, since they did not have a known past history of consanguinity (see below); and the 3 individuals for whom no linkage/mutation had been found. For the analysis of individuals with autosomal recessive disease whose parents were first cousins, we combined data from the 35 individuals from family set 1 (analyzed with SNPs) and the 65 individuals from family set 2 (analyzed with microsatellite markers and for whom full data were known). The mean size of the recessive disease–associated homozygous segment was 25.8 cM (n=100; range 5–70 cM; 95% CI 22.7–28.9 cM, under a χ2 distribution with 4 df). The distribution of size of the disease-associated homozygous segment is shown in figure 2A. Figure 2B shows that these disease-associated segments have a χ2 distribution with 4 df, as expected. The results obtained from the two data sources were 26.4 cM for SNPs and 25.5 cM for microsatellite markers. For the further 96 individuals of first-cousin marriages from set 2, the length of the homozygous segment was at least 4 cM. The longest homozygous segment found was 89 cM. When results were taken together for 196 individuals whose parents were first cousins, the homozygous disease-associated segment was always >4 cM.
Figure 1.
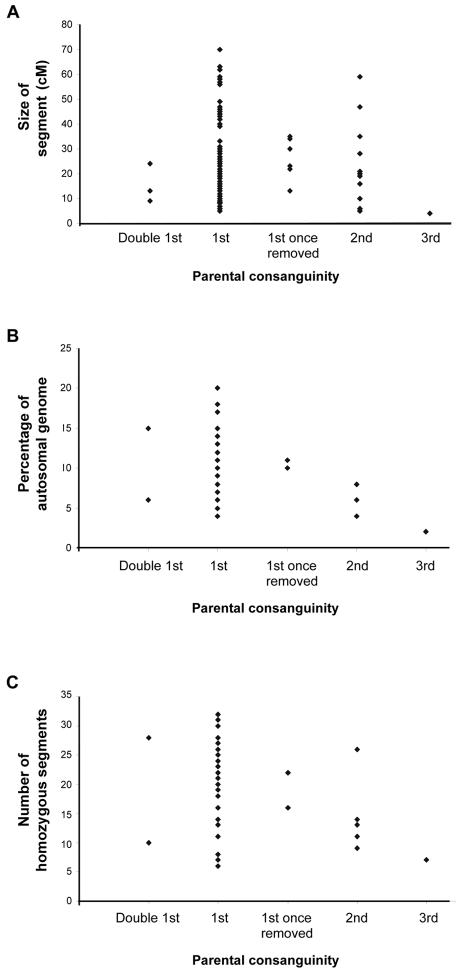
A, The size of the disease-associated homozygous segment in each of 100 individuals who have an autosomal recessive disease and whose parents are related. For these 100 individuals, the disease-associated homozygous segment was fully defined by flanking heterozygous markers. The degree of parental consanguinity is given for each individual on the X-axis. Offspring of double–first cousins would be expected to have 1/8 of the genome homozygous; first cousins, 1/16; first cousins once removed, 1/32; second cousins, 1/64; and third cousins, 1/256. B, The calculated total autosomal homozygosity for Pakistani and Arab individuals affected by autosomal recessive disease whose parents were related (n=48). C, The number of homozygous segments found in each of the Pakistani and Arab individuals shown in panel B.
Table 2.
Size of Disease-Causing Homozygous Segment in Individuals with Autosomal Recessive Disease Whose Parents Are First Cousins
|
Size of Disease-Causing Homozygous Segment(cM) |
|||
| Sample | Mean | Median | Range |
| All (n=100) | 26 | 22 | 5–70 |
| Individuals analyzed with: | |||
| SNP (n=35) | 30 | 27 | 5–63 |
| Microsatellite (n=65) | 24 | 20 | 5–70 |
Figure 2.
A, The distribution of size of disease-associated homozygous regions for 100 individuals whose parents were first cousins and who have an autosomal recessive disease for which the locus was known. The size of the segment is given in 5-cM increments. B, The ranked ordered lengths of homozygous segments, shown in panel A, surrounding the causal disease locus plotted against the corresponding quantiles of a χ2 distribution with 4 df. The line is calculated so that it passes through the median length. C, The distribution of size of all homozygous segments found in the 38 individuals from family set 1 whose parents were first cousins and who have an autosomal recessive disease. The size of the segment is given in 5-cM increments.
