Abstract
Genomes and antigenomes of many positive-strand RNA viruses contain 3′-poly(A) and 5′-poly(U) tracts, respectively, serving as mutual templates. Mechanism(s) controlling the length of these homopolymeric stretches are not well understood. Here, we show that in coxsackievirus B3 (CVB3) and three other enteroviruses the poly(A) tract is ∼80–90 and the poly(U) tract is ∼20 nt-long. Mutagenesis analysis indicate that the length of the CVB3 3′-poly(A) is determined by the oriR, a cis-element in the 3′-noncoding region of viral RNA. In contrast, while mutations of the oriR inhibit initiation of (−) RNA synthesis, they do not affect the 5′-poly(U) length. Poly(A)-lacking genomes are able to acquire genetically unstable AU-rich poly(A)-terminated 3′-tails, which may be generated by a mechanism distinct from the cognate viral RNA polyadenylation. The aberrant tails ensure only inefficient replication. The possibility of RNA replication independent of oriR and poly(A) demonstrate that highly debilitated viruses are able to survive by utilizing ‘emergence’, perhaps atavistic, mechanisms.
INTRODUCTION
The genomic RNA of many positive-strand RNA viruses is tailed with a 3′-poly(A). In contrast to cellular mRNAs, in which poly(A) is non-templated and is added post-transcriptionally, the viral poly(A) is believed to be template-coded (1). Accordingly, the negative strand contains a 5′-poly(U) tract, which is synthesized using the poly(A) of the positive viral RNA as a template. In turn, this poly(U) subsequently serves as a template for the synthesis of the poly(A) of progeny RNA. This alternate usage of the homopolymeric parts of the (+) and (−) RNA for each strand synthesis has not been studied in detail but it was observed that poly(U) in the negative RNA of poliovirus (2,3) and coronavirus (4) may be shorter than poly(A) in the viral RNAs. These observations pose intriguing questions about how a shorter poly(U) is synthesized on a longer poly(A) template and vice versa.
We addressed this conundrum using coxsackievirus B3 (CVB3), an enterovirus belonging to the picornavirus family as a model. Picornaviruses are small icosahedral viruses with a single-stranded 7–8 kb RNA genome of positive (mRNA-like) polarity. This family includes important human and animal pathogens such as poliovirus, hepatitis A virus, foot-and-mouth disease virus and others.
Three cis-acting elements are known to be involved in the replication of enterovirus RNA (5). The 5′-terminal cloverleaf structure (or oriL) is a multifunctional element that interacts, among other ligands, with viral 3CDpro protease and cellular poly(rC) binding proteins PCBP1 and PCBP2 (6–8) to form a ribonucleoprotein (RNP) complex. This complex is thought to be involved in the switch that represses translation and allows replication to commence (9), and is also required for the initiation of both (+)-strand (6) and (−)-strand RNA synthesis. The latter requires genome circularization mediated by an interaction between this RNP and the host poly(A)-binding protein (PABP) bound to 3′-poly(A) (10,11).
Another cis-acting replicative element (cre or oriI) is represented by an imperfect stem–loop located within the coding region of the enterovirus RNA (12–14). It ensures the covalent linkage of uridylates to the viral protein VPg via a two-step slide-back mechanism, thereby yielding the protein primer VPg-pU(pU) (15–18). Both (−)- and (+)-strand RNA 5′-termini have VPg covalently bound (5), but there is no consensus on whether the cre-mediated VPg-uridylylation is required for the initiation of only (+) RNA (19–21) or for the initiation of both strands (14). The former view implies that VPg uridylylation may occur directly on the 3′-poly(A) of (+) RNA, thus providing a primer for elongation into (−)-strands, in line with the finding that polyadenylates indeed support VPg-pUpU synthesis in vitro (22).
A third replicative cis-element, the oriR, is located in the 3′-noncoding region (3′NCR) of the viral RNA immediately upstream of the 3′-poly(A). It contains a multi-domain structure maintained by an intramolecular kissing interaction between helical elements X and Y (23–28). Mutations destroying this interaction or changing the mutual orientation of oriR helical elements resulted in a significant loss of fitness, as judged by the acquisition of temperature sensitive and quasi-infectious or lethal phenotypes. It was proposed that oriR is required for an efficient initiation of (−) RNA synthesis (24,26). However, partial and complete deletions of the 3′NCR still generated viable viruses challenging the essential role of 3′NCR and oriR in RNA replication (29–31).
The synthesis of a (−) RNA strand on the viral RNA template is believed to result in the formation of a genome-length double-stranded ‘replicative form’ (RF) RNA. Positive-strand RNA synthesis is initiated at the 3′-end of the (−) strand but the detailed mechanism is unknown. It has been proposed that destabilization and partial unwinding of this end of the duplex is required for efficient replication (32,33). Transcription of the (+)-strands from the (−)-RNA template results in partially double-stranded ‘replicative intermediate’ RNA molecules consisting of full-length (−) RNA and various numbers of incomplete (+) strands differing in length (5). Newly synthesized virion RNAs are thought to be displaced from the (−) strands by upcoming progeny strands through the duplex-unwinding activity of the viral RNA polymerase 3Dpol (34). However, a number of important issues pertaining to picornavirus genome replication remain contradictory or unsolved (35).
Here, we examined the mechanism for poly(A) and poly(U) synthesis and how their lengths are controlled. We found that the 3′-poly(A) of (+) RNA strands in CVB3 and some other enteroviruses is about 4-fold longer than the 5′-poly(U) of (−) strands and that the poly(A) length is controlled by the oriR. While ensuring efficient negative strand initiation, the oriR does not control the poly(U) length. Removal of the poly(A) (alone or together with the oriR) from transfected viral RNA resulted in regeneration of poly(A)-containing 3′-tails, which, however, ensured only inefficient viral reproduction. These results provide new insights into mechanisms and evolution of genome replication of positive-strand RNA viruses.
MATERIALS AND METHODS
Cells and media
Buffalo green monkey (BGM) cells were grown in minimal essential medium, MEM (Gibco), supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine (Sigma), 100 U penicillin per ml and 100 mg streptomycin per ml (Sigma). HeLa cells were grown in Dulbecco's MEM (Gibco) with the same supplements (but with streptomycin at 25 mg/ml). HeLa S3 cells (ATCC CCL 2.2) were grown either (i) in tissue culture flasks in Dulbecco's modified Eagle medium-nutrient mixture F-12 (Ham) (1:1) (Sigma) with the above supplements or (ii) in suspension using suspension MEM (Joklik modified) (Sigma) supplemented with l-glutamine, antibiotics and 10% newborn calf serum (Gibco).
Coxsackievirus cDNA mutant constructs
Randomization of 3′NCR
Mutations were introduced into the infectious pRibCB3/T7 construct (14) by PCR using synthetic oligonucleotides (Biolegio, The Netherlands). An oligonucleotide (Supplementary Data) containing 70 random residues downstream of the stop codon (N70) functioned as template in a PCR executed by ThermoPerfect DNA polymerase (Integro). Gel-purified products were inserted into the pGEM-T Easy vector (Promega) and selected clones were sequences as described below. The DraI and SalI restriction sites incorporated in the PCR primers were used to clone the insert into pRibCB3/T7-Stu (7310) digested with StuI and SalI. Sequence analysis verified the correct clones. Replicons in which the P1 region was replaced by the firefly luciferase gene were devised by a BssHII–SalI digestion (14).
