Abstract
The Snf2 family of helicase-related proteins includes the catalytic subunits of ATP-dependent chromatin remodelling complexes found in all eukaryotes. These act to regulate the structure and dynamic properties of chromatin and so influence a broad range of nuclear processes. We have exploited progress in genome sequencing to assemble a comprehensive catalogue of over 1300 Snf2 family members. Multiple sequence alignment of the helicase-related regions enables 24 distinct subfamilies to be identified, a considerable expansion over earlier surveys. Where information is known, there is a good correlation between biological or biochemical function and these assignments, suggesting Snf2 family motor domains are tuned for specific tasks. Scanning of complete genomes reveals all eukaryotes contain members of multiple subfamilies, whereas they are less common and not ubiquitous in eubacteria or archaea. The large sample of Snf2 proteins enables additional distinguishing conserved sequence blocks within the helicase-like motor to be identified. The establishment of a phylogeny for Snf2 proteins provides an opportunity to make informed assignments of function, and the identification of conserved motifs provides a framework for understanding the mechanisms by which these proteins function.
INTRODUCTION
Some 15 years ago Gorbalenya and Koonin (1,2) identified a large group of proteins sharing a series of short ordered motifs. The majority of members with known function were nucleic acid strand separating helicases so the sequences became known as helicase motifs and were labelled sequentially I, Ia, II, III, IV, V and VI. A number of additional conserved blocks with broad distributions within these helicase-like proteins have subsequently been identified, such as the TxGx (3) and Q motifs (4).
Proteins containing the helicase motifs are subdivided into several superfamilies on the basis of similarity. Structural characterizations have revealed that helicase-like superfamilies 1 and 2 (SF1 and SF2) are related with a common core of two recA-like domains (5). The helicase-like enzymes link ATP hydrolysis to a directed change in the relative orientation of these domains (6). Structural and mutagenesis studies have shown that each of the conserved motifs in the active site cleft between the recA-like domains plays a role in the transformation of chemical energy from ATP hydrolysis to mechanical motion. This enzymatic process has been suggested to represent one application of a more general mechanism used in many proteins containing a recA-like domain (7).
Proteins with a helicase-like region of similar primary sequence to Saccharomyces cerevisiae Snf2p comprise the Snf2 family within SF2 (Figure 1A). Indeed, Snf2p was specifically aligned within SF2 by Gorbalenya and Koonin (1). Many of the first identified Snf2 family members were ATPases within chromatin remodelling complexes and it was recognized that the presence of a core polypeptide related to Snf2p is a defining property of ATP-dependent chromatin remodelling (8). It is now apparent that the Snf2 family comprises a large group of ATP-hydrolysing proteins that are ubiquitous in eukaryotes, but also present in eubacteria and archaea.
Figure 1.
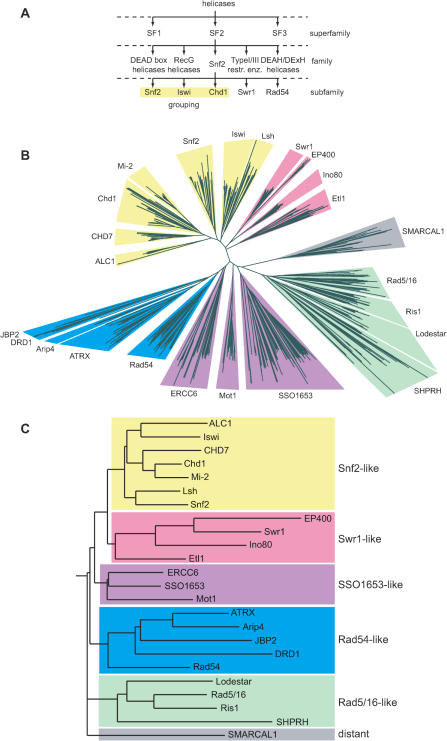
Tree view of Snf2 family. (A) Schematic diagram illustrating hierarchical classification of superfamily, family and subfamily levels. (B) Unrooted radial neighbour-joining tree from a multiple alignment of helicase-like region sequences excluding insertions at the minor and major insertion regions from motifs I to Ia and conserved blocks C–K for 1306 Snf2 proteins identified in the Uniref database. The clear division into subfamilies is illustrated by wedge backgrounds, coloured by grouping of subfamilies. Subfamilies DRD1 and JBP2 were not clearly separated, as discussed in text. (C) In order to illustrate the relationship between subfamilies, a rooted tree was calculated using HMM profiles for full-length alignments of the helicase regions. Groupings of subfamilies are indicated by colouring as in (B).
At least a subset of Snf2 family proteins act as ATP-dependent DNA translocases (9–12). Some of these proteins have also been found to be capable of generating unconstrained superhelical torsion in DNA (11,13–17), proposed to occur as a result of the translocation of DNA into constrained loops. This is substantiated by recent analysis of the action of RSC on single DNA molecules (18). In addition to distorting DNA, the ATP-dependent action of these proteins can disrupt chromatin as measured using a range of different assays (8), although other DNA–protein interactions can also be affected. For example, Rad54 promotes Rad51-dependent strand pairing (13), and Mot1 displaces the TATA-binding protein (TBP) from DNA (19). Thus although many Snf2 family proteins are likely to act to alter chromatin structure, this is not the case for all members of the family.
Early biochemical studies and sequence alignments suggested that members of the Snf2 family could be further subdivided into a number of subfamilies (20). These subfamilies have traditionally taken the name of the archetypal member, such as S.cerevisiae Snf2p (Snf2 subfamily), Drosophila melanogaster Iswi (Iswi subfamily), Mus musculus Chd1 (Chd subfamily) and S.cerevisiae Rad54p (Rad54 subfamily). Snf2p, therefore, lends its name to both the collective Snf2 family and a specific Snf2 subfamily (Figure 1A).
