Abstract
Risk of opioid dependence is genetically influenced. We recruited a sample of 393 small nuclear families (including 250 full-sib and 46 half-sib pairs), each with at least one individual with opioid dependence. Subjects underwent a detailed evaluation of substance dependence–related traits. As planned a priori to reduce heterogeneity, we used cluster analytic methods to identify opioid dependence–related symptom clusters, which were shown to be heritable. We then completed a genomewide linkage scan (with 409 markers) for the opioid-dependence diagnosis and for the two cluster-defined phenotypes represented by >250 families: the heavy–opioid-use cluster and the non–opioid-use cluster. Further exploratory analyses were completed for the other cluster-defined phenotypes. The statistically strongest results were seen with the cluster-defined traits. For the heavy–opioid-use cluster, we observed a LOD score of 3.06 on chromosome 17 (empirical pointwise P=.0002) for European American (EA) and African American (AA) subjects combined, and, for the non–opioid-use cluster, we observed a LOD score of 3.46 elsewhere on chromosome 17 (empirical pointwise P=.00002, uncorrected for multiple traits studied) for EA subjects only. We also identified a possible linkage (LOD score 2.43) of opioid dependence with chromosome 2 markers for the AA subjects. These results are an initial step in identifying genes for opioid dependence on the basis of a genomewide investigation (i.e., a study not conditioned on prior physiological candidate-gene hypotheses).
Opioid dependence (OD) is associated with serious medical, legal, social, and psychiatric problems. OD has a lifetime prevalence of 0.4%,1 and the combined lifetime prevalence of OD and opioid abuse is 0.7%.2 The magnitude of the disability and suffering attributable to OD has made the need for improved understanding and treatment of the disorder an important public health issue. Elucidating the genetic basis of OD would represent major progress toward understanding the etiology of this disorder.
Risk of OD, like the risk of many other forms of substance dependence, is influenced by genetic factors, as demonstrated by adoption studies (in the general case of substance dependence) and by twin studies. Tsuang et al.3 studied >3,000 Vietnam-era male twin pairs, among whom drug abuse was defined as at least weekly use of any of a variety of drugs. Significant pairwise concordance rates showed a familial basis for every drug considered. The difference in pairwise concordance rates for MZ and DZ twins was significant for the abuse of marijuana, stimulants, cocaine, and all drugs combined. For OD specifically, MZ twin tetrachoric correlation was 0.67±0.11, and DZ correlation was 0.29±0.21, with an estimated heritability of 0.43, an e2 (unique environment) of 0.31, and “nonadditive” genetic components accounting for an estimated 26% of the variance. MZ concordance was 13.3% (4/30), and DZ concordance was 2.9% (1/34). For most drugs studied (marijuana, stimulants, sedatives, heroin, and other opioids), there was evidence of both shared and specific (“unique”) risk factors; unique risk factors were most important for heroin abuse.
Kendler et al.4 also examined the specificity of genetic risk factors for substance dependence (involving cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates) in male twins. In this sample of nearly 1,200 male twin pairs, there were high levels of comorbidity for both drug use (tetrachoric correlations 0.60–0.85) and drug misuse (tetrachoric correlations 0.67–0.85 for substance abuse and/or dependence). Model fitting revealed that one common genetic factor exerted a relatively potent influence on risk of both use and misuse of all six substances. There was a modest effect of specific genetic factors on risk of substance use but not abuse or dependence. A common shared environmental factor was also found to exert an effect on risk of substance use and, to a lesser extent, on risk of abuse or dependence. OD, compared with the other substance-dependence phenotypes, showed the lowest loading on the common genetic factor (0.48) and the highest loading on the single unique environmental common factor (0.80). In this study, in contrast to the study of Tsuang et al.,3 OD showed no contribution from substancespecific additive genetic factors. Although Kendler et al.4 hypothesized that this difference might be the result of environmental characteristics of the Vietnam twin cohort studied by Tsuang et al.,3 the small number of individuals with OD in both studies means that either study could have had selection bias or inadequate statistical power. Karkowski et al.5 studied substance-dependence genetics in a cohort of >800 female twin pairs. The resolution of this sample for OD was quite small; this was reflected in the great difference in OD heritability estimated from a univariate model (0.52), compared with that from a multivariate model (0.10).
To our knowledge, there have been no previously published studies of OD with genomewide scope. However, published work supports population-specific association with certain candidate genes. OPRM1 encodes the μ-opioid receptor; markers at this locus have been reported to be associated with OD6 or with opioid and alcohol dependence combined.7 Xu et al.8 reported a highly significant association of DRD2 haplotypes with OD in Chinese subjects.
In the present study, we sought to identify the chromosomal location of genes that increase risk of OD and related traits identified using data reduction and cluster analytic approaches. The use of cluster analysis to identify subgroups that might increase genetic homogeneity for linkage was part of the originally planned approach9; we also applied the approach used here in a recently published linkage study of cocaine dependence (CD).10 We collected 634 small nuclear families at four sites in the eastern United States, with the recruitment condition that families have at least two siblings with either CD or OD.10 Of the families included, 393 had at least one subject with OD, and 235 had at least two individuals with OD. Those 393 informative families are included in the present study.
Subjects and Methods
Subject Recruitment and Assessment
There were four recruitment sites for the study: University of Connecticut Health Center (Farmington), Yale University School of Medicine (New Haven, CT; APT Foundation), McLean Hospital (Belmont, MA; Harvard Medical School), and Medical University of South Carolina (Charleston), hereafter referred to as UConn, Yale, McLean, and MUSC, respectively. Families were recruited on the basis of screening results suggesting that two siblings would meet diagnostic criteria for OD (at the UConn and Yale sites) or CD (at all sites). Some subjects with CD had comorbid OD and were informative for the present study. Subjects with a primary diagnosis of a major psychotic illness (schizophrenia or schizoaffective disorder) were excluded as probands. Once an affected sibling pair (ASP) was recruited, additional siblings and parents were recruited whenever possible, regardless of affection status.10 Demographic details and recruitment by site are presented in tables 1 and 2. This sample includes the subjects from our CD linkage study (when they were informative for OD) plus additional subjects.
Table 1.