Partial deletion constructs and kissing distortion mutants
The p53CB3/T7-ΔX, p53CB3/T7-ΔY, pRibCB3/T7-ΔXΔY, pRibCB3/T7-Δpoly(A), p53CB3/T7-Δ3′NCR-N70/A-Δpoly(A), p53CB3/T7-Δ3′NCR-N70/A-Δpoly(A)+AAUAAA, p53CB3/T7-Δpoly(A)+C9G, p53CB3/T7-Δpoly(A)+G23, pRibCB3/T7-Kissing distortion (KD) 6bp, pRibCB3/T7-KD 6bp+C, pRibCB3/T7-KD-4bp constructs were generated by PCR amplification using an infectious clone or p53CB3/T7-Δ3′NCR-N70/A (p53CB3/T7-Δ3′NCR-N70/A-Δpoly(A) and p53CB3/T7-Δ3′NCR-N70/A-Δpoly(A)+AAUAAA) as templates and amplification primers (Supplementary Data). PCR products were introduced into a p53- or pRibCB3/T7 infectious clone or luciferase replicon using the restriction sites mentioned above. With the exception of the Δpoly(A) constructs all other mutant constructs contained a poly(A) tail of 30 residues. The pRibCB3/T7-Δ3′NCR-Δpoly(A) was engineered using the complementary primer set 5′-AAATGGTTGGACTCCT4AG-3′ and 5′-TCGACTA4GGAGTCCAACCATTT–3′ already containing the correct overhang to be cloned in pRibCB3/T7-Stu(7310) as described above. For pRibCB3/T7-Δ3′NCR, pRibCB3/T7-Δ3′NCR+G15 and pRibCB3/T7-Δ3′NCR+C14G the same method was used (for primers see Supplementary Data). The p53CB3/T7-luc constructs varying in 3′-poly(A) size were PCR-generated using FPmut3NCR containing a DraI-site and RPA0-60 reverse oligonucleotides (Supplementary Data), containing a SalI restriction site. PCR products digested with DraI and SalI were cloned into the p53CB3/T7-luc-Stu (7310) digested with StuI and SalI, rendering constructs containing alternate 3′-poly(A) sizes after transcription.
Blocking the 3′-termini of transcript RNA with cordycepin-5′-triphosphate
Cordycepin (3′-deoxyadenosine, Ambion) was added to the 3′ end of in vitro transcribed RNA using yeast poly(A) polymerase (USB) according to the manufacturer's recommendations.
RNA transcription and transfection
SalI or MluI linearized cDNA served as templates for in vitro transcription by T7 RNA polymerase (Promega) (26). BGM cells were transfected in duplicate by a DEAE-dextran method (26) and incubated at 36°C and 5% CO2 until complete CPE. Then, the cultures were subjected to three cycles of freezing/thawing prior to storage at −80°C. If 6 days post-transfection no CPE was evident, the cells were lysed by three freeze/thaw cycles and 200 µl of cell suspension was transferred to a fresh culture (post-passage). The 50% tissue culture infective dose (TCID50) was determined by endpoint titration in BGM-cells (36).
Sequencing of recovered virus RNA
Viral RNA extraction was performed using the GenElute™ Mammalian Total RNA Miniprep kit (Sigma). RNA was reverse transcribed using Super RT (HT Biotechnology) and RT1 (Supplementary Data) and PCR-amplified using FPseq and RPseq. RT–PCR products were subcloned into pGEM-T easy vector (Promega). Two independent sequence reactions of selected clones were performed by capillary electrophoresis (3730 DNA Analyzer; Applied Biosystems) and Big Dye Terminator version 3 protocols (Applied Biosystems).
Virus purification
Two T150 flasks of either HeLa cells (for CVA21 strain Coe) or BGM cells (for CVB3 strain Nancy, EV71 strain BrCr and ECHO9strain Hill) were infected and incubated until complete CPE. After three freeze/thaw cycles, the cell debris was removed by centrifugation at 2500 r.p.m. Supernatants were collected in Beckmann centrifuge tubes containing 5 ml of 30% sucrose. Virions were pelleted at 140 000× g for 6 h (SW28 rotor, Beckman L8-70M centrifuge). The pellet was resuspended in 200 µl phosphate-buffered saline (PBS) and viral RNA was extracted as described above.
Tailing of the 3′-end of virion RNA
T4 RNA ligase (Promega) was used to attach an RNA oligonucleotide to virion RNA extracted from purified virus (Figure 1A). A complementary cDNA reverse primer (1) primed the RT reaction. The generated cDNA was amplified using primer 1 and virus-specific forward primers (Supplementary Data). In addition, a semi-nested-PCR was performed using primer 1 and semi-nested (SN) virus specific primers (Supplementary Data) to enhance specificity of the PCR and to increase the quantity of amplified product. The accumulated product was gel-purified and sequenced using the semi-nested PCR forward primers.
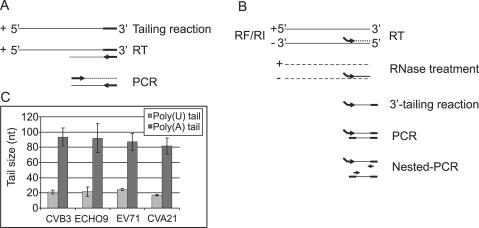
Figure 1.
(A) Schematic representation of the 3′-tailing reaction. An RNA ligase reaction was performed with purified virion RNA. A cDNA primer enabled reverse transcription (RT) followed by PCR. The 3′-poly(A) size was determined by sequencing. (B) Schematic representation of the 5′-RACE assay. RT was used to synthesize a cDNA strand after which RNases removed the RNA template. A 3′-tailing reaction added a homopolymeric tail to the cDNA. Amplification by PCR was followed by a nested PCR prior to sequencing. (C) The average 5′-poly(U)- and 3′-poly(A) sizes for four enteroviruses. For CVB3, ECHO9 and EV71 BGM-cells were infected, and for CVA21 HeLa-cells were used. The 3′-poly(A) columns represent the means ± SD of three independent experiments; the 5′-poly(U) columns represent the means ± SD of 10 values.
5′-RACE-assay
T25 flasks containing, depending on the virus strain (see above), BGM or HeLa cells were infected with an MOI of 10 TCID50/cell. Cells were lysed 6 h post infection (p.i.) and total RNA was obtained as described above. A 5′-rapid amplification of cDNA ends (5′-RACE)-PCR was performed using the 5′-RACE system (Invitrogen) according to the manufacturer's recommendations (Figure 1B). In brief, the RT-reaction on (−) RNA strands was executed by SuperScript™ II (Invitrogen) with virus-specific RT primers (Supplementary Data). After first strand cDNA synthesis, an RNase removed the original RNA template. Single-stranded cDNA was purified and terminal deoxynucleotidyl transferase (Invitrogen) added a 3′-poly(C) stretch. This cDNA was PCR-amplified using an AAP primer (Abridged Anchor Primer; 5′-RACE system, Invitrogen) and virus-specific forward primers (Supplementary Data), followed by a nested-PCR using an AUAP primer (Abridged Universal Amplification Primer) and virus-specific nested primers (Supplementary Data) to enhance specificity of the PCR and to increase the quantity of amplified product. The PCR background did not allow direct sequencing of the PCR-product, although the band of interest was clearly distinguishable. Therefore, PCR product was excised from the gel and ligated into a pGEM-T easy vector and the 5′-poly(U) size of 10 colonies was determined by sequencing using the nested forward primer (Supplementary Data).