Second, we determined the longest homozygous segment present in the 48 Pakistani or Arab individuals; all were from family set 1 (we excluded the Irish traveler family, since their consanguinity was only recent). In 8 of 48, the largest homozygous segment was the disease-associated homozygous segment (see appendix B [online only]).
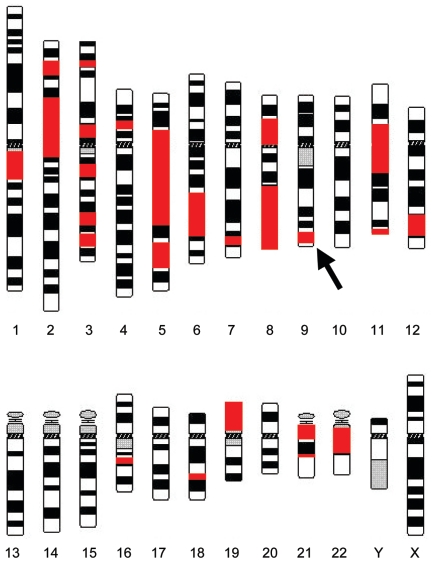
Next, we determined the extent of homozygosity in 38 individuals from set 1 whose parents were first cousins (fig. 1 and table 3). These individuals derive from pedigrees with multiple consanguineous marriages. The Irish travelers, with known consanguinity only in the most recent generation, were analyzed separately (see below). In Pakistanis and Arabs, the percentage of the genome that was homozygous and >3 cM in length was, on average, 11% (n=38; range 5%–20%; median 10%). They bore, on average, <20 homozygous segments (n=38; range 6–32; median 20); the average size was 19.5 cM (range 3–85 cM; median 16 cM). Figure 2C shows the distribution of size of all of the homozygous segments for the 38 offspring of first cousins. The average size of the longest autosomal homozygous segment was 54 cM (n=38; range 22–85 cM; median 57 cM). Figure 3 shows the distribution and size of all of the homozygous segments >3 cM for one illustrative study individual, plotted onto a chromosome ideogram.
Table 3.
Extent of Homozygosity of Set 1 Individuals Whose Parents Were First Cousins
|
AutosomalHomozygousRegion |
|||
|
Results for Set 1 (n=38) |
Genome Homozygosity (%) |
No. of Regions |
Largest Size (cM) |
| Mean | 11 | 20 | 54 |
| Median | 10 | 20 | 57 |
| Range | 5–20 | 6–32 | 22–85 |
Figure 3.
Ideogram of human chromosomes illustrating the position and size of the homozygous segments of one study individual. Homozygous segments are shown in red. The arrow indicates the recessive disease–associated homozygous segment. The individual was male, so the X and Y chromosomes would be expected to give homozygous results at all test SNP loci; this was the case for the X chromosome but was not assessed for the Y. Since this “homozygosity” between the X and Y chromosomes is the result of hemizygosity, it is not shown on the ideogram (University of Manitoba Department of Plant Science).
From set 1, 10 individuals resulted from degrees of inbreeding other than first cousins. These were two double–first cousins, two first cousins once removed, five second cousins, and one third cousin, all Pakistani or Arab. Grouped by degree of consanguinity, the percentage of their genome that was homozygous was, on average, 10%, 10%, 6%, and 2%, respectively (fig. 1B). These groups had 19, 19, 12, and 7 homozygous segments, respectively (fig. 1C). The average size of the longest autosomal homozygous segment in these groups was 69 cM, 55 cM, 33 cM, and 24 cM, respectively. Figure 1 plots the size of the disease-associated homozygous segment against the degree of parental consanguinity in sets 1 and 2 (full data given in appendices B and C [online only]). These data include 8 double–first cousins, 11 first cousins once removed, 13 second cousins, and 9 third cousins. For these groups, the average disease-associated homozygous segment sizes were 11 cM, 18 cM, 26 cM, and 4 cM, respectively. It appears that the proportion of genomic homozygosity shows some correlation with the degree of consanguinity, but, in this sample, the disease-associated homozygous segment does not show correlation, which perhaps reflects the unpredictable nature of crossover events.
We analyzed the Irish traveler family in which 10 individuals in three sibships had a neurodevelopmental disease. The parents were all first cousins, but there was no known further consanguinity in the 3 previous generations. Analysis showed that, on average, 6% of their genomes were homozygous and that the average size of the homozygous segment containing the disease locus was 21 cM. These figures are almost identical to those predicted for first cousins under the assumption of random mating, which suggests that the assumption of a history with absence of previous consanguinity was correct.