Demographics of Sample, by Site
|
Genotyped Subjects |
|||||
| Site | All | No. (%) Male |
Mean Age (years) |
AA | EA |
| Yale | 575 | 242 (42.1) | 39.1 | 357 | 218 |
| MUSC | 221 | 112 (50.7) | 38.0 | 109 | 112 |
| UConn | 578 | 285 (49.3) | 39.7 | 267 | 311 |
| McLean | 245 |
106 (43.3) |
40.9 | 114 |
131 |
| Total | 1,619 | 745 (46.0) | 39.4 | 847 | 772 |
Table 2.
Recruitment of Families, by Site
|
Families |
|||
| Site | All | AA | EA |
| Yale | 205 | 123 | 82 |
| MUSC | 98 | 45 | 53 |
| UConn | 231 | 101 | 130 |
| McLean | 100 |
45 |
55 |
| Total | 634 | 314 | 320 |
Subjects gave informed consent, as approved by the institutional review board at each clinical site, and a certificate of confidentiality for the work was obtained from the National Institute on Drug Abuse, National Institutes of Health. Subjects were interviewed using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) for psychiatric diagnosis, as described elsewhere.10,11 The reliability of SSADDA for the diagnosis of OD was excellent, with test-retest reliability of κ=0.94 (based on 120 subjects) and interrater reliability of κ=0.91 (based on 173 subjects).11 The diagnosis of OD was based on application of a computer algorithm that uses DSM-IV diagnostic criteria to the SSADDA data.12
Genotyping
In most cases, DNA was obtained from immortalized cell lines, but, for some subjects, DNA was obtained directly from blood or saliva. The Applied Biosystems mid-density linkage mapping set 2.5, augmented by 15 additional STR markers, was used as described elsewhere.10 We genotyped a total of 409 markers spanning the genome, including 391 autosomal markers and 18 X-chromosome markers. Marker map positions for analyses were based on the sex-averaged Marshfield map (Center for Medical Genetics Web site).
Phenotypic Cluster Analysis
We identified clusters of symptoms that could lead to valid OD subtyping, using binary symptom indicators from the opioid assessment section of SSADDA; this strategy is similar to the one we used for our study of CD.10 DSM-IV classification of OD may not represent the optimal OD-related phenotype for genetic mapping; other phenotypes might identify sets of subjects that are more genetically homogeneous. The strategy for the development of qualitative and quantitative traits included nonparametric data reduction, iterative two-stage clustering on the observed dimensions, and the assignment of probability of cluster membership in each cluster for each individual. Multiple correspondence analysis,13,14 a nonparametric data reduction method, was used to identify the underlying dimensions in the data. Each study participant with phenotypic data was assigned a score for each of the retained dimensions by use of a procedure that is conceptually similar to assigning factor scores. Next, a multistage clustering strategy was used to identify distinct subgroups. The first stage was an iterative k-means partitioning with the use of several different starting points and a larger-than-expected k (k=50). Nearest centroid sorting was used for the k-means clustering. The second stage involved cross-classification of the results of the k-means clustering and retention of the groups that consistently clustered together. These groups and the remaining observations were then used with an agglomerative hierarchical clustering to identify the final cluster structure. Ward’s method was used for the agglomerative hierarchical clustering. (This method avoids the idiosyncrasies that can occur with different starting seeds for k-means clustering. It also identifies intact groups for the hierarchical clustering that are joined before the agglomerative process. The intention is to retain the strengths of both types of clustering while mitigating their weaknesses.) A comparison of the within-to-between-group variation on variables other than those used to form the groups and group profiles provided the basis for selection of the final cluster solution. SPAD software (DECISIA) was used for both the multiple correspondence analysis and the clustering algorithms. SAS software15 was used for subsequent analyses, including cluster profiling. Binary logistic regression was used to estimate the probability of cluster membership for each study participant for each of the clusters. Variables selected for clustering were used in the estimation of the probability of cluster membership. The natural logarithm of the probability of membership in each group was the dependent measure in the quantitative-trait analyses. Heritability of the log of probability of group membership was computed using SOLAR.16 Although cluster assignment differentiates severity of opioid dependence (between some clusters), subjects assigned to different clusters differ in other features too (table 3), such as substance-use disorders and other comorbidities, demographic features, and mode of drug administration (e.g., injection vs. some other method). The clusters identified may be characterized briefly as follows.
Table 3.
Demographic Characteristics and Comorbid Psychiatric and Substance-Use Disorders[Note]
|
Whole Samplea(n=2,881) |
Sample withDSM-IV OD(n=666) |
Cluster E(n=346) |
Cluster A(n=730) |
|||||
| Characteristic | No. (%) of Subjects |
No. with Missing Data |
No. (%) of Subjects |
No. with Missing Data |
No. (%) of Subjects |
No. with Missing Data |
No. (%) of Subjects |
No. with Missing Data |
| Male sex | 1,492 (51.8) | 0 | 373 (56.0) | 0 | 197 (56.9) | 0 | 275 (37.7) | 0 |
| EA ethnicity | 1,331 (46.2) | 0 | 452 (67.9) | 0 | 253 (73.1) | 0 | 203 (27.8) | 0 |
| ASPD | 175 (12.0) | 1,421 | 87 (13.6) | 28 | 46 (13.8) | 13 | 72 (10.0) | 12 |
| Compulsive gambling | 135 (9.2) | 1,420 | 56 (8.8) | 30 | 24 (7.2) | 12 | 65 (9.1) | 12 |
| Panic disorder | 109 (7.4) | 1,412 | 73 (11.4) | 26 | 51 (15.1) | 9 | 29 (4.0) | 8 |
| Agoraphobia | 60 (4.1) | 1,426 | 26 (4.1) | 29 | 18 (5.4) | 11 | 27 (3.8) | 17 |
| PTSD | 149 (10.2) | 1,414 | 69 (10.8) | 26 | 39 (11.6) | 10 | 69 (9.6) | 10 |
| Major depression | 195 (13.4) | 1,431 | 96 (15.2) | 35 | 50 (15.2) | 17 | 87 (12.2) | 15 |
| Bipolar disorder | 77 (5.2) | 1,408 | 39 (6.1) | 24 | 26 (7.8) | 11 | 29 (4.0) | 5 |
| Tobacco dependence | 984 (65.4) | 1,376 | 492 (73.9) | 0 | 263 (76.0) | 0 | 422 (57.8) | 0 |
| Alcohol: | ||||||||
| Dependence | 653 (43.4) | 1,376 | 296 (44.4) | 0 | 164 (47.4) | 0 | 306 (41.9) | 0 |
| Abuse | 350 (23.3) | 1,376 | 149 (22.4) | 0 | 77 (22.3) | 0 | 169 (23.2) | 0 |
| Cocaine: | ||||||||
| Dependence | 1,238 (82.3) | 1,376 | 559 (83.9) | 0 | 294 (85.0) | 0 | 587 (80.4) | 0 |
| Abuse | 46 (3.1) | 1,376 | 28 (4.2) | 0 | 15 (4.3) | 0 | 15 (2.1) | 0 |
| Opioid: | ||||||||
| Dependence | 666 (44.3) | 1,376 | 666 (100.0) | 0 | 346 (100.0) | 0 | 0 (0) | 0 |
| Abuse | 31 (2.1) | 1,376 | 31 (4.7) | 0 | 0 (0) | 0 | 0 (0) | 0 |
Note.— ASPD = antisocial personality disorder; PTSD = posttraumatic stress disorder.