The only comprehensive analysis of the Snf2 family sequences to date was performed by Eisen et al. in 1995 (20). Although it has subsequently been revisited within the context of various biochemical studies (21–24), no broad survey has been conducted for over a decade.
To gain new insights into the Snf2 family, we have catalogued Snf2 family members by scanning for proteins containing spans with similarity in sequence over the helicase-like region, classifying them into subfamilies, analysing the distribution of these subfamilies in complete genomes, and mapping the common sequence characteristics onto the newly available three-dimensional structures. We have identified 24 distinct subfamilies, 11 with near ubiquitous representation in eukaryotic genomes. Many of these subfamilies correlate with known biological function, but there remain a significant number for which little information is currently available. The abundance of Snf2 family members in eukaryotes in comparison to archaea and eubacteria points to their diversification early in eukaryote radiation. This diversity and the currently known functional linkages suggest the Snf2 family helicase-like region is specifically adapted to perform distinct functions within different subfamilies. Underlying this, analysis of the conserved blocks of residues reveals a common core of structural features likely to be fundamental to the mechanism of the Snf2 family motors.
MATERIALS AND METHODS
Data and software sources
Swissprot/Uniprot (25) release 42 and Uniref100 (26) release 5 were downloaded from the European Bioinformatics Institute (ftp://ftp.ebi.ac.uk/pub/databases/swissprot/ and ftp://ftp.ebi.ac.uk/pub/databases/uniprot/, respectively). The sources and version details for the predicted protein complements of the 54 eukaryotic, 24 archaeal and 269 prokaryotic organisms surveyed are available at the webserver http://www.snf2.net/. Analyses were performed on a cluster of dual Pentium III microcomputers running a customized Debian Linux operating system.
Sequence data were manipulated with the EMBOSS suite version 2.8 (27). Multiple sequence alignments were created with Muscle version 3.0 (28) and MAFFT version 5.667 with parameters retree = 2 and maxiterate = 1000 (29) and visualized with Jalview version 2 (30). Phylogenetic and pairwise trees were constructed with neighbor, protdist and drawtree from the PHYLIP suite version 3.572 (31) and additionally visualized with ATV version 2.03 (32) and Hypertree version 1.0.0 (33). Hidden Markov model (HMM) construction, calibration and searching was performed with the HMMer suite versions 2.1.1 and 2.2g (34), and pairwise comparison of HMMs carried out with PRC version 1.5.3 in global-global mode (35). Sequence LOGOs were generated with WebLogo version 2.8.2 (36) and profile logos generated with logomat-p using the draw_logo method, version 0.71 (37). Protein structures were visualized with PyMol version 0.99 (38). Data were managed using mySQL (http://www.mysql.com) and PostgreSQL (http://www.postgresql.org) relational databases. Calculations were carried out using default parameters except where indicated. All other analyses used custom Perl or Python scripts written by the authors. All supplementary data for this report and an interactive database of the results are publicly available at the web server http://www.snf2.net/.
Experimental methods
A full technical description of the library construction and validation and details of the web server will be available elsewhere (D.M.A. Martin and A. Flaus, manuscript in preparation). The procedure used is summarized in outline below.
Global Snf2 family HMM construction
Twenty-eight biochemically characterized S.cerevisiae Snf2p-like chromatin remodelling proteins or close homologues were selected as seed sequences. The core helicase-like region spanning from 50 amino acids N-terminal to helicase motif I to 50 amino acids C-terminal of helicase motif VI was excised from each protein sequence and multiple alignments were created with Muscle using default parameters. An initial seed HMM was constructed following manual assessment of the alignment.
Swissprot 42 was searched using this profile, expanding the set of Snf2 family sequences to 620 candidates with matches up to E-values of 2 although some matches near this cut-off were helicase-like sequences which were not members of the Snf2 family. Further iterations of HMM construction and searching of Swissprot 42 and model organism databases, followed by curation of sequences to remove fragmented sequences and other artefacts not belonging to the Snf2 family, yielded a set of 948 manually curated sequences which were then aligned by MAFFT.
The resultant profile was employed in searching Uniref100 and identified 5046 sequences with a match of E-value 10 or better of which 3932 had E-values below 1 and 1879 had positive bit-scores. This cut-off may appear generous but was intentional to enable maximum possible inclusion of Snf2 family members. It is predicated by the considerable variation in sequence between helicase motifs III and IV giving rise to poor alignments to the general model in this region and consequently lower bit scores. The cut-offs selected were determined by manual inspection of the hit lists and alignments to include established Snf2 family-like proteins but exclude more distant relatives. The highest E-value for a sequence classified into a subfamily (see below) was 2.2 × 10−16.
Phylogenetic analysis of helicase-like sequences
A multiple sequence alignment with Muscle, followed by distance matrix calculation and neighbour-joining tree reconstruction allowed the curation of 2305 sequences into subgroupings in the Snf2 family based on the sequence of the helicase-like region. 1306 sequences were classified in 24 individual subfamilies within the Snf2 family (Table 1) after the exclusion of 436 which were fragmentary (did not span completely from helicase motifs I to VI) or contained unique large inserts or deletions. A further 220 were assigned to the prokaryotic rapA group and the remainder to more distantly related clusters or as highly truncated outliers which could not be reasonably aligned. An overview of a neighbour-joining tree constructed from a multiple alignment excluding the variable minor and major insertion regions (see text) and visualized using Hypertree demonstrates the clearly distinguishable division of subfamilies (Figure 1B). Each subfamily was individually examined for redundancy and further internal structure (data not shown).