We appreciate that there are deficiencies in our data. First, the families we studied may be a biased sample and may not represent all Pakistani and Arab consanguineous families; obviously, ascertainment for recessive disorders will introduce bias. Even so, these families are of great research and clinical significance, so our findings remain highly relevant. Second, our clinical approach necessarily overestimates, by 1–3 cM, the length of homozygous regions defined by SNPs, since some genotypes at the boundaries of a homozygous region may be uninformative rather than identical by descent. Third, the extent of whole-genome homozygosity would have been underestimated by the exclusion of homozygous regions of <3 cM or >3 cM but that did not contain 20 homozygous consecutive SNPs (there were, on average, two potentially homozygous regions >5 cM per person). Finally, we will have underestimated the size of homozygous regions defined by microsatellite markers by 2–4 cM, because of the greater distance between these markers.
One of the most important findings in our data was that the longest homozygous segment was the disease-associated segment in 8 (17%) of 48 individuals. Since, on average, each individual bore 20 homozygous segments >3 cM, it seems reasonable to initially suggest that any 1 of these 20 segments could contain the disease-gene mutation with similar likelihood. And, since the range of lengths would be greatly influenced by time (mutation and recombination) and place (chromosome and recombination), the disease-associated segment ought to have been the largest in only 5% of the time. This result—the disease-bearing homozygous segment being the longest segment more often than expected (P<.05)—may be explained by the segment being of more recent origin or by the fact that ascertainment involves greater parental consanguinity. If the two copies of a known locus carried by an individual are inherited identical by descent from a common ancestor G generations ago, then the size of the surrounding region that is inherited identical by descent should have a χ2 distribution with 4 df (see fig. 2B) and a mean of 100/G cM. This would suggest that, on average, the disease-associated homozygous segments are inherited from a common ancestor ∼4 generations ago (100/26), whereas the remaining other homozygous segments were inherited ∼5 generations ago (100/21). The finding that the longest homozygous segment was the disease-associated segment reflects a selection bias—the studied individuals were ascertained because they had a recessive disease. However, it is this group who comes to medical and research attention and for whom the results are valid, rather than other family members who do not have recessive disease.
The clinician managing recessive disease in consanguineous families is often presented with only one or two affected children, and seeking linkage to a known locus is frequently the clinician's first step. Although homozygosity at a disease locus does not prove disease linkage in a consanguineous family, it can add support and, if no overlapping homozygous segments are found in affected individuals, can permit exclusion. If there were more than one autosomal locus capable of causing a recessive disease phenotype with equal prevalence, then homozygosity at one test locus for one child born of first-cousin parents would not always indicate detection of the disease locus; a false-linkage result would occur 1/5 of the time if there were two loci, 1/3 of the time for five loci, and 1/2 of the time for eight loci (table 4). When locus prevalence is considered instead of heterogeneity, a locus causing 12% of recessive disease–load homozygosity would indicate true linkage in only 1/2 of cases, and, for a prevalence of 4%, just 1/4 of cases. However, for a sib pair in similar circumstances, homozygosity is more significant; when there are two loci of equal frequency, homozygosity in both siblings at one of these loci, even in a highly consanguineous population, would indicate correct linkage in 95% of cases, and, even when there are eight loci of equal frequency, homozygosity in both siblings would indicate correct linkage in 81% of cases. Another clinical problem often encountered involves individuals or families with more than one recessive disease. The degree of homozygosity revealed in the present study goes some way in explaining this phenomenon18 (fig. 3).
Table 4.
Calculated Chance of True and False Linkage[Note]
|
Chance of Linkage to a Test Locus(%) |
|||
|
Affected Sample and No. of Recessive Loci |
Locus Prevalence (%) |
True | Spurious |
| Single subject: | |||
| 1 | 100 | 100 | 0 |
| 2 | 50 | 82 | 18 |
| 3 | 33 | 75 | 25 |
| 4 | 25 | 69 | 31 |
| 5 | 20 | 64 | 36 |
| 6 | 17 | 60 | 40 |
| 7 | 14 | 56 | 44 |
| 8a | 12 | 52 | 48 |
| 25a | 4 | 26 | 74 |
| Sib pair: | |||
| 1 | 100 | 100 | 0 |
| 2 | 50 | 95 | 5 |
| 3 | 33 | 92 | 7 |
| 4 | 25 | 90 | 10 |
| 5 | 20 | 88 | 12 |
| 6 | 17 | 86 | 14 |
| 7 | 14 | 84 | 16 |
| 8a | 12 | 81 | 19 |
| 25a | 4 | 59 | 41 |
Note.— Chance of true and false linkage was calculated for families in which a child has a recessive disease, the parents are first cousins, the disease phenotype can be caused by mutation at multiple recessive loci, and the child is homozygous for one of the loci. Calculation was done using the Bayes theorem, with use of locus prevalence and an 11% chance of being homozygous at any autosomal locus.