Single-cycle growth analysis
Confluent BGM cell monolayers were infected with an MOI of five TCID50/cell and incubated at 36°C for 2, 4, 6 and 8 h (26). Each growth curve was performed in triplicate and viruses were released by three cycles of freezing/thawing. Virus titers were determined by endpoint titration (36).
Plaque-assay
Six-well dishes with >90% confluent monolayer of BGM cells were incubated with 10-fold virus dilutions in MEM for 1 h and rocked every 15 min. After absorption, the cells were overlaid with 2.5 ml of M199 (Sigma) containing 3% FBS, 1% glutamine, 0.5% gentamicine, 0.5% penicillin, 0.5% Funguzone, 1% MgCl2 and 1.3% Na-bicarbonate in 0.6% agar (1:1 mixture with 2×M199). At 48 or 96 h p.i., cells were fixed by methanol-acetic acid (3:1, vol/vol) for 10 min and stained with a 0.5% crystal violet in 20% ethanol.
Luciferase assay
BGM cells seeded in 6-well plates were transfected using the DEAE-dextran method (26) with 4 µg of replicon RNA. Cells were washed twice with PBS and lysed using 200 µl of CCLR lysis buffer (Promega). The luciferase assay system (Promega) was used to measure the luciferase activity on a BioOrbit 1251 Luminometer.
In vitro translation and replication assay
HeLa cell S10 extract and initiation factors were prepared as described (37). Negative strand RNA synthesis was analyzed as described (38) with minor modifications: 2 µg of CVB3 RNA transcript were mixed with 30 µl of HeLa S10 extract, 2 µl of initiation factors, 5 µl of 10× NTP/energy mix (37) and 1 µl of 100 mM guanidine-HCl in a volume of 50 µl. All other steps were executed as described (38).
RESULTS
The 5′-poly(U) of enterovirus negative strands is significantly shorter than the 3′-poly(A) of the positive RNA strands
Using 3′-tailing and 5′-RACE reactions, we determined the lengths of the 3′-poly(A) of virion RNA and the 5′-poly(U) in (−) RNA for four different enteroviruses; CVB3, ECHO9, enterovirus 71 (EV71) and coxsackievirus A21 (CVA21) (Figure 1A and B). In all cases, the average size of the poly(A) was 80–90 nt, whereas the length of the poly(U) was only ∼20 nt (Figure 1C). The poly(A) of CVB3 RNA from purified virions recovered from different cell cultures (BGM, HeLa and COS) was of a similar average size (data not shown).
OriR controls the initiation efficiency of negative RNA synthesis and the poly(A) length of virion RNA but not the size of the poly(U) of negative strands
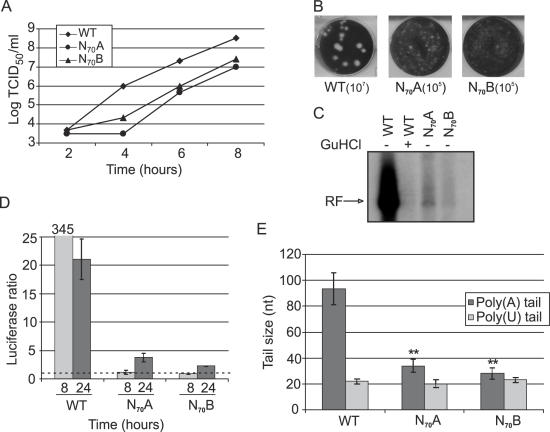
A simple explanation for the smaller size of the 5′-poly(U) tract compared with its 3′-poly(A) template is that (−) strand RNA synthesis is initiated on an internal position of the poly(A) template and there exists a spatial mechanism controlling the choice of this position. One can hypothesize that this mechanism involves the oriR. To examine this, two mutant RNAs, N70/A and N70/B, were generated, in which oriR was replaced by random sequences of 70 nt (Table 1). These RNAs proved to be viable but exhibited a reduced rate of replication [cytopathic effect (CPE) developed 4–6 days after transfection versus 2 days in the case of wild-type (wt) transcripts]. Sequencing the virus pools showed that N70/A had retained the engineered mutations while an adenosine insertion was found in N70/B within an internal oligo(A) tract downstream of the UAG stop codon (shown in bold, Table 1). The recovered viruses showed impaired replication (≥10-fold decrease in the single-cycle growth yield at 8 h p.i.) and small-plaque phenotypes (Figure 2A,B). Luciferase-expressing CVB3 replicons with the 3′NCR replaced by the randomized RNA segments were assayed for RF accumulation in a cell-free system. The assay was performed with RNA transcripts containing two 5′-terminal non-viral G residues (originating from T7 RNA polymerase promoter), which suppressed (+) strand RNA synthesis, allowing thereby estimation of the (−) strand synthesis (14,38). Randomization of oriR resulted in a severe deficiency in RF accumulation (Figure 2C), suggesting impairment of the (−) strand initiation. OriR randomization also inhibited RNA replication in BGM cells, as determined by using luciferase-expressing CVB3 replicons (Figure 2D). The replicon RNAs used to transfect BGM cells contained authentic 5′-ends (owing to the presence of a ribozyme sequence), improving replication kinetics (14,38). The viral RNAs extracted from virions harboring randomized oriRs possessed a markedly shortened 3′-poly(A) (30–35 nt on average) but their 5′-poly(U) on (−) RNA retained a wt length of ∼20 nt (Figure 2E).
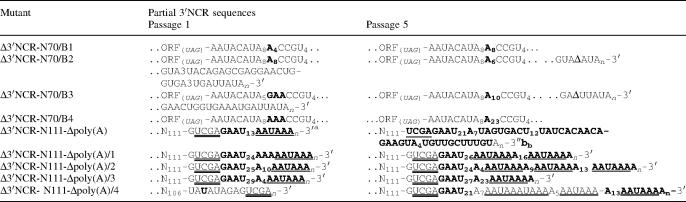
Table 1.
3′-terminal sequences of some engineered CVB3 mutants and of the viruses recovered after transfection
The stop codon of the open reading frame (ORF) is shown in italics, the sequences acquired after transfection are in bold, the canonical cellular polyadenylation signal, AAUAAA, is underlined and the partial SalI site is double-underlined. The last trinucleotides of the ORF or wt 3′NCR are given as subscripts in parenthesis. N63 corresponds to the first 63 bases of the N70/A sequence and N111 corresponds to 5′-GAGGAAUAACAGAUGA5GGU4CAGGGCAGAUAGAAUUAGGUAAUUGCCUAUGUUGAUAUGCGUUUCCUGCAGGCGGCCGCGAAUUCACUAGUGAUUGUACAUAGAG-3′.
aindicates the use of a ribozyme-containing construct for these particular mutants.
Figure 2.
Characterization of CVB3 3′NCR randomization mutants using single-cycle growth curves (A), in which the 50% tissue culture infective dose (TCID50) is set against the time of incubation (time), plaque assay, in which the dilution factor is indicated underneath the depicted well (B), and in vitro replication assay (C). GuHCl, guanidinium hydrochloride. (D) Luciferase activity of wt or mutant RNA determined 8 and 24 h post-transfection in BGM cells expressed as the fold difference between cells incubated in the presence and absence of GuHCl. (E) 3′-Poly(A) and 5′-poly(U) sizes of the Δ3′NCR-N70/A (N70A) and Δ3′NCR-N70/B (N70B) mutants. The 3′-poly(A) columns represent the means ± SD of three independent experiments; the 5′-poly(U) columns represent the means ± SD of 10 values. ** corresponds to P < 0.01.