Table 1.
Summary of subfamilies
| Subfamily | Archetype gene | Assigned from Uniref | Other names associated with members |
|---|---|---|---|
| Snf2 | S.cerevisiae SNF2 | 117 | Snf2p, Sth1p, snf21, SMARCA4, BRG1, BAF190, hSNF2beta, SNF2L4, SMARCA2, hBRM, hSNF2a, SNF2L2, SNF2LA, SYD, splayed, psa-4, brahma |
| Iswi | D.melanogaster Iswi | 83 | Isw1p, Isw2p, SMARCA1, SNF2L, SNF2L1, SNF2LB, SMARCA5, hSNF2H |
| Lsh | M.musculus Hells | 35 | YFR038W, SMARCA6, HELLS, LSH, PASG, DDM1, cha101 |
| ALC1 | Homo sapiens CHD1L | 19 | SNF2P |
| Chd1 | M.musculus Chd1 | 96 | CHD2, CHD-Z, hrp1, hrp3 |
| Mi-2 | H.sapiens CHD3 | 88 | CHD3, Mi-2a, Mi2alpha, ZFH, PKL, pickle, CHD4, Mi-2b, Mi2beta, let-418, CHD5 |
| CHD7 | H.sapiens CHD7 | 53 | CHD6, RIGB, KISH2, Kis-L, kismet, CHD8, HELSNF1, DUPLIN |
| Swr1 | S.cerevisiae SWR1 | 44 | SRCAP, Snf2-related CBP activator protein, dom, domino, PIE1 |
| EP400 | H.sapiens EP400 | 27 | E1A binding proten p400, TNRC12, hDomino |
| Ino80 | S.cerevisiae INO80 | 34 | |
| Etl1 | M.musculus Smarcad1 | 44 | SMARCAD1, hHEL1, Fun30p, snf2SR |
| Rad54 | S.cerevisiae RAD54 | 76 | Rad54l, hRAD54, RAD54A, Rdh54p, RAD54B, Tid1, okr, okra, mus-25 |
| ATRX | H.sapiens ATRX | 52 | XH2, XNP, Hp1bp2 |
| Arip4 | M.musculus Srisnf2l | 23 | ARIP4 |
| DRD1 | Arabidopsis thaliana DRD1 | 12 | |
| JBP2 | T.brucei JBP2 | 4 | |
| Rad5/16 | S.cerevisiae RAD5, RAD16 | 61 | rhp16, rad8, SMARCA3, SNF2L3, HIP116, HLTF, ZBU1, RNF80, RUSH-1alpha, P113, MUG13.1 |
| Ris1 | S.cerevisiae RIS1 | 35 | |
| Lodestar | D.melanogaster Lodestar | 40 | LDS, TTF2, hLodestar, HuF2,factor 2 |
| SHPRH | H.sapiens SHPRH | 44 | YLR247C |
| Mot1 | S.cerevisiae MOT1 | 45 | TAFII170, TAF172, BTAF1, Hel89B |
| ERCC6 | H.sapiens ERCC6 | 71 | rad26, rhp26, CSB, csb-1, RAD26L |
| SSO1653 | S.solfataricus SSO1653 | 149 | SsoRad54like |
| SMARCAL1 | H.sapiens SMARCAL1 | 54 | HARP, DAAD, ZRANB3, Marcal1 |
Listing subfamily name from prevailing protein name for first characterized member, archetype organism and official gene name, number of subfamily members identified in Uniref as in Materials and Methods, and a non-exhaustive list of alternative names for archetype and other subfamily members.
Construction and use of subfamily profiles
Each subfamily sequence set was realigned by MAFFT, manually curated with Jalview and an HMM profile constructed. These profiles were aligned with PRC in an all-against-all comparison. Although these profile comparisons do not give a true phylogenetic tree, the scores obtained from the pairwise profile alignments can be used to construct a representational tree (Figure 1C), indicating the relationship between the HMM profiles to be consistent with the sequence-based tree (Figure 1B). It was also observed that the subfamilies could be aggregated into some broad groupings that correlate with functional properties, where known (Figure 1).
All 24 subfamily profiles were combined in one HMM library and the hmmpfam application employed in searching individual genomic datasets to provide phylogenomic information about the taxonomic distribution of Snf2-like proteins. The subfamily hit with maximal bitscore >100 was used to assign membership in a semi-automatic procedure. With very few exceptions, classification was extremely clear with strong discrimination between the top hit and the second best hit (data not shown).
RESULTS AND DISCUSSION
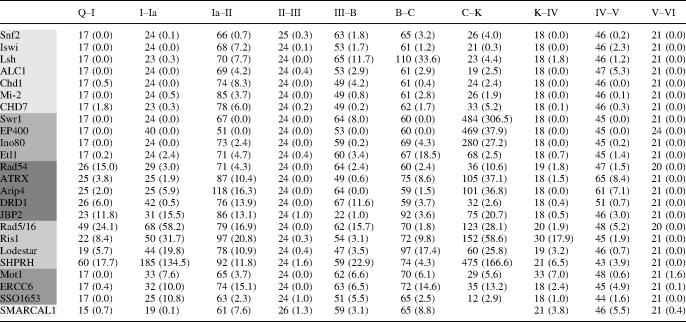
Starting from a seed set of helicase-like region sequences from 28 demonstrated Snf2p-related proteins or close homologues, we have carried out a broad survey of Snf2 family proteins. This was achieved by iterative cycles of manual curation of multiple alignments and neighbour-joining trees to identify Snf2 proteins by similarity, construction of an HMM profile from the multiple alignments of identified proteins, and scanning of global and model organism protein databases using the HMM profile to uncover further sequences for curation. Our current global Snf2 family profile scan revealed 3932 sequences with E-value under 1 (1879 with bitscore > 0) in Uniref100 release 5 [2.4 million entries (26)]. Of these, 1306 sequences were identified as belonging to the Snf2 family and to span the full helicase region from motifs I to VI without introducing large unique insertions or deletions. A further 220 sequences fall within the rapA group, while other hits appear to belong to more distantly related groups (see below) or were too highly truncated to be aligned. Neighbour-joining trees from multiple alignments of the set of 1306 sequences revealed a well-defined branching structure (Figure 1B) and enabled their assignment to 24 distinct subfamilies (Table 1).