Illustration of real examples: autosomal recessive primary microcephaly, for which there are 8 loci known, and retinitis pigmentosa, for which there are 25 recessive loci.
For researchers mapping recessive loci in consanguineous families, excess homozygosity as the result of unexpected or hidden consanguinity has been reported as a practical problem in homozygosity mapping.19 Similar problems exist for the researcher wanting to include small consanguineous families in linkage analysis. Should all families apparently linked to a locus have mutations in the candidate gene? Our data suggest not (see table 4). Furthermore, it may be of use to determine the size of the apparent disease-associated homozygous segment in an affected individual from a small family; a homozygous disease-associated segment of <3 cM would be unusual in an offspring of first cousins.
In conclusion, we found that the amount of homozygosity is greater than expected (11% observed vs. 6% expected) in individuals with autosomal recessive disease whose parents are first cousins and who come from communities that frequently practice consanguineous marriage. First-cousin offspring had as much homozygosity as would have been expected for double–first cousin offspring. The method we used, analyzing 10,000 SNPs spread throughout the genome, seems capable of generating a “coefficient of inbreeding” for an individual by observation rather than by inferential methods. Although we have fewer data for individuals other than the children of first cousins, the degree of homozygosity in the data we do have is consistently greater than expected. The cause of this excess homozygosity is unclear from our data: it is unlikely that the pedigree information is significantly incorrect, since data were ascertained from multiple individuals in each family; recent previous first-cousin unions in the pedigree have little effect on the inbreeding coefficient. Ascertainment for a recessive disorder will, of course, preferentially ascertain the more-inbred parents. The effect may be the consequence of an effective small founder population or relative selection for haplotypes carrying no mutation compared with those that bear deleterious mutations. Empirically, the scientists and clinicians involved with consanguineous families should expect considerably more homozygosity than is predicted by simple calculations that are based on the known pedigree.
Acknowledgments
We thank Andrew Jackson, who wrote Neurobase, the database we used for the organization and analysis of families and markers. Neurobase is available from Dr. Andrew Jackson, Medical Research Council Human Genetics Unit, Western General Hospital, Crewe Road, Edinburgh, United Kingdom. We also thank the Wellcome Trust (grants 057964, 062444, 06424, 073243, and 073477) and Yorkshire Eye Research for funding. We acknowledge and thank Geneservice for their assistance in SNP analysis. Finally, we thank Prof. J. H. Edwards, Department of Biochemistry, University of Oxford, for enthusiasm and help at all stages of this project.
Appendix A: Rationale for the Use of 20 Consecutive SNPs to Detect Homozygous and Heterozygous Autosomal Genomic Segments
Homozygous segments were detected and defined by the use of at least 20 consecutive homozygous SNP allele calls. This figure was used as a compromise between the size of the segment to be detected, ideally >3 cM, and the probability of being able to detect a homozygous segment on a background of heterozygosity. The human genome is expected to be predominantly heterozygous.4 For the Affymetrix 10K-SNP chip 2, the allele call rate was 93%, on average, for the 9,857 autosomal SNPs. The declared average heterozygosity for each SNP was 0.38, and the mean genetic distance between SNPs was 0.36 cM. Thus, a heterozygous autosomal region of the genome containing 20 consecutive SNPs would yield consecutive homozygous SNP allele calls with a frequency of 0.38.20 The region would be 20×0.36 cM=7.2 cM. This must be multiplied by the number of consecutive runs of 20 observable SNPs and is based on the average call rate of 93%, which is 9,167. This result is the chance of observing each run of 20 consecutive homozygous SNPs per person per autosomal genome and is 0.000036, or log10−4.4. A homozygous region of the genome containing 20 SNPs would yield homozygous SNP allele calls by definition, a result that would occur by chance20 0.62×9,167 times—that is, 0.65 times per genome per individual. Thus, a homozygous run of 20 consecutive SNPs would be very rare in a heterozygous genome and would occur approximately once by chance per two individuals analyzed.