“Whole sample” includes nongenotyped parents.
Clusters with no or low opioid use (A and B)
Cluster A (“non–opioid users”) was the largest cluster, containing slightly more than half of the sample. None of the members of this cluster received a diagnosis of OD. Of cluster B members (“low-opioid users”), 38% received a diagnosis of OD. Demographic characteristics differentiate the clusters. Cluster A contained a higher proportion of African Americans (AAs) and women. Although individuals in cluster A and the majority of individuals in cluster B were not opioid dependent, the majority of individuals in these groups were cocaine dependent (80% and 90% of clusters A and B, respectively). Of cluster A subjects, 5% were dependent on substances other than cocaine, whereas 20% of cluster B subjects were.
Clusters with moderate or heavy opioid use (C, D, and E)
Clusters C (“moderate-opioid users”), D (“heavy-opioid and mixed-substance users”), and E (“heavy-opioid users”) comprised 10%, 7%, and 23% of the sample, respectively. Again, there were demographic differences among the clusters. More than 40% of clusters C and D was AA, whereas only 28% of cluster E was AA. All three clusters had comorbid CD, at prevalences of 72%, 93%, and 85%. The reported average number of lifetime episodes of opioid drug use exceeded 4,000 for all three clusters and was >4,700 for clusters D and E. For all subjects in these three clusters, the reported duration of time during which they used opioids most heavily was >3 years (on average), and >95% of these individuals were opioid dependent (100% of cluster E subjects). A majority of all three groups (57%, 72%, and 79% in clusters C, D, and E, respectively) reported injecting opioids. A majority of cluster D members (61%) reported that they “got higher and stayed higher longer than others” when they first started to use opioids (compared with 47% of cluster C and 48% of cluster E). Cluster D also had higher rates of abuse and dependence on other substances, including alcohol, marijuana, stimulants, sedatives, tobacco, and other substances. Additionally, cluster D members were more likely to have major depression and cocaine-induced paranoia than any other group. Members of cluster E were users of both cocaine and opioids but were less likely to use other drugs than were members of cluster D. More than 95% of cluster E reported that they had made a failed attempt to quit use of opioids.
Sample characteristics, including cluster assignments, are presented in table 4. (Detailed information about cluster analysis procedures and results will be presented in a separate publication.) We present linkage results here for clusters A and E only, because they were the largest clusters (>250 informative families each) and therefore provided reasonable power for linkage analysis. Exploratory analyses were completed for the other clusters (data not shown).
Table 4.
Pedigree Characteristics
| Characteristic | Total | AA | EA |
| No. of genotyped subjects | 1,619 | 847 | 772 |
| Mean age (years) | 39.4 | 40.9 | 37.9 |
| No. of pedigrees analyzed | 634 | 314 | 320 |
| No. of pedigrees with: | |||
| 1 Subject with OD | 158 | 80 | 78 |
| 2 Subjects with OD | 204 | 48 | 156 |
| 3 Subjects with OD | 26 | 10 | 16 |
| 4 Subjects with OD | 5 | 1 | 4 |
| No. of pedigrees with at least 1 subject: | |||
| With OD | 393 | 139 | 254 |
| In cluster A | 389 | 251 | 138 |
| In cluster B | 115 | 48 | 67 |
| In cluster C | 127 | 52 | 75 |
| In cluster D | 90 | 32 | 58 |
| In cluster E | 254 | 76 | 178 |
| No. of full-sib pairs with OD | 250 | 56 | 194 |
| No. of half-sib pairs with OD | 46 | 33 | 13 |
The heritability of the natural logarithm of the probability of group membership was computed using SOLAR,16 with adjustment for sex and European American (EA) ethnicity. It should be noted that, because this sample was ascertained using an ASP strategy (although for OD and CD diagnosis, not for cluster membership), heritability estimates are potentially inflated. Heritability for the log of the probability of cluster membership for each of the clusters is shown in table 5.
Table 5.
Cluster Heritability
| Cluster | Heritability | P |
| A (non–opioid users) | .61 | 7.1 × 10−22 |
| B (low-opioid users) | .32 | 4.7 × 10−6 |
| C (moderate-opioid users) | .63 | 1.1 × 10−19 |
| D (heavy-opioid and mixedsubstance users) | .65 | 2.0 × 10−25 |
| E (heavy-opioid users) | .40 | 4.4 × 10−9 |
Linkage Analysis
Valid assignment of subjects to populations
There are known differences in patterns of use and in substance-dependence risk between AA and EA individuals.17–20 Also, different populations could contain different important risk loci, and the contribution of a set of risk loci could vary by population. It is thus important to evaluate linkage separately by population, but self-reported racial background might not always identify population groups optimally. Therefore, as in our study of CD, we used a Bayesian model-based clustering method to assign individuals to populations21 on the basis of ancestry proportions inferred from genotype data (table 6). This method is implemented in the program STRUCTURE. For STRUCTURE runs, we used 100,000 iterations for the burn-in, with a run length of 100,000. Included in the analyses were 390 STR markers from the genomewide scan.
Table 6.
Cluster Assignment Compared with Self-Described Ethnicity
|
ClusterAssignment(%) |
||
|
Self-Reported Ethnicity |
AA | EA |
| Native American/American Indian | 1.3 | .4 |
| Asian | .0 | .0 |
| Pacific Islander | .4 | .0 |
| African American/black: | ||
| Not of Hispanic origin | 90.4 | 2.4 |
| Of Hispanic origin | 3.8 | 4.9 |
| Caucasian/white: | ||
| Not of Hispanic origin | .9 | 68.2 |
| Of Hispanic origin | .0 | 17.9 |
| Other | 3.3 |
6.3 |
| Totala | 100.1 | 100.1 |
Percentages add up to >100 because of rounding.