These data confirmed the oriR involvement in the replication of (−) strands and uncovered its role in controlling the poly(A) length of the viral RNA. On the other hand, oriR has little, if any, effect on the poly(U) length in (−) strands.
Structural requirements of oriR that control the poly(A) size
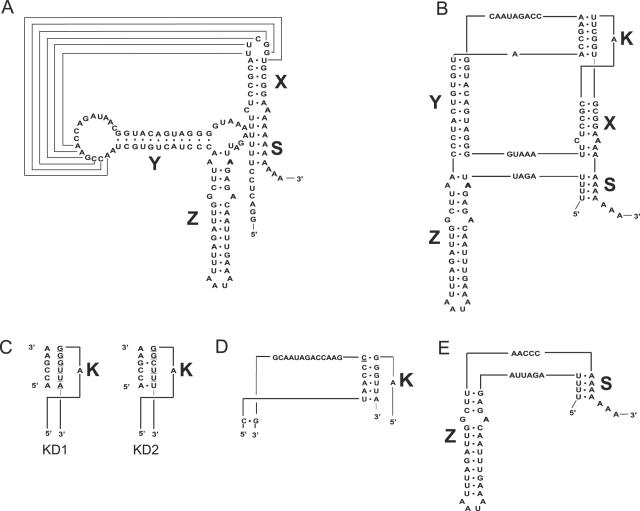
Previous studies indicated that the enterovirus oriR has a conserved structure composed of helical domains joined by a kissing interaction (K) between the loops of the X- and Y-domains (Figure 3A and 3B). Disruption of this interaction by alteration of 6 nt (mutant KD1) or 4 nt (KD2) in the 3′-terminal component of the kissing interaction (Figure 3C) resulted in quasi-infectious transcripts: after a delayed CPE, viable viruses were recovered from the transfected cultures but none of them faithfully retained the engineered structure. The pseudoreversions exhibited by the viruses could be grouped into two types. The representative of one class acquired a mutation restoring potential for a stable, though altered, kissing interaction. Thus, a C insertion at position 7350 could potentially support an alternative 6 nt kissing interaction (Figure 3D). When this mutation was engineered into the KD1 RNA, a virus (KD1+C) exhibiting a nearly wt phenotype was obtained, as judged by plaque size, single-cycle growth, cell-free RF formation and luciferase expression in transfected BGM cells (data not shown). The poly(A) of the KD1+C viral RNA was ∼70 nt-long, close to the wt value (data not shown). Besides this C-insertion no other mutations were found thus far that could potentially restore the kissing interaction.
Figure 3.
Secondary and tertiary structures of wild type and mutant CVB3 3′NCRs. Secondary (A) and tertiary (B) structure of the wt CVB3 3′NCR. The 3′NCR consists of three hairpin domains, X, Y and Z. The structure can be closed by an interaction between the poly(A) with a 4 nt U-stretch overlapping the oriR and the 3D-coding region. The X domain can be stacked to the tertiary ‘kissing’ (K) interaction to form a coaxial helical element, which is connected by single-stranded nucleotide stretches to a second coaxial helical domain Z-Y. (C) Schematic representation of the kissing distortion mutants, KD1 and KD2, in which 6 and 4 bp, respectively, were mutated (underlined). (D) Partial 3′NCR sequence of a virus recovered after transfection with the KD1 mutant. The inserted cytosine residue in the recovered mutant is underlined. (E) The entire 3′NCR sequence of a virus recovered after transfection with the KD2 mutant.
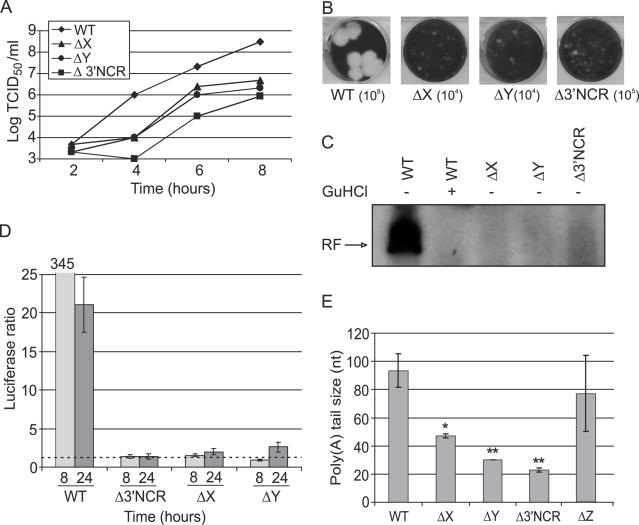
The second class was represented by mutants with spontaneous deletion of both X and Y domains (Figure 3E). To investigate the significance of these domains, deletions of either X or Y, or both, or the entire heteropolymeric 3′NCR were engineered into the CVB3 RNA. The separate deletions of X or Y were engineered into p53CB3/T7 RNA, whereas more extended deletions were introduced into pRibCB3/T7, which was found to be more infectious owing to the presence of the hammerhead ribozyme sequence (14,38). All constructs tested had a poly(A) of 30 nt and generated CPE in transfected culture, though with a significant delay (Table 2). Recovered viruses retained the engineered mutations and did not acquire any additional changes within the 3′NCR. In the Δ3′NCR and ΔXY mutants, the entire genome and P3 region were sequenced, respectively, and no additional mutations were found. All these mutants exhibited a marked deficiency in the virus reproduction (Figure 4A) and a small plaque-phenotype (Figure 4B) as well as inefficient cell-free RF accumulation (Figure 4C) and RNA replication in BGM cells (Figure 4D). The mutations were also accompanied with a severe reduction in poly(A) length, to ∼45 nt (ΔX), ∼30 nt (ΔY) and ∼20 nt (Δ3′NCR) (Figure 4E). The deletion of the Z-domain alone had no significant impact on poly(A) length.
Table 2.
Time to CPE after transfection or infection with CVB3 oriR mutants
| Construct/mutation | Days before CPE | |
|---|---|---|
| Post transfection | Post passage | |
| p53CB3/T7 (wildtype) | 1 | nd |
| p53CB3/T7-ΔX | — | 4 |
| p53CB3/T7-ΔY | — | 4 |
| pRibCB3/T7 (wildtype) | <1 | nd |
| pRibCB3/T7-ΔXΔY | 4/5 | nd |
| pRibCB3/T7-Δ3′NCR | 5 | nd |
A total of 5 × 106 cells were transfected with ∼4µg of RNA. ‘—’ means not detectable CPE; nd means not done.
Figure 4.
Characterization of CVB3 3′NCR deletion mutants using single-cycle growth curves (A), plaque assay (B) and in vitro replication assay (C). Δ indicates a deletion. (D) Luciferase activity of wt or mutant RNA determined 8 and 24 h post transfection in BGM (see legend of Figure 2 for details). (E), 3′-Poly(A) size. * and ** correspond to P < 0.05 and P < 0.01, respectively. The columns represent the means ± SD of three independent experiments.
These results confirmed that efficient viral RNA replication requires an oriR with a sufficiently stable kissing interaction, although the sequence of the K-domain could vary. Also, the efficient initiation of the (−) strand and generation of long poly(A) tails in the progeny (+)RNA could not be supported by its individual domains.