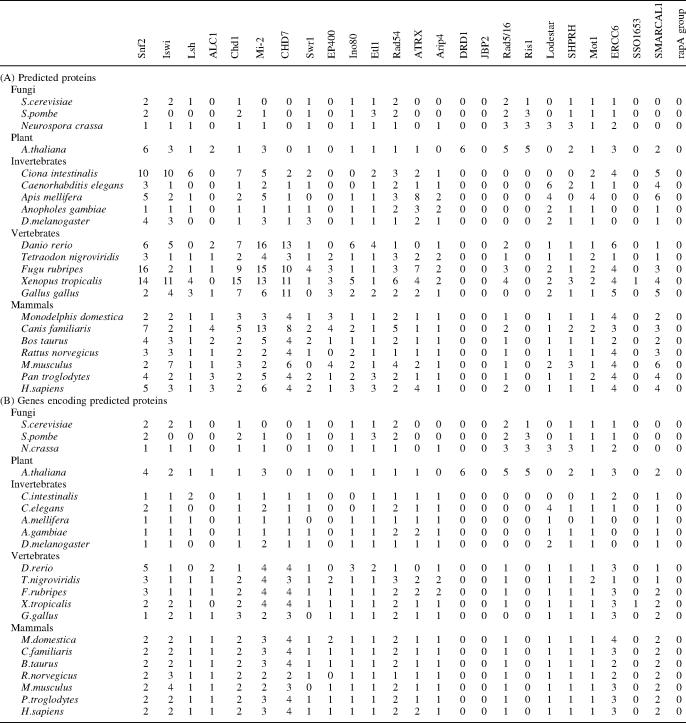
Subfamily-specific HMM profiles were constructed from these assignments and used to characterize the Snf2 family complement for 54 complete eukaryote genomes. The counts of predicted proteins and unique encoding genes for 21 selected genomes are listed in Table 2, part A and B, respectively (see Supplementary Table S1A for full analysis of eukaryotic genomes, and Supplementary Table S2 for gene IDs by subfamily for seven common model organisms). In addition, 24 complete archaeal and 269 bacterial genomes were scanned (Supplementary Tables S1B and S1C).
Table 2.
Subfamily occurrences in selected complete eukaryotic genomes
A: Counts by genome of predicted proteins assigned to each subfamily, for assignments based on highest positive bitscore for protein against each subfamily HMM profile in turn, where maximum bitscore >100. For a complete list of 54 genomes see Supplementary Table S1A. B: Counts of unique genes encoding the predicted proteins listed in part A. Single gene encoding each protein assumed for fungal genomes. Gene names for seven model organisms are tabulated in Supplementary Table S2, including official protein names where assigned.
Subfamilies within the Snf2 family
The clear distinction and significant number of subfamilies based on the helicase-like region (Figure 1B and Table 1) reflects both a remarkable breadth and specificity in the Snf2 family. An additional level of similarity distinguishes apparent groupings of subfamilies (Figure 1), which echo current understanding of their functional diversity (Table 3). Most of the best studied Snf2 family proteins fall into a grouping of ‘Snf2-like’ subfamilies including proteins such as S.cerevisiae Snf2p, D.melanogaster Iswi, mouse Chd1 and human Mi-2, which are core subunits of the well-known ATP-dependent chromatin remodelling complexes. A separate ‘Swr1-like’ grouping encompasses the Swr1, Ino80, EP400 and Etl1 subfamilies. The ‘Rad54-like’ grouping contains the Rad54 subfamily, relatives such as ATRX and Arip4, and also includes the recently recognized DRD1 and JBP2 proteins. A further, unexpected, ‘Rad5/16-like’ grouping links several poorly studied subfamilies, three of which contain RING finger insertions within the helicase-like region (see below). The ‘SSO1653-like’ grouping of Mot1, ERCC6 and SSO1653 is notable because all three subfamilies are thought to have non-chromatin substrates. Finally, we have labelled SMARCAL1 proteins as ‘distant’ because they lack several otherwise conserved sequence hallmarks of the Snf2 family (see below). Although some groupings are clear, further investigations will be required to verify those where the boundaries are less distinct.
Table 3.