Estimation of marker-allele frequencies
The results of ASP linkage analysis are sensitive to marker-allele frequencies, especially when little parental information is available, as for our sample. In this situation, identity by descent (IBD) must be estimated on the basis of observed identity by state and population allele frequencies; population allele frequencies generally differ between EAs and AAs. If we had analyzed our sample of AA and EA small families as a single group, inaccurate IBD estimates would have resulted. Therefore, in all stratified analyses and in the combined analyses, all linkage results were obtained using IBD values that were calculated using the appropriate population allele frequencies.
Inconsistency checking
Inconsistency checking was accomplished (as described elsewhere10) by using PedCheck,22 and Merlin,23 to identify Mendelian inconsistencies and probably-incorrect genotypes on the basis of estimation of the probability of double-crossover events. We checked for errors in specified family relationships, using PREST (pedigree relationship statistical test).24 When a potential pedigree error was detected, ALTERTEST,24 was used to determine relationships compatible with the observed genotype data. After correction, PREST was run again to confirm that family relationship reassignments were consistent with estimated IBD patterns.
Parametric linkage analysis
We performed parametric linkage analysis, using the package FASTLINK.25,26 All linkage analyses were two-point, unless stated otherwise. We tested four different models: recessive and dominant models with high (0.75) and low (0.25) penetrances. These values were chosen in an attempt to limit the number of models while covering as much of the parameter space as possible. Susceptibility-allele frequency was set at 0.01 for dominant models and 0.14 for recessive models. The frequency of 0.14 for recessive models corresponds to the 0.01 frequency for dominant models under the assumption of a constant population prevalence—that is, q based on the frequency of cases under a recessive model (q2) versus a dominant model (p2+2pq). Multilocus linkage analyses were performed for chromosomal regions showing genomewide evidence of linkage with individual markers or suggestive evidence of linkage with adjacent markers. These analyses, comprising up to three markers at a time, were performed using the program MLMAP, a modification of the MFMAP program.27 Unlike in MFMAP—in which several other models of inheritance are tested on the basis of the initial model—in MLMAP, the model of inheritance is fixed to the one model that gave the optimal two-point LOD score.
Nonparametric linkage analysis (model free)
We performed a model-free analysis using Genehunter-Plus.28 This analysis includes affected subjects only, a robust but (especially with a data set such as ours, which includes many discordant sib pairs) conservative approach. IBD status was estimated on the basis of observed identity-by-state status for both population subsets (i.e., EAs and AAs) separately, by use of observed allele frequencies for each group. The data were also analyzed for the population groups separately, to detect areas of interest that might be apparent in one subgroup only, and for the population groups combined, to detect loci where both subgroups contribute to linkage. In the combined analysis results, the IBD files were combined before calculation of the nonparametric LOD, also called the “Kong and Cox allele-sharing LOD.”29
Estimation of empirical P values
Type I error rates for LOD scores >3 were determined by analyzing 50,000 replicates of a fully informative simulated unlinked marker in the pedigrees under the null hypothesis of no linkage by use of the software SLINK.30–32 The specific set of pedigrees, missing data, and model parameters for each simulation corresponded to those used in the actual analyses yielding the significant LOD scores. Each replicate was analyzed using the program FASTLINK.25,26 The proportion of the replicates with a LOD score exceeding the observed values from analyses of the actual data was considered to be an empirical P value.
Results
Population Group Assignment
Using STRUCTURE, we stratified our total data set of 1,619 individuals in 634 families into 314 AA families and 320 EA families, with 847 and 772 genotyped subjects, respectively. As noted elsewhere,10 Hispanic families did not cluster into a distinct third population group; instead, they were allocated into the AA or EA group. All subjects who reported that they were of white Hispanic origin clustered in the EA group, whereas subjects who reported that there were of black Hispanic origin clustered into the AA and EA groups in almost equal proportions (table 6).
Pedigree information is summarized in table 4. Of this total set of families ascertained for ASPs with CD and/or OD, there were 235 families with two or more members with OD (including a total of 250 informative full-sib pairs and 46 half-sib pairs; see “Inconsistency checking” in the “Subjects and Methods” section for discussion of sibship assignment). By population group, there were a total of 59 AA and 156 EA families with two or more members with OD, including a total of 56 AA and 194 EA full-sib pairs and 33 AA and 13 EA half-sib pairs. (Half-sib pairs are more informative for linkage analysis than full-sib pairs.33) Thus, although there were more AA than EA subjects with CD,10 the OD-affected part of the sample shows a predominance of EA subjects. In addition, there were 158 families (80 AA and 78 EA) who had one member with OD and an unaffected sibling. These discordant sib pairs were informative for parametric linkage analysis.
Relationship Assignment
On the basis of the results of PREST, we reclassified 367 individuals; four sibships were reclassified as parent-child pairs, 123 individuals who were thought to be full sibs were reclassified as half sibs, and 240 individuals were reclassified as unrelated and were excluded from the study; these included 38 duplicates and 24 MZ twins.
Linkage Analysis
DSM-IV OD
The highest LOD score (2.43) for the trait of DSM-IV–classified OD was obtained from parametric analyses (by use of a dominant-inheritance and high-penetrance [0.75] model) at position 221.1 cM on chromosome 2 in AAs. Two additional suggestive-linkage findings were observed for OD: a LOD score of 1.99 at 82 cM on chromosome 5 in AAs, under a dominant model with low penetrance (0.25), and a LOD score of 1.93 at 9.2 cM on chromosome 6 in EAs, under a recessive model with high penetrance. We also observed a LOD score of 1.75 on chromosome 16 in EAs, under a recessive model with high penetrance. Finally, we observed a LOD score of 1.76 on chromosome 17 at 103.5 cM, in the combined (EA and AA) sample, under a recessive model with high penetrance. (Lander and Kruglyak35 proposed a LOD score of 1.9 as the value for “suggestive” linkage.)
Analysis of Clusters
In analyses of cluster traits, we observed two LOD scores >3.0, one of which exceeded 3.3 and thus met the level of genomewide significance. Three other LOD scores were between 1.9 and 3.0 (table 7) and met the level of suggestive linkage.35
Table 7.