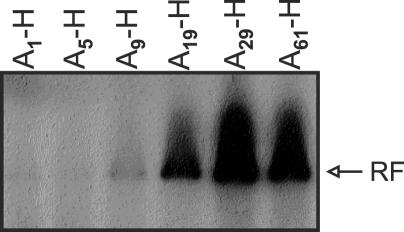
Effect of poly(A) length on the initiation efficiency of negative-strand RNA synthesis
As assayed by cordycepin-treated transcript RNA (see Materials and Methods), the minimal poly(A) size ensuring efficient cell-free RF RNA synthesis was ∼20–30 nt (Figure 5). These results are in line with previous reports showing that a poly(A) of ∼20 nt is sufficient for the efficient replication of poliovirus (also an enterovirus) (39), whereas shorter poly(A) resulted in a dead or highly debilitated virus (2,10,31,40).
Figure 5.
The minimal 3′-poly(A) size required for efficiently (−)-strand RNA synthesis in vitro. CVB3 replicons, P1 region exchanged for a luciferase gene, were used varying in 3′-poly(A) size. To prevent 3′-end elongation in vitro, RNA transcripts were cordycepin-treated. The RF accumulation was visualized by autoradiography.
Thus, the minimal length of the poly(A) in the (+) RNA ensuring efficient replication is ∼20 nt, coinciding with the poly(U) length in the (−) RNA.
Regeneration of RNA 3′-tails in viruses recovered from poly(A)-lacking genomes
To better understand the role of poly(A) in RNA replication, mutant viral cDNAs were generated, in which poly(A) was either missing [Δpoly(A)] or replaced by other sequences, such as G23 or C9G [Δpoly(A)-G23 and Δpoly(A)-C9G]. The Δpoly(A) construct contained 3′-terminal TCGA derived from a SalI site at the linearization locus. Transcripts of all these constructs exhibited a CPE delayed for 2–3 days and generated viruses with newly acquired 3′-poly(A), as evidenced by the ability of oligo(dT)18 to serve as a reverse primer in PCR-based sequencing. The RNA of the recovered Δpoly(A) mutant corresponded to its DNA construct, whereas the poly(G) moiety of the G23 mutant lost 21–22 G-residues and the C9G moiety of the respective mutant was replaced by a CCUUU sequence (Table 1). No alterations within the heteropolymeric part of 3′NCR was detected. The recovered viruses exhibited a nearly wt plaque phenotype at 36°C but formed somewhat smaller plaques at 39°C (Table 3). In BGM-cells, the poly(A)-lacking RNA of Δpoly(A) construct generated, at 10 h post-transfection, luciferase levels equal to ∼10% of that of wt RNA, whereas both G23 or C9G constructs produced luciferase levels similar to that found in the cells transfected with wt RNA in the presence of a potent inhibitor of viral reproduction guanidine-HCl (data not shown).
Table 3.
Plaque phenotypes of CVB3 oriR mutants
| Virus | Plaque size (mm)a | |
|---|---|---|
| 36°C | 39°C | |
| Wild-type CVB3 | 4.1 ± 0.9 | 4.3 ± 1.4 |
| G23 | 3.6 ± 0.7 | 3.1 ± 0.9 |
| C9G | 4.0 ± 1.0 | 2.8 ± 0.4 |
| Δ3′NCR+Δpoly(A) G15 | <0.5 | <0.5 |
| Δ3′NCR+Δpoly(A)C14G | <0.5 | <0.5 |
| Δ3′NCR+Δpoly(A) | <0.5 | <0.5 |
| Δ3′NCR-N111+Δpoly(A) P1 | <0.5 | <0.5 |
| Δ3′NCR-N111+Δpoly(A) P5 | <0.5 | <0.5 |
| N70+Δpoly(A)+AAUAAA | <0.5 | <0.5 |
aThe size was measured on the second day p.i., except for plaques with a diameter <0.5 mm, which became discernible only on the third day p.i.
In view of oriR involvement in polyadenylation of viral RNA, the effect of simultaneous deletion of heteropolymeric 3′NCR and poly(A) was investigated. When applied to the p53CB3/T7-based RNA, such a damage resulted in a nonviable genome. However, the equivalent ribozyme-containing transcript with a genuine 5′-end, Δ3′NCR-Δpoly(A), was viable, though exhibited a delayed CPE of 5 days after transfection. The viral genome acquired a 3′-poly(A) but no other modifications in the 3′-terminal portion of the molecule (Table 1).
To better understand the requirements for polyadenylation of poly(A)-lacking RNA, viral genomes were generated in which the 3′NCR was replaced by various structures. The poly(A)-lacking RNA with the oriR replaced by 111 random nucleotides, Δ3′NCR-N111-Δpoly(A), acquired a 3′-poly(A) preceded by an AU-rich stretch containing a U13 element and an AAUAAA hexamer corresponding to the cellular polyadenylation signal, (Table 1). A ribozyme-lacking N70-Δpoly(A) mutant was dead. However, insertion of an AAUAAA-containing oligonucleotide into the randomized N70 sequence produced a viable virus with 3′-poly(A) preceded by a long AU-rich region containing multiple AAUAAA hexamers (Table 1). The oriR-lacking RNAs terminated with G15 or C14G (together with a SalI site-derived UCGA) also generated viable genomes in which 3′-poly(A) preceded by relatively long poly(U) tracts and an oligo(A) stretch interspersed with several single U residues, again generating AAUAAA polyadenylation-like signals (Table 1). All the recovered poly(A)-lacking viruses with 3′NCR deletion/substitution generate minute plaques (Table 3), suggesting that they had a severe defect in viral replication.
The results indicated that poly(A)-lacking viral RNA can be viable but that 3′-poly(A) is readily regenerated. This poly(A) is usually, but not always, preceded by newly emerged AU-rich sequences.
Genetic instability of aberrant 3′NCR in viral RNA
To understand whether the aberrant 3′NCRs could be functionally ‘improved’ by natural evolution, viruses containing such 3′NCR were subjected to additional passages. The 3′NCR of Δ3′NCR-N70/A RNA remained unchanged after five passages, although the plaque size was somewhat increased (data not shown), possibly due to alterations elsewhere in the viral RNA. Four clonal derivatives of Δ3′NCR-N70/B RNA exhibited several differences compared with the original recovered viruses and underwent further changes upon passages (Table 4). Modifications included a marked increase in the length of internal oligo(A) blocks (e.g. from 3 to 23 nt in Δ3′NCR-N70/B4) and deletions of 13 or 29 nt-long heteropolymeric segments. The virus recovered after transfection with Δ3′NCR-N111-Δpoly(A) as well as its four clonal derivatives exhibited, upon passaging, in addition to lengthening of internal poly(A), also lengthening of poly(U) and acquisition of relatively extended AU-rich sequences containing the AAUAAA hexamers (Table 4). None of these changes were accompanied by a significant increase in the plaque size.
Table 4.
Genetic instability during passages of viral RNA containing aberrant 3′NCR
The stop codon is shown in italics and given as subscripts in parenthesis, the sequences acquired after transfection are in bold, the cellular polyadenylation signal, AAUAAA, is underlined, Δ means deletion and the partial SalI site is double underlined. For the mutant N70/B cDNA and N111 sequences see Table 1.
aRecovered virus.
bPassage 10.