Functional and sequence characteristics of subfamilies
| Grouping | Subfamily | Functional characteristics |
|---|---|---|
| Snf2-like | Snf2 | The archetype of the Snf2 subfamily, and the entire Snf2 family, is the S.cerevisiae Snf2 protein, originally identified genetically (mutations that were Sucrose Non Fermenting or defective in mating type SWItching). However, these genes were later found to play roles in regulating transcription of a broader spectrum of genes and to catalyse alterations to chromatin structure. Subsequently, the proteins were purified as a non-essential 11 subunit multi-protein complex capable of ATP-dependent chromatin disruption termed the SWI/SNF complex (8) |
| Close sequence homologues have also been identified in many model organisms, including the paralogue RSC (60,61) and the orthologues D.melanogaster Brahma (62), and human hBRM (63) and BRG1 (64). Many of these have been shown to alter the structure of chromatin at the nucleosomal level and to be involved in transcription regulation, although other nucleosome-related roles have also been identified (8). Recent hypotheses have centred on Snf2 subfamily members performing a generally disruptive function on nucleosomes leading either to sliding of the nucleosome (65,66) alterations of histone DNA contacts (67) or to partial or complete removal of the histone octamer components (68,69) | ||
| Homologues such as BRG1 and hBRM are components of megadalton-sized complexes containing other proteins that are also related to components of the yeast SWI/SNF complex (70). However, Snf2 subfamily members have also been reported to interact with additional proteins including histone deacetylases (71), methyl DNA-binding proteins (72), histone methyl transferases (73), the retinoblastoma tumor suppressor protein (74,75), histone chaperones (76), Pol II (77,78) and cohesin (79). These complexes may be recruited to specific regions of the genome through interactions with sequence-specific DNA-binding proteins [reviewed by (80)] or specific patterns of histone modifications (81,82) | ||
| ISWI | Iswi (Imitation SWI2) protein was identified in D.melanogaster by similarity to Snf2p (83) and is at the catalytic core of both the NURF and the ACF/CHRAC chromatin remodelling complexes (84–86). Biochemical studies favour the ability of Iswi proteins to reposition rather than disrupt nucleosomes. Significantly, all Iswi subfamily proteins require a particular region of the histone H4 tail near the DNA surface as an allosteric effector (87–89) | |
| Iswi subfamily members participate in a variety of complexes and functional interactions. For example, human SNF2H has been found as part of RSF, hACF/WCRF, hWICH, hCHRAC, NoRC and also associated with cohesin, while SNF2L is the catalytic subunit of human NURF [summarized in (90)]. Such complexes are involved in a variety of functions including activation/repression of the initiation and elongation of transcription, replication and chromatin assembly [reviewed in (90–93)]. Similar to the Snf2 subfamily, Iswi subfamily members appear to be adaptable subunits for complexes related to the alteration of nucleosome positioning (90) | ||
| Lsh | Despite its name, the archetypal mouse Lsh (lymphoid-specific helicase) protein (94) is widely expressed and without detectable helicase activity. Lsh and its human homologue are alternatively known as PASG, SMARCA6 or by the official gene name HELLS (Helicase Lymphoid Specific). Mutants lead to premature aging with cells exhibiting replicative senescence (95). Importantly, global loss of CpG methylation is observed in both mammalian mutant cell lines and the Arabidopsis thaliana homologue, DDM1 (96,97). Consistent with a direct role in DNA methylation, Lsh is localized to heterochromatic regions (98). Evidence has been presented that A.thaliana DDM1 can slide nucleosomes in vitro (99). The S.cerevisiae subfamily member at locus YFR038W has no assigned name and deletion strains are viable | |
| Lsh subfamily members are detected over a very broad range of eukaryotes including not only fungi, plants and animals, but also protists where their function is likely to be independent of DNA methylation. Furthermore, our genome scans also did not identify Lsh subfamily members in a number of lower animals, or in S.pombe. This may represent functional redundancy or difficulties in assigning distant homologues relative to other subfamilies in the grouping | ||
| ALC1 | The ALC1 subfamily derives its name from the observation that the human gene is ‘Amplified in Liver Cancer’ (100). Two alternative but potentially confusing names, CHD1L [CHD1-like (101)] and SNF2P [SNF2-like in plants (102)], have also been used to refer to subfamily members. ALC1 subfamily members contain a helicase-like region which is relatively similar to the nucleosome-active Snf2, Iswi and Chd1 subfamilies, but which is coupled ahead to a macro domain implicated in ADP–ribose interactions (103). ALC1 subfamily members can be identified in both higher animals and plants, but not in lower animals | |
| Chd1 | The archetypal ‘Chd’ protein is mouse Chd1, named after the presence of ‘Chromodomain, Helicase and DNA binding’ motifs (104). The characteristic chromodomain motifs can in principle bind diverse targets including proteins, DNA and RNA (105). Although Snf2 family proteins containing chromodomains are often referred to as a single ‘Chd’ subfamily, it has previously been recognized they fall into the same three distinct subfamilies (106,107) which we have distinguished in this analysis | |
| Mouse Chd1 protein is the archetype of the first chromodomain-containing subfamily. Chd1 proteins have been purified as single subunits (108,109) although associations between Chd1 and other proteins have been identified subsequently (110,111). Yeast Chd1 has been implicated in transcription elongation and termination (58,112), and the human CHD1 and CHD2 proteins and D.melanogaster dChd1 have been linked to transcriptional events (113). S.pombe Chd1 subfamily member hrp1 (helicase related protein 1) has been linked with both transcription termination (58) and chromosome condensation (114), whereas the paralogous hrp3 has been linked with locus-specific silencing (115) | ||