Appendix B
Table B1.
Results for Set 1 Individuals
| Family and Individual (Largest Homozygous Segment, in cM) | Parental Relationship (Cousins) |
Ethnicity | No. of Homozygous Segmentsa | Total Size (cM) |
Percentage of Autosomal Genomeb | Size of Linked Segmentc (cM) |
| 1: | ||||||
| 1 (82) | First | Pakistani | 24 | 683 | 20 | 25 |
| 2 (59) | First | Pakistani | 19 | 419 | 12 | 15 |
| 3 (56) | First | Pakistani | 20 | 292 | 8 | 56 |
| 2: | ||||||
| 1 (64) | Double first | Pakistani | 28 | 520 | 15 | 9 |
| 2 (74) | Double first | Pakistani | 10 | 210 | 6 | 13 |
| 3: | ||||||
| 1 (69) | First | Pakistani | 16 | 242 | 7 | ? |
| 2 (68) | First | Pakistani | 32 | 644 | 18 | ? |
| 3 (81) | First | Pakistani | 21 | 591 | 17 | ? |
| 4: | ||||||
| 1 (27) | Second | Pakistani | 9 | 137 | 4 | 19 |
| 2 (55) | First once removed | Pakistani | 16 | 366 | 10 | 22 |
| 3 (55) | First once removed | Pakistani | 22 | 376 | 11 | 13 |
| 4 (31) | Second | Pakistani | 26 | 288 | 8 | 16 |
| 5: | ||||||
| 1 (43) | First | Pakistani | 23 | 428 | 12 | 43 |
| 2 (42) | First | Pakistani | 14 | 302 | 9 | 16 |
| 3 (75) | First | Pakistani | 11 | 313 | 9 | 15 |
| 6: | ||||||
| 1 (60) | First | Pakistani | 31 | 536 | 15 | 29 |
| 2 (60) | First | Pakistani | 25 | 452 | 13 | 17 |
| 3 (50) | First | Pakistani | 27 | 491 | 14 | 14 |
| 7: | ||||||
| 1 (24) | Third | Pakistani | 7 | 62 | 2 | ? |
| 8: | ||||||
| 1 (57) | First | Pakistani | 26 | 394 | 11 | 57 |
| 2 (63) | First | Pakistani | 13 | 231 | 7 | 63 |
| 9: | ||||||
| 1 (57) | First | Pakistani | 27 | 388 | 11 | 57 |
| 10: | ||||||
| 1 (68) | First | Arab | 22 | 376 | 11 | 33 |
| 2 (33) | First | Arab | 18 | 263 | 8 | 23 |
| 3 (39) | First | Arab | 19 | 326 | 9 | 33 |
| 11: | ||||||
| 1 (50) | Second | Arab | 14 | 289 | 8 | 5 |
| 2 (34) | Second | Arab | 13 | 218 | 6 | 28 |
| 3 (24) | Second | Arab | 11 | 151 | 4 | 21 |
| 12: | ||||||
| 1 (42) | First | Arab | 6 | 141 | 4 | 42 |
| 2 (58) | First | Arab | 7 | 159 | 5 | 58 |
| 3 (41) | First | Arab | 8 | 245 | 7 | 27 |
| 4 (49) | First | Arab | 13 | 212 | 6 | 49 |
| 13: | ||||||
| 1 (49) | First | Pakistani | 13 | 247 | 7 | 31 |
| 2 (32) | First | Pakistani | 19 | 301 | 9 | 5 |
| 3 (82) | First | Pakistani | 23 | 494 | 14 | 19 |
| 4 (71) | First | Pakistani | 21 | 457 | 13 | 28 |
| 14: | ||||||
| 1 (32) | First | Pakistani | 24 | 349 | 10 | 16 |
| 2 (40) | First | Pakistani | 30 | 530 | 15 | 6 |
| 3 (39) | First | Pakistani | 28 | 467 | 13 | 15 |
| 15: | ||||||
| 1 (32) | First | Arab | 21 | 317 | 9 | 17 |
| 2 (31) | First | Arab | 18 | 315 | 9 | 17 |
| 3 (85) | First | Arab | 20 | 408 | 12 | 12 |
| 16: | ||||||
| 1 (58) | First | Arab | 16 | 278 | 8 | 21 |
| 2 (66) | First | Arab | 22 | 481 | 14 | 42 |
| 3 (22) | First | Arab | 11 | 143 | 4 | 7 |
| 4 (70) | First | Arab | 16 | 435 | 12 | 43 |
| 5 (59) | First | Arab | 14 | 344 | 10 | 42 |
| 6 (45) | First | Arab | 24 | 425 | 12 | 42 |
| 17: | ||||||
| 1 (30) | First | Irish traveler | 17 | 186 | 5 | 21 |
| 2 (40) | First | Irish traveler | 8 | 151 | 4 | 21 |
| 3 (36) | First | Irish traveler | 25 | 267 | 8 | 21 |
| 4 (45) | First | Irish traveler | 11 | 235 | 7 | 21 |
| 5 (35) | First | Irish traveler | 15 | 262 | 8 | 21 |
| 6 (38) | First | Irish traveler | 8 | 127 | 4 | 21 |