DISCUSSION
Evidence is presented here that the mutual templating of the 3′-poly(A) of (+) RNA strands and 5′-poly(U) of (−) strands in viruses with positive RNA genomes, exemplified by enteroviruses, is not straightforward. In CVB3 (and three other enteroviruses), poly(U) is ∼20 nt-long, whereas poly(A) is ∼4 times longer. Destruction of the oriR, a cis-element in the 3′NCR of genomic RNA, significantly reduces the poly(A) size and decreases the efficiency of (−) RNA synthesis but does not affect the poly(U) length. Even though efficient genome replication requires the presence of a poly(A) of at least ∼20 nt, its complete removal did not kill the virus because of the poly(A) capacity to regenerate.
Mechanism and control of viral RNA polyadenylation
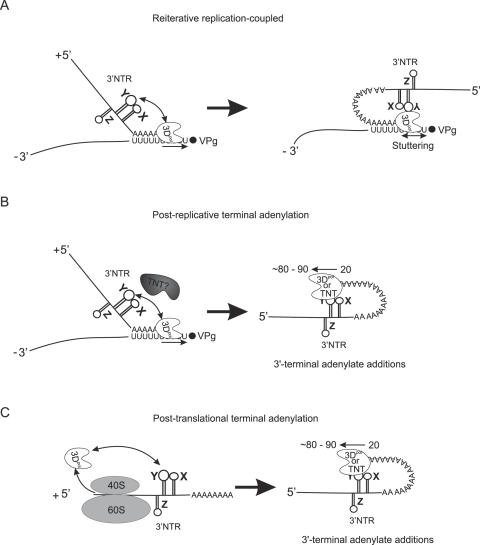
The existence of a longer poly(A) compared with its poly(U) template indicates that polyadenylation of viral RNA is not limited to the faithful copying of the template. Theoretically, additional polyadenylation may be because of either reiterative usage of the poly(U) template or a non-templated reaction catalyzed by a terminal nucleotide transferase (TNT)-like enzyme or poly(A) polymerase. Since the poly(A) is markedly shortened in the absence of oriR, the mechanism should involve this cis-element.
The reiterative hypothesis (Figure 6A) assumes that the synthesis of a longer poly(A) is due to a kind of ‘stuttering’ of the viral polymerase, common to negative-strand RNA viruses (41,42). The oriR role may be envisioned as follows. The nascent oriR, just released from the RNA-dependent RNA polymerase (RdRP) molecule engaged in the 5′-poly(U) copying, interacts with this RdRP in cis (43) and ‘encourages’ it to repeatedly use the poly(U) template. The RdRP/oriR interaction may be aided by other cellular (44) or viral proteins (45). Cases of increase in the length of internal oligo(A) in poliovirus (23,46) and foot-and-mouth disease virus (47) RNAs support the possibility of reiterative copying during picornavirus RNA replication. In this model, additional polyadenylation of viral RNA is replication-coupled, which is compatible with relative inefficiency of the RdRP complementation in trans (48,49).
Figure 6.
3′-Polyadenylation models. (A) The reiterative replication-coupled model. During the synthesis of a (+) RNA strand on a (−) RNA template, the nascent oriR just released from the RdRP molecule interacts with this RdRP in cis and ‘encourages’ it to repeatedly use the poly(U) template. (B) The post-replicative terminal adenylation model. The oriR of a newly synthesized (+) RNA [having a ∼20 nt-long poly(A)] binds and activates an enzyme with the adenylate transferase activity of viral or host origin, which accomplishes poly(A) elongation. (C) The post-translational terminal adenylation model. Viral RNA with a short poly(A), being translated, generates, among other proteins, the RdRP, which binds in cis the nearby oriR just liberated from the ribosome. This binding activates the enzyme intrinsic adenylate transferase activity.
On the other hand, poly(A) elongation may result from 3′-terminal adenylate additions. In this case, the nascent viral RNAs has only ∼20 nt-long poly(A). Further polyadenylation is accomplished by a TNT-like enzyme activated by binding to oriR. This job may be performed by a host poly(A) polymerase (50) or the terminal adenylate transferase activity may be an intrinsic property of the viral RdRP, as reported for the poliovirus enzyme by Neufeld et al. (51). A TNT activity has also been detected in RdRP of hepatitis C virus (52).
The oriR-dependent poly(A) additions to the ‘immature’ viral RNA may be post-replicative (i.e. occurring after completion of nascent positive strand transcription) (Figure 6B) or post-translational (i.e. accomplished by the nascent RdRP molecule or TNT-like enzyme) (Figure 6C). The post-translational hypothesis would explain why translation of viral RNA is a prerequisite for its replication (53). The ‘maturation’ of viral RNA through its further polyadenylation is required for its usage as template for the next round of RNA replication and also contributes to its stabilization.
Also it cannot be excluded that the oriR itself controls the poly(A) length just by its stabilization.
Although the above hypotheses are speculative, each of them makes specific predictions amenable for experimental testing.
Control of initiation of negative strands
Although oriR is not essential for the (−) strand synthesis (29,31; this study), it is important for its efficiency. Again, several mechanisms of such a control can be considered. One may suppose that binding of the viral RdRP (within an RNP complex or otherwise) to the oriR is a prerequisite for efficient initiation of (−) RNA synthesis or that the very presence of oriR makes the poly(A) tail more accessible for the initiation complex. Either of these notions can easily be combined with the models of poly(A) elongation considered above.
Remarkably, transcripts harboring certain oriR mutations produced progeny in which these mutations were not retained but either a wt-like oriR structure was regained or this structure was completely destroyed (see KD1 and KD2). Hence, it could be more harmful for the virus to possess certain oriR modifications than to lack this element altogether. This may indicate that some oriR modifications do not permit formation of a functional initiation complex, resulting in a dead genome.
Even though oriR controls the efficiency of (−) RNA synthesis, it does not appear to affect the site at which this synthesis is initiated. The fixed location of this site could be explained by unavailability of other loci on the poly(A), due for example to masking by PABP (10,11), other components of the replicase complex or a higher-order RNA structure (54). Nevertheless, it is difficult to envision how the choice of the initiation site could be independent of the poly(A) size, if it is merely determined by any sort of template masking. Rather the results seem to suggest the existence of a tool able to measure the appropriate distance (∼20 nt) downstream of the heteropolymeric part of the viral RNA regardless of its sequence or structure. The nature of this tool is yet to be elucidated, but a possible role of the CRE(2C), the site of VPgpUpU synthesis utilized for (−) strand RNA synthesis, cannot be excluded (14). One may also speculate that a putative cis-element in the RdRP-coding part of the viral RNA (31) may be involved in such measurements.
There is still another possibility for maintaining a constant poly(U) length. Following the nidovirus example, where the (−) RNA strand is synthesized discontinuously (55,56), one may contemplate that VPg-dependent synthesis of enterovirus (−) strands starts at or near the 3′-end of the poly(A) template but after synthesizing ∼20 nt-long poly(U), the viral RdRP together with the nascent poly(U) jumps to the heteropolymer/homopolymer boundary to continue the elongation. As yet, such a mechanism for picornaviruses is not supported by any evidence.
Emergency tailing of positive strands
Efficient enterovirus replication requires the presence of a poly(A) tail in viral RNA with a minimal length of ∼20 nt. Nevertheless, most poly(A)-lacking constructs proved to be (quasi)-infectious, generating genomes with newly-acquired AU-rich tails terminating with poly(A). Such nucleotide preference might reflect peculiarities of a yet-to-be-defined tailing reaction or merely resulted from selection because other types of tails might confer a lesser fitness. Mechanisms of elongation of the RNAs lacking poly(A) [or poly(A) and 3′NCR] are not necessarily the same as those operating in the case of wt viruses.