Linkage Results for Cluster Phenotypes[Note]
|
ParametricAnalysis |
NonparametricAnalysis |
|||||
|
Chromosome and Location (cM) |
Trait | Ethnic Group(s) | Type, Modela | LOD | Type | LOD |
| Chromosome 3: | ||||||
| 177.8 | Cluster A | AA | Two-point, R, HP | 2.04 | Multipoint | 1.69 |
| 177.8 | Cluster A | AA and EA | Two-point, R, HP | 1.91 | Multipoint | 2.10 |
| Chromosome 11: | ||||||
| 16.7 | Cluster A | EA | Two-point, D, LP | 1.69 | Multipoint | 2.20 |
| Chromosome 17: | ||||||
| 75.0 | Cluster A | EA | Two-point, R, LP | 3.46 | Two-point | 1.98 |
| 103.5 | Cluster E | AA and EA | Two-point, R, HP | 3.06 | Two-point | 1.88 |
| 103.5 | Cluster E | EA | Two-point, R, HP | 2.18 | Multipoint | 1.51 |
| 117.8 | Cluster E | AA and EA | Two-point, R, HP | .72 | Multipoint | 2.27 |
Note.— Listed are locations for which parametric linkage analysis reached a LOD score of 1.9 and/or model-free nonparametric linkage analysis reached a LOD score of 2.2, meeting the criteria for suggestive linkage evidence proposed by Lander and Kruglyak.35
D = dominant; HP = high penetrance (0.75); LP = low penetrance (0.25); R = recessive.
Cluster E (heavy-opioid users)
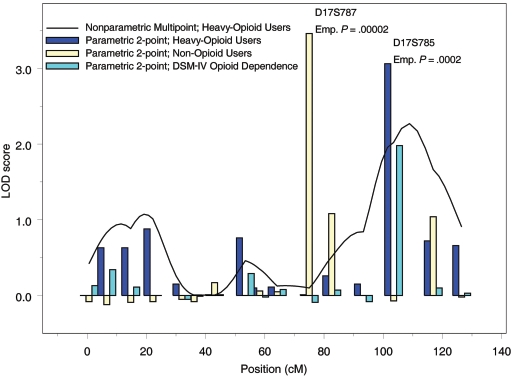
For cluster E, we observed results of interest under both parametric and nonparametric models, on chromosome 17 (fig. 1). At 103.5 cM, we observed a LOD score of 3.06 for EA and AA subjects combined, under a recessive, high-penetrance model. At the same location, nonparametric analysis resulted in a LOD score of 1.88. (Note that this is the same location where we observed a LOD score of 1.76 for the DSM-IV–classified OD trait under the same model.) At 117.8 cM, the multipoint parametric LOD score was 2.27.
Figure 1.
Linkage analysis results for chromosome 17 in EA and AA subjects, including subjects with DSM-IV–classified OD (teal bars) and subjects in cluster A (yellow bars) and cluster E (blue bars and black line) (see main text for cluster characteristics). Results of nonparametric multipoint analysis and parametric two-point linkage analyses with a recessive, high-penetrance model are shown. Emp. = empirical.
Cluster A (non–opioid users)
Interestingly, the highest LOD score for this study (3.46) was observed for cluster A, which included no subjects with OD; however, most of these subjects were dependent on substances other than opioids (80% were dependent on cocaine, 58% tobacco, 42% alcohol, and 25% marijuana). For this trait, we observed a multipoint parametric LOD score of 2.20 on chromosome 11 at 16.7 cM. On chromosome 17 at 75 cM, we observed a LOD score of 3.46 in EAs under a recessive, low-penetrance model; at the same location, we observed a nonparametric LOD score of 1.98.
Computer simulations were used to evaluate empirically the significance of the two LOD scores >3. The point empirical P value corresponding to the LOD score of 3.46 for linkage to cluster A (non–opioid users) in 313 EA families was .00002. The pointwise empirical P value corresponding to the LOD score of 3.06 for linkage to cluster E (heavy-opioid users) in EA and AA families combined was .0002.
Discussion
We present evidence, from linkage analyses based on ASPs assessed because they share OD or CD, that strongly supports a risk locus for a trait defined by symptoms related to heavy opioid use on chromosome 17 (LOD score 3.06; P=.0002). At this location, we also observed a LOD score of 1.76 for DSM-IV OD. We identified a putative risk locus elsewhere on chromosome 17 (LOD score 3.46; P=.00002) for a trait (cluster A) generally defined by the presence of substance dependence other than OD. This may represent a locus that protects against OD specifically, in the context of general substance-dependence susceptibility. This signal is ∼22 cM from a possible linkage observed for conduct-disorder symptoms in adolescents34 and could plausibly represent the same underlying locus. However, our observed linkage signal extends distal from the peak (fig. 1), and the conduct-disorder signal is more proximal. For this cluster, which is not opioid dependent but is dependent on other drugs, the second highest LOD score observed was on chromosome 11p, in proximity to a region where linkage to or association with several forms of substance dependence—alcohol dependence,36 habitual cigarette smoking,37 and polysubstance dependence38—has been reported. Also, for this same trait, we observed LOD scores consistent with possible linkage at 177.8 cM on chromosome 3. This location is consistent with several previous linkage or association reports for related traits34,36,38 and was also observed in a subset of this same sample for the trait of CD.10 Further, we observed a LOD score of 2.43 for DSM-IV–classified OD on chromosome 2 at 221.1 cM (D2S126) in AAs. We had observed a similar LOD score in the same population (AAs) at the same location for a CD-related trait (“heavy cocaine use, later onset”).10 Thus, some of these results are consistent with previous reports of linkage to substance dependence other than OD.