| 7 (41) | First | Irish traveler | 13 | 204 | 6 | 21 |
| 8 (46) | First | Irish traveler | 15 | 235 | 7 | 21 |
| 9 (40) | First | Irish traveler | 16 | 212 | 6 | 21 |
| 10 (40) | First | Irish traveler | 11 | 172 | 5 | 21 |
Only genomic segments constituting ⩾20 homozygous SNPs were scored.
With use of a human genome length of 3,488 cM.20
The size of the linked homozygous autosomal segment in the family and the individual. Results are given only for individuals from families with proven linkage (a LOD score >3.5) or homozygous mutations.
Appendix C
Table C1.
Results for Set 2 Individuals
| Family and Affected Family Member | Size of Minimal Linked Region(cM) | Parental Relationship(Cousins) |
| 1: | ||
| 1 | 22 | First |
| 2 | 49 | First |
| 3 | >89 | First |
| 4 | >71 | First |
| 5 | 19 | First |
| 6 | 46 | First |
| 2: | ||
| 1 | 8.5 | First |
| 2 | >8.5 | First |
| 3: | ||
| 1 | >10 | First |
| 4: | ||
| 1 | >10 | First |
| 2 | >10 | First |
| 3 | >10 | First |
| 5: | ||
| 1 | >7 | First |
| 6: | ||
| 1 | >4 | First |
| 2 | >4 | First |
| 7: | ||
| 1 | >7 | First |
| 8: | ||
| 1 | >7 | First |
| 9: | ||
| 1 | >7 | First |
| 2 | >4 | First |
| 10: | ||
| 1 | >2 | First |
| 11: | ||
| 1 | >7 | First |
| 12: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 13: | ||
| 1 | >7 | First |
| 2 | >2 | First |
| 3 | >2 | First |
| 14: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 15: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 3 | >7 | First |
| 4 | >7 | First |
| 5 | >7 | First |
| 6 | >7 | First |
| 7 | >7 | First |
| 8 | >7 | First |
| 9 | >7 | First |
| 10 | >7 | First |
| 11 | >7 | First |
| 12 | >7 | First |
| 16: | ||
| 1 | >6 | First |
| 2 | >3 | First |
| 3 | >3 | First |
| 4 | >3 | First |
| 5 | >3 | First |
| 17: | ||
| 1 | >7 | First |
| 18: | ||
| 1 | >15 | First |
| 19: | ||
| 1 | >11 | Double first |
| 2 | >3 | Double first |
| 20: | ||
| 1 | 22 | First |
| 2 | 21 | First |
| 3 | 59 | First |
| 4 | 39 | First |
| 21: | ||
| 1 | 7 | First |
| 2 | 7 | First |
| 3 | 7 | First |
| 22: | ||
| 1 | 30 | First once removed |
| 2 | 35 | First once removed |
| 3 | 59 | Second |
| 4 | 35 | Second |
| 23: | ||
| 1 | 29 | First |
| 2 | >20 | First |
| 3 | 10 | First |
| 4 | >8 | First |
| 5 | >19 | First |
| 6 | 18 | First |
| 7 | 19 | First |
| 24: | ||
| 1 | 5 | First |
| 2 | 6 | First |
| 3 | 7 | First |
| 4 | 7 | First |
| 5 | 18 | First |
| 6 | >13 | First |
| 7 | >15 | First |
| 8 | 5 | First |
| 9 | 11 | First |
| 25: | ||
| 1 | >4 | First |
| 2 | >3 | Second |
| 3 | >3 | Second |
| 26: | ||
| 1 | >9 | First |
| 2 | >9 | First |
| 27: | ||
| 1 | >9 | First |
| 2 | 5 | First |
| 28: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 29: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 3 | >7 | First |
| 4 | >7 | First |
| 5 | >7 | First |
| 30: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 3 | >12 | First |
| 31: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 3 | >7 | First |
| 32: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 3 | >6 | First |
| 33: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 34: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 35: | ||
| 1 | >7 | First |
| 2 | >7 | First |
| 3 | >4 | First |
| 36: | ||
| 1 | 9 | First |
| 2 | 9 | First |
| 3 | 24 | First |
| 4 | 24 | Double first |