| Mi-2 | The second of the chromodomain-containing subfamilies is Mi-2, whose name derives from the Mi-2α and Mi-2β proteins which are the commonly used names for the human CHD3 and CHD4 gene products, respectively. Mi-2 was isolated as an autoantigen in the human disease dermatomyositis (116). Subsequently, the two proteins and their homologues in D.melanogaster and Xenopus have been recognized as core subunits of NuRD complexes which link DNA methylation to chromatin remodelling and deacetylation (117). The chromodomains in D.melanogaster Mi-2 are required for activity on nucleosome substrates (118). Human Mi-2α differs from Mi-2β principally by its additional C-terminal domain which directs complexes containing it for a specific transcriptional repression role (119). Since Mi-2 proteins are widely expressed but have specific roles, it has been suggested they may be directed by incorporation of different targeting subunits (117,120) | |
| An additional human member of the Mi-2 subfamily, CHD5, may have a role in neural development and neuroblastomas (121,122) although its biochemical associations are unknown. The A.thaliana subfamily member, PKL (swollen roots of mutants resemble a pickle), has been shown to play a role in repressing embryonic genes during plant development (123) | ||
| CHD7 | The third chromodomain-containing CHD7 subfamily includes four human genes, CHD6–CHD9. CHD7 has recently been linked to CHARGE syndrome which is a common cause of congenital abnormalities (124) with most linked mutations resulting in major nonsense, frameshift or splicing changes (125). There is little functional information available about CHD6 [originally known as CHD5 (126)], CHD8 or CHD9 [also known as CReMM (127)]. The most studied member of the CHD7 subfamily is the product of the D.melanogaster gene kismet. The enormous 574 kDa KIS-L (but not the ‘smaller’ 225 kDa KIS-S form) contains a Snf2 family helicase-like region (128). Although identified as a trithorax family gene acting during development, a recent report suggests that KIS-L may play a global role at an early stage in RNA pol II elongation (129) | |
| Swr1-like | Swr1 | The archetype of the Swr1 subfamily is Swr1p (SWI/SNF-related protein) from S.cerevisiae which is part of the large SWR1 complex that exchanges histone H2A.Z-containing for wild-type H2A-containing dimers (41–43,130). Three other characterized proteins belong to the subfamily: PIE1 is involved in the A.thaliana vernalization regulation [Photoperiod-Independent Early flowering (131)] through a pathway intimately linked with histone lysine methylation events (132). D.melanogaster Domino is an essential, development-linked protein (133) and alleles can suppress position effect variegation, implying they may be linked to heterochromatin functions. Domino participates in a complex that combines components of the homologous yeast SWR1 and NuA4 complexes (134), and has been shown to function in acetylation dependent histone variant exchange within the TRRAP/TIP60 complex (135). The human member SRCAP [Snf2p-related CBP activating protein (136)] acts as a transcriptional co-activator of steroid hormone dependent genes and has recently been shown to be a component of a human TRRAP/TIP60 complex (137) and other complexes (see EP400 below). It also interacts with several coactivators including CBP (136,138). SRCAP can rescue D.melanogaster domino mutants (139), implying functional homology |
| EP400 | The EP400 subfamily archetype, E1A-binding protein p400, appears to have a role in regulation of E1A-activated genes (140–142). EP400 has been shown to interact strongly with ruvB-like helicases TAP54α/β in the TRRAP/TIP60 histone acetyl transferase complex (142) | |
| The complex patterns of similarities and distinctions between EP400 and Swr1 subfamilies suggest a close functional relationship (Supplementary Figure S3A). Our HMM profiles clearly distinguish EP400 from Swr1 members, and show that EP400 members are restricted to vertebrates whereas Swr1 subfamily members are found in almost all eukaryotes (Table 2 and Supplementary Table S1A). Although most vertebrate genomes contain a gene each for an Swr1 and EP400 member, some have only one of the pair. The consensus for the helicase-like regions of the subfamilies shows 50% identity and animal members of both contain large proline and serine/threonine rich insertions at the major insertion site (Supplementary Figure S3A; see below). Members of the two subfamilies also contain overlapping combinations of accessory domains outside the helicase-like region: D.melanogaster DominoA (Swr1 subfamily) and human EP400 (EP400 subfamily) both contain SANT domains, whereas human SRCAP (Swr1 subfamily) instead contains an AT hook (139). This has led to some confusion, with human EP400 being referred to as hDomino although D.melanogaster Domino has higher similarity to human SRCAP (142) and SRCAP can complement Domino mutants (139). In addition to the complexity in primary sequence relationships, complexes of potentially overlapping composition exist involving human EP400 and SRCAP including the NCoR-1 histone deacetylase (143), TRRAP/TIP60 histone acetylase (137,142,144) and DMAP1 complex (145). The confusing overlap of the mammalian Swr1 and EP400 subfamily members may stem from multiple roles for Swr1 subfamily members in lower animals and fungi. For example, it has been suggested that D.melanogaster alternative splice isoforms DominoA and DominoB are functional homologues of EP400 and SRCAP, respectively (139), and that S.cerevisiae Eaf1p is a functional homologue of human EP400 (134) although it lacks both Snf2-related helicase-like and extended proline-rich regions | ||
| Ino80 | The archetype of the Ino80 subfamily is the Ino80 protein from S.cerevisiae. Further members have been identified by sequence similarity in fungi, plants and animals (24). Ino80p was first isolated through its role in transcriptional regulation of inositol biosynthesis (146,147) and forms part of the large Ino80.com complex (148). This complex is notable not only because it can reposition nucleosomes, but also because it is the only known Snf2 family-related complex able to separate DNA strands in a traditional helicase assay (148). However, the Ino80 complex contains two RuvB-like helicase subunits which may assist in this. The human INO80 complex has recently been shown to contain many proteins homologous to Ino80.com subunits, including the RuvB-like helicases, and to be capable of mobilizing mononucleosomes (149). S.cerevisiae INO80-deleted strains are sensitive to DNA damaging agents, and recent studies have implicated Ino80p directly in the events of double-stranded break repair (150,151), perhaps for the eviction of nucleosomes in the vicinity of the break (152) although other remodelling complexes such as Swr1, RSC and SWI/SNF may also participate in this repair pathway [reviewed in (153)] | |