As in our study of cocaine, we detected the statistically strongest linkage evidence with phenotypes derived using cluster analysis of opioid-related items on the SSADDA interview. Our interpretation of this result is that, by means of cluster analysis, we were able to identify subgroups of OD-affected individuals with increased genetic homogeneity, reducing “noise” that might obscure linkage signals. The chromosome 17q results are consistent with this hypothesis. Interestingly, we observed a LOD score of 1.76 for DSM-IV–classified OD on chromosome 17 for the EA and AA populations combined, but when we used the cluster E diagnosis definition (i.e., the cluster characterized by the most-severe OD symptoms), the evidence for linkage increased by more than an order of magnitude, to LOD score 3.06. Genes identified by this method would be expected to be relatively specific to subjects who meet the definition of the linked cluster. Such possible linkages might be more vulnerable to being spurious, because they relied on subsets of the sample; however, the sample size involved was still reasonably large. Replication in additional samples or verification by identification of genes associated with the trait is particularly important for these cluster-defined traits. We infer that the cluster strategy actually reduced genetic heterogeneity because (1) the clusters have less intragroup variation than between-group variation on the items of interest and, (2) empirically, even in subsets of our sample, we have significant linkage and association results for the clusters. Our results support the idea that this is a powerful and useful refinement for OD diagnosis.
There are, to our knowledge, no previous genomewide studies of OD, but there have been numerous candidate-gene studies. The most widely studied candidate gene to date for OD is the gene encoding the μ-opioid receptor, OPRM1.6,7,39 Elsewhere, we report that variants in each of the two observed haplotype blocks at this locus are associated with drug (including opioid) and alcohol dependence.40 This gene maps to 6q24-q25 and is not in close proximity to any of our strongest linkage signals.
Although the nonparametric analysis results are not identical to the results from parametric analysis—because the analyses are based on different algorithms, and they use different aspects of the family data—in most cases, the results of the different analyses are consistent. Whereas nonparametric analysis uses information from only affected subjects, parametric analysis uses data from both affected and unaffected subjects, and thus requires classification of affection. We feel that this is a very useful classification in the present sample, for several reasons.10 First, because most affection-status information in this sample derives from siblings, we assume that environmental exposures (such as exposures to opioid drugs) are correlated. Second, to the extent that this was not the case, the lack of exposure should be at least partially genetically determined. For sib pairs who share affection status for some form of substance dependence other than OD, there should be an increased likelihood that their discordance for OD reflects genetic factors specific to OD risk, rather than to general substance-dependence risk.
On the basis of demographic characteristics and opioid-use histories, the cluster analytic approach yielded phenomenologically distinct OD-related clusters, confirmed by estimates of the heritability of the trait embodied in cluster membership. We used this approach for clinical subtyping in our study of CD.10 In that study and the present one, because the choice and implementation of the clustering algorithm as well as data quality and the stability of the underlying groups can influence the validity of cluster analysis results, we applied a multistage, iterative approach to identify cluster membership. This effectively repeated the clustering to achieve stable groups. The advantages of the modified strategy are that the clustering is repeated with several starting points, and two different methods are used to obtain stable clusters; this should increase the generalizability of the cluster results.
Our analysis involved a parametric approach, in which we tested dominant and recessive parametric models with high and low penetrance. Although the issue of applying multiple models, and then correcting for the resulting multiple (usually correlated) statistical tests, remains a controversial issue, it is often the case that adding parametric models increases the power to detect linkage.41 Consistent with the theoretical reasons to test several models, there is already evidence, specifically in the context of substance-dependence genetics, that some loci act to increase risk in a way that presently can be best explained through a recessive mechanism.42,43 For our parametric analyses, we tested a limited number of models. A Bonferroni correction for multiple tests is clearly too conservative, since these models are not independent, but our results should still be viewed in the context of these tests. Major peaks were observed in the different analyses and in multiple models. In addition, we considered the EA and AA parts of the sample separately, but this does not constitute multiple testing in the conventional sense. First, we could not make the assumption that we could perform valid analyses on a combined data set because, as discussed above, the marker-allele frequencies are different in different populations (which could lead to false-negative and false-positive results). Second, there is no a priori reason to assume that the risk loci are the same in genetically different populations; for many phenotypes, it has been shown empirically that they are not.10,44 This difference underlies part of the theoretical basis of the technique of admixture mapping.45 The results in EA and AA subsamples can be appropriately viewed as reflecting two separate studies (the results of which sometimes confirm each other and sometimes do not), although the data were collected and analyzed concurrently.
Because performing more tests increases type I error, we used simulations of an unlinked marker to test the type I error rate directly in the data. Applying this method, we found that the chance that the reported LOD score of 3.46 represents a type I error is 1 in 50,000 (P=.00002). Consideration of multiple traits and models in different samples introduces a concern about whether the LOD score results fully account for an increased probability of type I error. This concern is predicated on the assumption that multiple independent tests were performed. Although assignment of affection status across the OD cluster traits is mutually exclusive, these traits are highly correlated with DSM-IV drug-dependence diagnoses and are not independent. Neither are the parametric models independent; for a given mode of inheritance, the models in our study differed by only the penetrance assumption (0.25 vs. 0.75). The parametric and nonparametric analyses test different null hypotheses and do not require correction for multiple testing. When one is able to confirm a significant linkage finding for a trait by analyzing the same trait with different methods, it clearly makes this finding more robust, not less. In recognition of the growing awareness that the definition of multiple testing, as well as the appropriate corrections to use when the tests are not independent, are not clear, we addressed this issue by implementing a point simulation method that enabled us to derive empirical estimates of significance for any particular result.
Our approach simulated 50,000 replicates of a fully informative marker, which is equivalent to simulating ∼167 genome scans based on the assumption of 300 independent marker tests. (The number of independent tests is difficult to quantify.) Our empirical P value estimates are robust, considering that the estimates obtained from a simulation of a genome scan are based on many markers that are much less informative than the fully informative ones specified in our simulation. Under the assumption that there are ∼300 linkage groups and without correction for the increased information from the simulated markers (a conservative approach), the linkage result for non–opioid users has an approximate genome-based empirical significance of 0.003, and the result for heavy-opioid users yields a genome-based significance of ∼0.03, uncorrected for multiple analytic models and diagnosis definitions.