| 5 | 9 | Double first |
| 6 | 9 | Double first |
| 7 | 9 | Double first |
| 8 | 11 | First |
| 9 | 11 | First |
| 10 | 7 | First |
| 11 | 7 | First |
| 12 | 9 | Double first |
| 13 | 9 | Double first |
| 14 | 9 | First |
| 37: | ||
| 1 | 13 | First |
| 2 | 26 | First |
| 3 | 26 | First |
| 4 | 10 | First |
| 5 | 11 | First |
| 6 | 10 | First |
| 7 | >24 | First |
| 8 | >24 | First |
| 38: | ||
| 1 | 20 | First |
| 2 | 62 | First |
| 3 | 62 | First |
| 4 | 25 | First |
| 5 | >30 | First |
| 39: | ||
| 1 | >14 | First |
| 2 | >14 | First |
| 3 | >14 | First |
| 40: | ||
| 1 | 33 | First |
| 2 | 27 | First |
| 3 | 33 | First |
| 41: | ||
| 1 | 40 | First |
| 2 | 40 | First |
| 3 | 21 | Second |
| 42: | ||
| 1 | 4 | Third |
| 2 | 4 | Third |
| 3 | 4 | Third |
| 4 | 4 | Third |
| 5 | 4 | Third |
| 6 | 4 | Third |
| 7 | 4 | Third |
| 8 | 4 | Third |
| 9 | 4 | Third |
| 43: | ||
| 1 | 23 | First once removed |
| 2 | 23 | First once removed |
| 3 | 23 | First once removed |
| 4 | 23 | First once removed |
| 5 | 23 | First once removed |
| 44: | ||
| 1 | 33 | First |
| 2 | 33 | First |
| 3 | 44 | First |
| 45: | ||
| 1 | >17 | First |
| 2 | >17 | First |
| 3 | >17 | First |
| 4 | >17 | First |
| 5 | >17 | First |
| 46: | ||
| 1 | 15 | First |
| 2 | >17 | First |
| 3 | >17 | First |
| 4 | 8 | First |
| 5 | 8 | First |
| 47: | ||
| 1 | 45 | First |
| 2 | 45 | First |
| 3 | 20 | Second |
| 48: | ||
| 1 | 47 | Second |
| 2 | 35 | First once removed |
| 3 | 47 | First |
| 4 | 34 | First once removed |
| 49: | ||
| 1 | 70 | First |
| 2 | 70 | First |
| 3 | 47 | First |
| 50: | ||
| 1 | 47 | Second |
| 2 | 30 | First |
| 51: | ||
| 1 | 26 | First |
| 2 | 26 | First |
| 3 | 31 | First |
| 52: | ||
| 1 | 20 | First |
| 2 | 20 | First |
| 53: | ||
| 1 | >9 | First |
| 2 | >12 | First |
| 3 | >12 | First |
| 4 | >3 | First |
| 5 | >10 | First |
| 6 | >10 | First |
| 7 | >10 | First |
| 8 | >10 | First |
| 9 | >10 | First |
| 10 | >12 | Second |
| 11 | 10 | Second |
| 12 | 6 | Second |
| 13 | >12 | Double first |
Web Resources
The URLs for data presented herein are as follows:
- Affymetrix, http://www.affymetrix.com/support/technical/datasheets/10k2_datasheet.pdf (for 10K SNP chip data)
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html (for Marshfield genetic maps for polymorphic microsatellite genetic locations)
- Geneservice, http://www.geneservice.co.uk/home/ (formerly MRC Geneservice, now an independent company)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Alstrom syndrome, Fuhrmann syndrome, Lebers amaurosis, nonsyndrome mental retardation, oro-facial-digital syndrome, retinitis pigmentosa, primary microcephaly, Seckel syndrome, and Warburg micro syndrome)
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for the Human Genome Browser and for physical order and location of markers and SNPs)
- University of Manitoba Department of Plant Science, http://www.umanitoba.ca/afs/plant_science/COURSES/CYTO/l17/Idiogram_panel3.jpg (for Chromosome ideogram
References
- 1.Smith CAB (1974) Measures of homozygosity and inbreeding in populations. Ann Hum Genet 37:377–391 [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- 3.Wright AF, Teague PW, Bruford E, Carothers A (1997) Problems in dealing with linkage heterogeneity in autosomal recessive forms of retinitis pigmentosa. In: Edwards JH, Pawlowitzki IH, Thompson E (eds) Genetic mapping of disease genes. Academic Press, London, pp 255–272 [Google Scholar]