| Etl1 | Mouse Etl1 (Enhancer Trap Locus 1) derives its name from identification in an expression screen for loci having interesting properties in early development (154). Although members are present in all but the lowest eukaryotes, including the human homologue SMARCAD1 (SMARCA containing DEAD box 1) (155) and S.cerevisiae FUN30 (Function Unknown 30) (156), very little attention has been focussed on these proteins. Etl1 is very widely expressed but non-essential, although deletion causes a variety of significant developmental phenotypes (157). FUN30 deletions are viable although temperature sensitive (158), and mutants show decreased sensitivity to UV radiation (159) | |
| Rad54-like | Rad54 | The archetype of the Rad54 subfamily is the Rad54 protein from S.cerevisiae which was isolated because its inactivation leads to increased sensitivity to ionizing radiation. Rad54p and its homologues in other organisms play an important but as yet incompletely understood role in homologous recombination by stimulating Rad51-mediated single strand invasion into the target duplex, and subsequent steps in the process (160,161). Many organisms also contain a second subfamily member, such as S.cerevisiae Rdh54p or S.pombe tid1p. These are frequently implicated in mitotic repair and meiotic crossover (162), although the role of the human homologue RAD54B is unclear (163) |
| Rad54 proteins have been extensively studied in vitro. They have been shown to be able to generate local changes in DNA topology in supercoiled plasmids (13,164,165), to translocate along DNA by biochemical (11) and other methods, and to alter the accessibility of nucleosomal DNA (11,166,167). However, this latter activity appears inefficient compared to purified complexes from the Snf2 and Iswi subfamilies. The crystal structure of the zebrafish Rad54 helicase-like region has been determined recently (47) and is discussed in more detail in the text | ||
| ATRX | The ATRX subfamily derives its name from the Alpha Thalassemia/Mental Retardation syndrome, X-linked genetic disorder caused by defects in the activity of the human member, ATRX (168). This protein is localized to centromeric heterochromatin (169), and purified complexes have been shown to increase the accessibility of nucleosomal DNA although with only moderate efficiency (12). ATRX has been implicated both in the regulation of transcription and heterochromatin structure (170), although the mechanism by which it acts is unclear | |
| Arip4 | Mouse androgen receptor interacting protein 4 (171) can bind to DNA and generate ATP-dependent local torsion (16). Although it can also bind nucleosomes, Arip4 does not appear to be able to alter their nuclease sensitivity, leading to the conclusion that nucleosome mobilization may not be its primary role (172). Interestingly, mutation of the six lysine sumoylation sites in the protein destroyed DNA binding and ATPase activity (172) | |
| DRD1 | A.thaliana DRD1 is named from its phenotype ‘Defective in RNA-directed DNA methylation’ (173). DRD1 functions together with an atypical RNA polymerase IV to establish and also remove non-CpG DNA methylation as part of an RNA interference mediated pathway (174,175) | |
| JBP2 | The JBP2 subfamily takes its name from the T.brucei J Binding Protein 2 which regulates insertion of an unusual glycosylated thymine-derived base, J, which marks silenced telomeric DNA (176) | |
| Rad54-like (cont.) | Both DRD1 and JBP2 are involved in processes which target modifications at the C5 position of the pyrimidine ring which will be exposed in the major groove. JBP2 and DRD1 members show sequence similarity, but have been conservatively assigned to separate subfamilies due to their distinct evolutionary ranges and the limited numbers of members available for building HMM profiles. The identification of a DRD1 subfamily member in Dictyostelium suggests that the subfamilies may be more widespread than indicated by the current small sample set. The L.major, T.brucei and T.cruzi genomes with JBP2 subfamily members also contain proteins assigned to the Arip4 subfamily whose members are otherwise found in higher fungi and animals, but not in DRD1-containing plants. It is possible that a relationship encompasses not only DRD1 and JBP2, but also Arip4 | |
| Rad5/16-like | Rad5/16 | S.cerevisiae Rad5 and Rad16 proteins are distinct but dual archetypes for this subfamily and both are intimately involved in DNA repair pathways |
| Rad5p acts with the Ubc13p–Mms2p E2 ligase complex via its RING finger in one fate of the Rad6 pathway of replication linked DNA damage bypass to poly-ubiquitylate PCNA in (177). It has also been suggested that Rad5p participates in double-stranded break repair in a role dependent on its helicase-like region but not its RING finger (178). A clear function for the helicase-like motor in either role has not been suggested | ||
| Rad16p acts in complex with Rad7p and Elc1p as the NEF4 nucleotide excision repair factor (179,180), possibly scanning along chromatin for lesions as part of non-transcribed strand repair (179) or by distorting DNA to expose the lesion for processing (17). Although the basis is not known, the RING finger of Rad16p influences the stability of the Rad4 protein responsible for recognizing the lesion (180) | ||
| Paradoxically, no DNA repair link has been reported for the single member of the Rad5/16 subfamily present in each mammalian genome such as human SMARCA3 (see also Lodestar and ERCC6 sections of this table). Instead, under the name RUSH1alpha, some have been reported as steroid regulated transcriptional regulators (181) and, under the name HLTF, to be silenced in cancers (182) | ||
| Ris1 | The Ris1 protein from S.cerevisiae interacts with Sir4p and has a role in mating type silencing (183) Members are found in all fungi and plant genomes, but not in animals or lower eukaryotes | |