This study has several limitations that should be noted. Foremost among these is the limited statistical power for mapping a complex trait with a set of 393 informative families (158 of which were informative for parametric analyses only, because they included only one affected subject). Because the cluster analyses identified subgroups of subjects, the samples for those analyses were smaller, and the need to analyze separately by population group engendered a further loss of power. Finally, the conformation of the pedigrees—most included an ASP and an additional sibling or one parent at best—limited the genetic information because of a lack of direct observation of IBD (as a result of the general lack of availability of both parents). In practical terms, the latter limitation is almost impossible to remedy through collection of both parents, because of the medical and social consequences of OD in the United States—for example, its destructive effects on family relationships, the subjects’ wish to avoid involving their parents in the study, and deaths of relatives as a consequence of their own substance dependence. However, this limitation could plausibly be addressed directly through study of OD in other societies in which OD-affected families might be less fragmented. It could also be addressed indirectly through an increase in genetic information via an increase in marker density—for example, by using a relatively dense panel of SNP markers rather than a moderately dense panel of ∼400 STR markers. The information increment is estimated to be close to 75% in families for which parental genotypes are unavailable but to be <50% in families for which parental genotype information is available.46 Evans and Cardon47 also emphasized that very dense SNP maps provide the greatest increment in linkage information when parental genotypes are unavailable. For example, Schaid et al.48 compared genomewide microsatellite data with SNP data from the 10K Affymetrix chip in a study of prostate cancer susceptibility and found that, with the SNP data, average information content increased from 41% to 61%. This resulted in the discovery of some previously unseen putative linkage peaks. Since the average family size for that study (2.8 affected genotyped individuals per family) is comparable to ours, we would expect a comparable increase in information from the use of a denser SNP map. These studies consistently showed an increase in empirically useful information from the high-density SNP panel, in the form of more and (sometimes) narrower linkage peaks and greater statistical significance.
Elucidating the genetic basis of OD could represent major progress in understanding the etiology of OD and could contribute to the development of biologically based treatments for the disorder. Our 10-cM genomewide linkage scan identified several genomic regions deserving further study that, we conclude, are likely to contain risk loci for OD or related clinical syndromes. Further research will be required to identify diseaseinfluencing loci that map within these genomic regions. Previous research has demonstrated the possibility of remarkable success in identifying risk loci for substance-use disorders on the basis of genetic linkage data—for example, the GABRA2 gene was identified as a risk-influencing locus for alcohol dependence,49 a finding that was replicated,50,51 and the CHRM2 locus was identified as a risk locus for alcohol dependence and depression,52 a finding that also was replicated.42 We are hopeful that similar progress can be made in identifying risk-modifying genes for OD.
Acknowledgments
This work was supported by National Institute on Drug Abuse (NIDA) grants R01 DA12690 and R01 DA12849. We thank Dr. David Curtis for sharing the MLMAP program. Greg Kay, Lingjun Zuo, and Ann Marie Lacobelle provided excellent technical assistance. We appreciate the efforts of the following individuals, who conducted SSADDA interviews: Michelle Slivinsky, Michelle McKain, Deborah Pearson, and Kevin Young (UConn); Alisha Pollastri, Yari Nunez, Matthew Madura, and Melyssa Pokrywa (Yale); Catherine Cogley (McLean); and Heather Remy (MUSC). Jennifer Blesso and John Farrell provided excellent database support. Alisa Manning and Deborah Cebrik assisted in statistical analyses and data management. We also thank the Rutgers University Cell and DNA Repository, the contractor for the NIDA Center for Genetic Studies, codirected by Dr. Jay Tischfield and Dr. John Rice.
Web Resources
The URLs for data presented herein are as follows:
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/
- FASTLINK, http://www.ncbi.nlm.nih.gov/CBBresearch/Schaffer/fastlink.html
- Genehunter-Plus, http://galton.uchicago.edu/genehunterplus/Current/INSTALL.ghp
- Merlin, http://www.sph.umich.edu/csg/abecasis/Merlin/
- PedCheck, http://watson.hgen.pitt.edu/register/docs/pedcheck.html
- PREST and ALTERTEST, http://galton.uchicago.edu/~mcpeek/software/prest/
- SLINK, http://linkage.rockefeller.edu/ott/SLINK.htm
- SOLAR, http://www.sfbr.org/solar/index.html
- STRUCTURE, http://pritch.bsd.uchicago.edu/software.html
References
- 1.Anthony JC, Warner LA, Kessler RC (1994) Comparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants: basic findings from the National Comorbidity Survey. Exp Clin Psychopharmacol 2:244–268 10.1037/1064-1297.2.3.244 [DOI] [Google Scholar]
- 2.Regier D, Farmer M, Rae D, Locke BZ, Keith SJ, Judd LL, Goodwin FK (1990) Comorbidity of mental disorders with alcohol and other drug abuse: results from the Epidemiologic Catchment Area (ECA) Study. JAMA 264:2511–2518 10.1001/jama.264.19.2511 [DOI] [PubMed] [Google Scholar]
- 3.Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True Lin NW, Meyer JM, Toomey R, Farone SV, Eaves L (1996) Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet 67:473–477 [DOI] [PubMed] [Google Scholar]
- 4.Kendler KS, Jacobson KC, Prescott CA, Neale MC (2003) Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry 160:687–695 10.1176/appi.ajp.160.4.687 [DOI] [PubMed] [Google Scholar]
- 5.Karkowski LM, Prescott CA, Kendler KS (2000) Multivariate assessment of factors influencing illicit substance use in twins from female-female pairs. Am J Med Genet 96:665–670 [DOI] [PubMed] [Google Scholar]
- 6.Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, Kreek MJ (2004) Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry 9:547–549 10.1038/sj.mp.4001504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo X, Kranzler HR, Zhao H, Gelernter J (2003) Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European Americans. Am J Med Genet B Neuropsychiatr Genet 120:97–108 10.1002/ajmg.b.20034 [DOI] [PubMed] [Google Scholar]
- 8.Xu K, Lichtermann D, Lipsky RH, Franke P, Liu X, Hu Y, Cao L, Schwab SG, Wildenauer DB, Bau CH, Ferro E, Astor W, Finch T, Terry J, Taubman J, Maier W, Goldman D (2004) Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. Arch Gen Psychiatry 61:597–606 10.1001/archpsyc.61.6.597 [DOI] [PubMed] [Google Scholar]
- 9.Basu D, Ball SA, Feinn R, Gelernter J, Kranzler HR (2004) Typologies of drug dependence: comparative validity of a multivariate and four univariate models. Drug Alcohol Depend 73:289–300 10.1016/j.drugalcdep.2003.11.004 [DOI] [PubMed] [Google Scholar]