At least for the cases, where the engineered RNA did not possess an appropriate oligoadenylate to bind, or to generate, the VPg-pUpU primer [e.g. Δ3′NCR-N70-Δpoly(A) and Δ3′NCR-N111-Δpoly(A)], it is likely that some tailing of the viral RNA occurred prior to its use as a template for the (−) strand. Such tailing may involve a TNT-like enzyme of viral or host origin or non-replicative recombination (46,57,58), or both. Participation of recombination is supported by the observed inclusion of apparently host-derived sequences identical to a part of the human heterogeneous nuclear RNP U mRNA (residues 2408–2437) into the acquired 3′-tails upon passages (data not shown).
Passages of viruses with altered 3′-tails resulted in further modifications, e.g. elongation of internal poly(U) and poly(A) stretches. One may hypothesize that in addition to the terminal nucleotide transfer and recombination, reiterative synthesis using either 3′ end-adjacent sequences of (+) strand or 5′ end-adjacent sequences of (−) strand as templates may be involved. It seems likely that similar aberrant tailing can occur during the wt virus infection as well, but low-fitness viruses thereby generated are readily outcompeted and become undetectable.
Not surprisingly, the AU-rich sequences often contain one or more canonical cellular nuclear polyadenylation signals, AAUAAA. It is unknown whether they appear just by chance or are selected because they promote recruitment of the host polyadenylation machinery. A possible contribution of the AAUAAA signals to the appearance of oligo(A) stretches in the aberrant 3′-tails of the viral RNA is supported by the finding that the addition of this signal restored viability of Δ3′NCR-N70-Δpoly(A) RNA (Table 2). Also in Bamboo Mosaic Virus it was shown that this hexameric sequence is involved in genomic RNA polyadenylation (59). Furthermore, treatment of human cells with siRNAs specific for the cellular polyA polymerase resulted in downregulation of the enzyme and inhibition of poliovirus replication (C. Polacek and R. Andino, unpublished data). The origin of the poly(U) blocks in the (+) RNA molecules is enigmatic. It cannot be excluded that they are also due to a TNT-like activity. Remarkably, these segments are often located upstream of the 3′-poly(A). Hence, if tailing occurs at the level of viral RNA, the 3′-poly(A) may be added by snap-back synthesis on poly(U) blocks. However, the scarcity of experimental data makes such notion rather speculative.
Mechanisms of the (−) RNA initiation on the abnormal 3′-tails of the genomic RNA are yet to be determined, but our data suggest that viral RNA undergoes polyadenylation prior to replication.
Robustness of the viral genome
Illuminating examples of the robustness of the enteroviral genomes are accumulating. The oriR, a highly conserved cis-element, controls polyadenylation of the progeny RNA and initiation of the (−) strands but is nevertheless dispensable for viral viability [(29–31); this study]. Removal of poly(A), an element required for efficient genome replication, especially in combination with oriR inactivation, inflicted a severe damage to the viral fitness, though again usually not fatal. The poly(A) is functionally replaced by newly acquired AU-rich poly(A)-terminated 3′-tails potentially able to accommodate the VPg primer. Such a relatively unspecific regeneration of damaged 3′-tails of viral RNAs is not unique for enteroviruses (60).
Several mechanisms may be responsible for the viral robustness. First, the error-prone viral RdRP generates a variety of neutral or slightly detrimental mutants, not accumulating under normal conditions. However, ‘at emergency’, when the viral genome is severely invalidated, some of such mutations may result in a fitness increase, as exemplified by apparent restoration of the functional oriR structure after destruction of the kissing interaction [(24); this study] or changing mutual orientation of the oriR helical elements (25). Second, oriR belongs to a class of cis-elements, which are important but not essential. It is tempting to assume that such elements could have been acquired at some steps of enterovirus evolution in order to increase viral fitness. If so, the oriR-lacking mutants could be regarded as relying on ancient, ‘atavistic’ replicative tools. Regeneration of poly(A) after its complete deletion suggests the existence of unspecific mechanisms, which may ‘revive’ nearly dead viruses. Some of these mechanisms perhaps involve host enzymes.
Although certain RNA damages may generate rather crippled viruses, their very survival gives them chances to acquire by recombination either their lost element(s) or structurally unrelated but functionally active element from other viruses or hosts. Such loss of an element and acquisition of its functional substitute from an outside source may be an important path of viral evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
This research was supported by grants of the Council of Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW) grant 98008 to W.M., and both the European Communities INTAS 2012 and NWO-RFBR 047.017.023 to W.M. and V.I.A. V.I.A. was also supported by Russian Foundation for Basic Research and the Program for Support of Leading Scientific Schools, and R.A. by NIH grant AI40085. Funding to pay the Open Access publication charges for this article was provided by the authors.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dorsch-Hasler K., Yogo Y., Wimmer E. Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded. J. Virol. 1975;16:1512–1517. doi: 10.1128/jvi.16.6.1512-1517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spector D.H., Baltimore D. Polyadenylic acid on poliovirus RNA IV. Poly(U) in replicative intermediate and double-stranded RNA. Virology. 1975;67:498–505. doi: 10.1016/0042-6822(75)90450-x. [DOI] [PubMed] [Google Scholar]
- 3.Larsen G.R., Dorner A.J., Harris T.J., Wimmer E. The structure of poliovirus replicative form. RNA. 1980;8:1217–1229. doi: 10.1093/nar/8.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawicki D., Wang T., Sawicki S. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 2001;82:385–396. doi: 10.1099/0022-1317-82-2-385. [DOI] [PubMed] [Google Scholar]
- 5.Paul A.V. Possible unifying mechanism of picornavirus genome replication. In: Semler B.L., Wimmer E., editors. Molecular Biology of Picornaviruses. Washington DC: ASM Press; 2002. pp. 227–246. [Google Scholar]
- 6.Andino R., Rieckhof G.E., Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 7.Andino R., Rieckhof G.E., Achacoso P.L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blyn L.B., Swiderek K.M., Richards O., Stahl D.C., Semler B.L., Ehrenfeld E. Poly(rC) binding protein 2 binds to stem–loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl Acad. Sci. USA. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes. Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herold J., Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton D.J., O'Donnell B.J., Flanegan J.B. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKnight K.L., Lemon S.M. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodfellow I., Chaudhry Y., Richardson A., Meredith J., Almond J.W., Barclay W., Evans D.J. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Ooij M.J.M., Vogt D.A., Paul A.V., Castro C., Kuijpers J., van Kuppeveld F.J.M., Cameron C.E., Wimmer E., Andino R., Melchers W.J.G. Structural and functional characterization of the Coxsackievirus B3 CRE(2C): role of CRE(2C) in negative and positive strand RNA synthesis. J. Gen. Virol. 2006;87:103–113. doi: 10.1099/vir.0.81297-0. [DOI] [PubMed] [Google Scholar]
- 15.Gerber K., Wimmer E., Paul V.A. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2A(pro) J. Virol. 2001;75:10979–10990. doi: 10.1128/JVI.75.22.10979-10990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul A.V., Rieder E., Kim D.W., van-Boom J.H., Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul A.V., Yin J., Mugavero J., Rieder E., Liu Y., Wimmer E. A ‘slide-back’ mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J. Biol. Chem. 2003;278:43951–43960. doi: 10.1074/jbc.M307441200. [DOI] [PubMed] [Google Scholar]