- 4.Broman KW, Weber JL (1999) Long homozygous chromosomal segments in reference families from the Centre d’Étude du Polymorphisme Humain. Am J Hum Genet 65:1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaber L, Merlob P, Bu X, Rotter JI, Shohat M (1992) Marked parental consanguinity as a cause for increased major malformations in an Israeli Arab community. Am J Med Genet 44:1–6 10.1002/ajmg.1320440102 [DOI] [PubMed] [Google Scholar]
- 6.Bundey S, Alam H (1993) A five-year prospective study of the health of children in different ethnic groups, with particular reference to the effect of inbreeding. Eur J Hum Genet 1:206–219 [DOI] [PubMed] [Google Scholar]
- 7.Hoodfar E, Teebi AS (1996) Genetic referrals of Middle Eastern origin in a western city: inbreeding and disease profile. J Med Genet 33:212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel F, Motulsky AG (1997) Human genetics: problems and approaches, 3rd ed. Springer-Verlag, New York, pp 482–485 [Google Scholar]
- 9.Ober C, Hyslop T, Hauck WW (1999) Inbreeding effects on fertility in humans: evidence for reproductive compensation. Am J Hum Genet 64:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad WI (1994) Reflections on the consanguinity and birth outcome debate. J Public Health Med 16:423–428 [DOI] [PubMed] [Google Scholar]
- 11.Audinarayana N, Krishnamoorthy S (2000) Contribution of social and cultural factors to the decline in consanguinity in South India. Soc Biol 47:189–200 [DOI] [PubMed] [Google Scholar]
- 12.Bittles A (2001) Consanguinity and its relevance to clinical genetics. Clin Genet 60:89–98 10.1034/j.1399-0004.2001.600201.x [DOI] [PubMed] [Google Scholar]
- 13.Boucher W (1988) Calculation of the inbreeding coefficient. J Math Biol 26:57–64 [DOI] [PubMed] [Google Scholar]
- 14.Woods CG, Valente EM, Bond J, Roberts E (2004) A new method for autozygosity mapping using single nucleotide polymorphisms (SNPs) and EXCLUDEAR. J Med Genet 41:e101 10.1136/jmg.2003.016873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- 16.Darr A, Modell B (1988) The frequency of consanguineous marriage among British Pakistanis. J Med Genet 25:186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.al-Gazali LI, Bener A, Abdulrazzaq YM, Micallef R, al-Khayat AI, Gaber T (1997) Consanguineous marriages in the United Arab Emirates. J Biosoc Sci 29:491–497 10.1017/S0021932097004914 [DOI] [PubMed] [Google Scholar]
- 18.Guy R, Forsyth JM, Cooper A, Morton RE (2001) Co-existence of lysosomal storage diseases in a consanguineous family. Child Care Health Dev 27:173–181 10.1046/j.1365-2214.2001.00165.x [DOI] [PubMed] [Google Scholar]
- 19.Collins A, Frezal J, Teague J, Morton NE (1996) A metric map of humans: 23,500 loci in 850 bands. Proc Natl Acad Sci USA 93:14771–14775 10.1073/pnas.93.25.14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miano MG, Jacobson SG, Carothers A, Hanson I, Teague P, Lovell J, Cideciyan AV, Haider N, Stone EM, Sheffield VC, Wright AF (2000) Pitfalls in homozygosity mapping. Am J Hum Genet 67:1348–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]