- 10.Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR (2005) Genomewide linkage scan for cocaine dependence and related traits: linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet Neuropsych Genet 136:45–52 10.1002/ajmg.b.30189 [DOI] [PubMed] [Google Scholar]
- 11.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR (2005) Diagnostic reliability of the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA). Drug Alcohol Depend 80:303–312 10.1016/j.drugalcdep.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 12.American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Press, Washington, DC [Google Scholar]
- 13.Greenacre M (1984) Theory and applications of correspondence analysis. Academic Press, New York [Google Scholar]
- 14.Lebart L, Morineau A, Warwick KM (1984) Multivariate descriptive statistical analysis: correspondence analysis and related techniques for large matrices. John Wiley, New York [Google Scholar]
- 15.SAS Institute (2001) SAS release 8.02, Cary, NC [Google Scholar]
- 16.Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Institute on Drug Abuse (2003) Drug use among racial/ethnic minorities, revised. National Institutes of Health document 03-3888. US Department of Health and Human Services, Bethesda, MD [Google Scholar]
- 18.Vega WA, Zimmerman RS, Warheit GJ, Apospori E, Gil AG (1993) Risk factors for early adolescent drug use in four ethnic and racial groups. Am J Public Health 83:185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellickson PL, Morton SC (1999) Identifying adolescents at risk for hard drug use: racial/ethnic variations. J Adolesc Health 25:382–395 10.1016/S1054-139X(98)00144-X [DOI] [PubMed] [Google Scholar]
- 20.Turner RJ, Lloyd DA (2003) Cumulative adversity and drug dependence in young adults: racial/ethnic contrasts. Addiction 98:305–315 10.1046/j.1360-0443.2003.00312.x [DOI] [PubMed] [Google Scholar]
- 21.Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connell JR, Weeks DE (1998) PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 24.McPeek MS, Sun L (2000) Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 66:1076–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- 27.Curtis D, Sham PC (1995) Model-free linkage analysis using likelihoods. Am J Hum Genet 57:703–716 [PMC free article] [PubMed] [Google Scholar]
- 28.Kruglyak L, Lander ES (1998) Faster multipoint linkage analysis using Fourier transforms. J Comput Biol 5:1–7 [DOI] [PubMed] [Google Scholar]
- 29.Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]
- 32.Weeks DE, Lathrop M, Ott J (2001) SLINK: a general simulation program for linkage analysis (http://linkage.rockefeller.edu/ott/SLINK.htm) (accessed March 14, 2006)
- 33.Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- 34.Stallings MC, Corley RP, Dennehey B, Hewitt JK, Krauter KS, Lessem JM, Mikulich-Gilbertson SK, Rhee SH, Smolen A, Young SE, Crowley TJ (2005) A genome-wide search for quantitative trait loci that influence antisocial drug dependence in adolescence. Arch Gen Psychiatry 62:1042–1051 10.1001/archpsyc.62.9.1042 [DOI] [PubMed] [Google Scholar]
- 35.Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- 36.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221 [DOI] [PubMed] [Google Scholar]
- 37.Gelernter J, Liu X, Hesselbrock V, Page GP, Goddard A, Zhang H (2004) Results of a genomewide linkage scan: support for chromosomes 9 and 11 loci increasing risk for cigarette smoking. Am J Med Gen B Neuropsychiatr Genet 128:94–101 10.1002/ajmg.b.30019 [DOI] [PubMed] [Google Scholar]
- 38.Uhl GR, Liu QR, Walther D, Hess J, Naiman D (2001) Polysubstance abuse–vulnerability genes: genome scans for association, using 1,004 subjects and 1,494 single-nucleotide polymorphisms. Am J Hum Genet 69:1290–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan EC, Tan CH, Karupathivan U, Yap EP (2003) Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport 14:569–572 10.1097/00001756-200303240-00008 [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, Zvartau E, Gelernter J (2006) Association between two μ-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Molec Genet 15:807–819 10.1093/hmg/ddl024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abreu PC, Greenberg DA, Hodge SE (1999) Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet 65:847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J (2005) CHRM2 gene predisposes to alcohol dependence, drug dependence, and affective disorders: results from an extended case-control structured association study. Hum Mol Genet 14:2421–2434 10.1093/hmg/ddi244 [DOI] [PubMed] [Google Scholar]
- 43.Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J (2005) ADH4 gene variation is associated with alcohol and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology (http://www.nature.com/npp/journal/vaop/ncurrent/abs/1300925a.html) (electronically published October 12, 2005; accessed February 27, 2006) [DOI] [PubMed] [Google Scholar]
- 44.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A (2005) Admixture mapping for hypertension loci with genome-scan markers. Nat Genet 37:177–181 10.1038/ng1510 [DOI] [PubMed] [Google Scholar]
- 45.Stephens JC, Briscoe D, O’Brien SJ (1994) Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am J Hum Genet 55:809–824 [PMC free article] [PubMed] [Google Scholar]
- 46.The International Multiple Sclerosis Genetics Consortium (2004) Enhancing linkage analysis of complex disorders: an evaluation of high-density genotyping. Hum Mol Genet 13:1943–1949 10.1093/hmg/ddh202 [DOI] [PubMed] [Google Scholar]
- 47.Evans DM, Cardon LR (2004) Guidelines for genotyping in genomewide linkage studies: single-nucleotide–polymorphism maps versus microsatellite maps. Am J Hum Genet 75:687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaid DJ, Guenther JC, Christensen GB, Hebbring S, Rosenow C, Hilker CA, McDonnell SK, Cunningham JM, Slager SL, Blute ML, Thibodeau SN (2004) Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer–susceptibility loci. Am J Hum Genet 75:948–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe R, Goate A, Hesselbrock V, Jones K, Kwon J, Li T-K, Nurnberger J, O’Connor S, Reich T, Rice J, Schuckit M, Porjesz B, Foroud T, Begleiter H (2004) Variations in GABRA2, encoding the α2 subunit of the GABAA receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet 74:705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR (2004) Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet 129:104–109 10.1002/ajmg.b.30091 [DOI] [PubMed] [Google Scholar]
- 51.Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg L, Covault J, Kranzler HR, Krystal JH, Gelernter J (2005) Association between alcoholism and γ-amino butyric acid α2 receptor subtype in a Russian population. Alcohol Clin Exp Res 29:493–498 10.1097/01.ALC.0000158938.97464.90 [DOI] [PubMed] [Google Scholar]
- 52.Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J, Jones K, Nurnberger JI Jr, Tischfield J, Porjesz B, Edenberg HJ, Hesselbrock V, Crowe R, Schuckit M, Begleiter H, Goate AM, Bierut LJ (2004) Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet 13:1903–1911 10.1093/hmg/ddh194 [DOI] [PubMed] [Google Scholar]