- 18.Rieder E., Paul A.V., Kim D.W., van-Boom J.H., Wimmer E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 2000;74:10371–10380. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodfellow I.G., Polacek C., Andino R., Evans D.J. The poliovirus 2C cis-acting replication element-mediated uridylylation of VPg is not required for synthesis of negative-sense genomes. J. Gen. Virol. 2003;84:2359–2363. doi: 10.1099/vir.0.19132-0. [DOI] [PubMed] [Google Scholar]
- 20.Morasco B.J., Sharma N., Parilla J., Flanegan J.B. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 2003;77:5136–5144. doi: 10.1128/JVI.77.9.5136-5144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray K.E., Barton D.J. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 2003;77:4739–4750. doi: 10.1128/JVI.77.8.4739-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul A.V., van-Boom J.H., Filippov D., Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 23.Pilipenko E.V., Maslova S.V., Sinyakov A.N., Agol V.I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992;20:1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilipenko E.V., Poperechny K.V., Maslova S.V., Melchers W.J., Bruins-Slot H.J., Agol V.I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing’) interactions. EMBO. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 25.Melchers W.J., Bakkers J.M., Bruins-Slot H.J., Galama J.M., Agol V.I., Pilipenko E.V. Cross-talk between orientation-dependent recognition determinants of a complex control RNA element, the enterovirus oriR. RNA. 2000;6:976–987. doi: 10.1017/s1355838200000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melchers W.J., Hoenderop J.G., Bruins-Slot H.J., Pleij C.W., Pilipenko E.V., Agol V.I., Galama J.M. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirmomeni M.H., Hughes P.J., Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Bakkers J.M., Galama J.M., Bruins-Slot H.J., Pilipenko E.V., Agol V.I., Melchers W.J. Structural requirements of the higher order RNA kissing element in the enteroviral 3′UTR. Nucleic Acids Res. 1999;27:485–490. doi: 10.1093/nar/27.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todd S., Towner J.S., Brown D.M., Semler B.L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohll J.B., Moon D.H., Evans D.J., Almond J.W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown D.M., Cornell C.T., Tran G.P., Nguyen J.H., Semler B.L. An authentic 3′ noncoding region is necessary for efficient poliovirus replication. J. Virol. 2005;79:11962–11973. doi: 10.1128/JVI.79.18.11962-11973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunner J.E., Nguyen J.H., Roehl H.H., Ho T.V., Swiderek K.M., Semler B.L. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 2005;79:3254–3266. doi: 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma N., O'Donnell B.J., Flanegan J.B. 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J. Virol. 2005;79:3565–3577. doi: 10.1128/JVI.79.6.3565-3577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho M.W., Richards O.C., Dmitrieva T.M., Agol V.I., Ehrenfeld E. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J. Virol. 1993;67:3010–3018. doi: 10.1128/jvi.67.6.3010-3018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agol V.I., Paul A.V., Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. doi: 10.1016/s0168-1702(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 36.van Kuppeveld F.J., Galama J.M., Zoll J., Melchers W.J. Genetic analysis of a hydrophobic domain of coxsackie B3 virus protein 2B: a moderate degree of hydrophobicity is required for a cis-acting function in viral RNA synthesis. J. Virol. 1995;69:7782–7790. doi: 10.1128/jvi.69.12.7782-7790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barton D.J., Black E.P., Flanegan J.B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herold J., Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silvestri L.S., Parilla J.M., Morasco B.J., Ogram S.A., Flanegan J.B. Relationship between poliovirus negative-strand RNA synthesis and the length of the 3′poly(A) tail. Virology. 2005 doi: 10.1016/j.virol.2005.10.019. (in press) [DOI] [PubMed] [Google Scholar]
- 40.Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelan S.P., Barr J.N., Wertz G.W. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 42.Neumann G., Brownlee G.G., Fodor E., Kawaoka Y. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 2004;283:121–143. doi: 10.1007/978-3-662-06099-5_4. [DOI] [PubMed] [Google Scholar]
- 43.Meredith J.M., Rohll J.B., Almond J.W., Evans D.J. Similar interactions of the poliovirus and rhinovirus 3D polymerases with the 3′ untranslated region of rhinovirus 14. J. Virol. 1999;73:9952–9958. doi: 10.1128/jvi.73.12.9952-9958.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mellits K.H., Meredith J.M., Rohll J.B., Evans D.J., Almond J.W. Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J. Gen. Virol. 1998;79:1715–1723. doi: 10.1099/0022-1317-79-7-1715. [DOI] [PubMed] [Google Scholar]
- 45.Harris K.S., Xiang W., Alexander L., Lane W.S., Paul A.V., Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 46.Gmyl A.P., Belousov E.V., Maslova S.V., Khitrina E.V., Chetverin A.B., Agol V.I. Nonreplicative RNA recombination in poliovirus. J. Virol. 1999;73:8958–8965. doi: 10.1128/jvi.73.11.8958-8965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Escarmis C., Gomez-Mariano G., Davila M., Lazaro E., Domingo E. Resistance to extinction of low fitness virus subjected to plaque-to-plaque transfers: diversification by mutation clustering. J. Mol. Biol. 2002;315:647–661. doi: 10.1006/jmbi.2001.5259. [DOI] [PubMed] [Google Scholar]
- 48.Bernstein H.D., Sarnow P., Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J. Virol. 1986;60:1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teterina N.L., Zhou W.D., Cho M.W., Ehrenfeld E. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 1995;69:4245–4254. doi: 10.1128/jvi.69.7.4245-4254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dickson K.S., Thompson S.R., Gray N.K., Wickens M. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem. 2001;9:41810–41816. doi: 10.1074/jbc.M103030200. [DOI] [PubMed] [Google Scholar]
- 51.Neufeld K.L., Galarza J.M., Richards O.C., Summers D.F., Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J. Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranjith-Kumar C.T., Gajewski J., Gutshall L., Maley D., Sarisky R.T., Kao C.C. Terminal nucleotidyl transferase activity of recombinant flaviviridae RNA-dependent RNA polymerases: Implication for viral RNA replication. J. Virol. 2001;75:8615–8623. doi: 10.1128/JVI.75.18.8615-8623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novak J.E., Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes. Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 54.Kuznetsov Y.G., Daijogo S., Zhou J., Semler B., McPherson A. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 2005;347:41–52. doi: 10.1016/j.jmb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 55.van der Bron E., Posthuma C.C., Gultyaev A.P., Snijder E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79:6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawicki S.G., Sawicki D.L. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 2005;87:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gmyl A.P., Korshenko S.A., Belousov E.V., Khitrina E.V., Agol V.I. Nonreplicative homologous RNA recombination: promiscuous joining of RNA species? RNA. 2003;9:1221–1231. doi: 10.1261/rna.5111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gallei A., Pankraz A., Thiel H.J., Becher P. RNA recombination in vivo in the absence of viral replication. J. Virol. 2004;78:6271–6281. doi: 10.1128/JVI.78.12.6271-6281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen I.H., Chou W.J., Lee P.Y., Hsu Y.H., Tsai C.H. The AAUAAA motif of bamboo mosaic virus RNA is involved in minus-strand RNA synthesis and plus strand RNA polyadenylation. J. Virol. 2005;79:14555–14561. doi: 10.1128/JVI.79.23.14555-14561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raju R., Hajjou M., Hill K.R., Botta V., Botta S. In vivo addition of poly(A) tail and AU-rich sequences to the 3′ terminus of the Sindbis virus RNA genome: a novel 3′-end repair pathway. J. Virol. 1999;73:2410–2419. doi: 10.1128/jvi.73.3.2410-2419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]