| Lodestar | This subfamily is the only one within the Rad5/16 grouping which does not contain RING fingers in the major insertion site (Figure S3B). D.melanogaster Lodestar protein was first identified as an essential cell-cycle regulated protein localizing to chromosomes during mitosis (184). Subsequently, the human homologue TTF2 was shown to terminate elongating RNA pol I and pol II complexes independently of transcript length, possibly by directly clearing Pol II from the template at the entry to mitosis (185). TTF2 may also have a role in interphase termination, and in repair (185). This suggestion is interesting because no clear functional homologue of S.cerevisiae Rad5p has been identified in the higher eukaryote genomes which Lodestar is restricted to (see also ERCC6 section of this table). TTF2 has been observed to rescue RNA polymerases stalled at lesions (186) | |
| SHPRH | SHPRH proteins derive their name from the ordered sequence of domains Snf2_N, Linker_Histone (i.e. H1), PHD finger, Zf_C3HC4 (i.e. e RING finger), Helicase_C in the human member (187) (Supplementary Figure S3B). The Linker_Histone and PHD finger motifs are located adjacent to each other at the minor insertion site between motifs I and Ia, whereas the RING finger domain is located at the major insertion site. The linker histone-related domain in human SHPRH corresponds to the globular winged helix structure of histone H1 (188) and transcription factor HNF3 (189,190). The PHD finger motifs are specialized zinc finger structures which occur in a range of proteins involved in chromatin-mediated transcriptional regulation but whose exact function is unclear (191,192) | |
| Fungal SHPRH subfamily members typically do not contain the linker histone-related motif, although they do contain the PHD and RING finger domains. Animal SHPRH members contain an additional ∼50 kDa polypeptide sequence immediately upstream of the RING finger domain within the major insertion region (Supplementary Figure S3B). Although lacking an identifiable motif, this region has a number of cysteines suggestive of a zinc finger type coordination and has some 30% charged residues. Fungal members also contain a significant region of charged residues ahead of the RING finger | ||
| SSO1653-like | Mot1 | S.cerevisiae Mot1 protein (Modifier of Transcription) (193) and homologues with highly conserved helicase-like regions are present across fungi and all higher eukaryotes, where they are known as BTAF1 or TAF172 (194). In vitro and in vivo studies suggest that Mot1p interacts intimately with TBP (195), probably acting to recycle it from DNA-bound states (196). Mot1p is therefore thought to be a Snf2 family enzyme whose role is not to manipulate nucleosome structure, although a possible direct involvement with chromatin has also been proposed (197) |
| ERCC6 | Human ERCC6 protein (198), also known as Cockayne Syndrome B (CSB), and S.cerevisiae homologue Rad26p (199) were initially regarded as repair proteins due to effects on transcription coupled nucleotide excision repair. However, it has more recently been suggested that the function of these proteins may be to assist transcribing RNA polymerases to either pass or dissociate from blocking DNA lesions (200). Such a role would not directly involve chromatin. The consequent barriers to transcription elongation and sensitivity to DNA damage for non-functional mutants would explain features of Cockayne syndrome and is analogous to the role of the non-Snf2 family Mfd DNA translocase from Escherichia coli (201) | |
| Most higher animal genomes contain three separate genes assigned to the ERCC6 subfamily, along with single Lodestar and Rad5/16 subfamily members. Conversely, fungal genomes typically encode a single ERCC6 member, no Lodestar subfamily member, but at least two Rad5/16 members. This may reflect divergent strategies for accomplishing transcription-coupled repair | ||
| A number of mutations in the helicase region which result in Cockayne syndrome have been identified and these map to interesting locations in the Snf2 family crystal structures (46,198). In vitro, purified ERCC6 protein can alter nuclease sensitivity and spacing of nucleosomes in an ATP-dependent manner (202). ERCC6 can also bind and negatively supercoil DNA in the presence of non-hydrolysable ATP analogues (203) | ||
| SSO1653 | SSO1653, the sole Snf2 family member in archaeal S.solfataricus, is the archetype for the uniquely archaeal and eubacterial subfamily most similar to the eukaryotic Snf2 proteins (see text). It is encoded in the P2 strain genome by juxtaposing SSO1653 and SSO1655 genes, which are punctuated by transposase SSO1654 inserted into the second recA domain immediately upstream of motif V. Although it is highly unlikely that a Snf2 family enzyme would be functional with a 40 kDa transposase insertion in this conserved part of the protein, the enzyme with transposase removed can generate DNA torsion in an ATP-dependent analogous to eukaryotic Snf2 family proteins and was used successfully for structure determination (46). Since a full-length gene can be cloned with appropriate screening (M. F. White, personal communication), the SSO1654 transposase must be active and we refer to the re-fusion of SSO1653 and SSO1655 for simplicity as SSO1653. No information is available for the biological role of any member of the subfamily, although a role in an archaeal chromatin remodelling can be excluded because S.solfataricus lacks archaeal histone-like proteins | |
| SSO1653-like (cont.) | The SSO1653 subfamily helicase-like region also shows close linkage with a zinc finger SWIM motif that may bind to nucleic acids (204,205). For example, coordinately regulated SSO1656 immediately downstream of SSO1653–1655 encodes a 26 kDa basic protein containing the SWIM motif. An SSO1653 subfamily member is present in all Bacillus and Streptococcus genomes (21), and many of these polypeptides also carry a SWIM motif in the same polypeptide. Polypeptides with a Snf2 family helicase-like region but lacking a SWIM motif appear to be in the same operon as a second smaller protein which carries the SWIM motif instead (204). Although the SWIM motif also occurs in eukaryotes, it has not been linked to any of the eukaryotic Snf2 family proteins (204) | |
| Distant | SMARCAL1 | The human SMARCAL1 (SMARCA-Like 1) protein and homologues, also known as HARP (22), are unusual within the analysis because they include two subtypes with highly similar helicase-like regions that are flanked by completely different auxiliary domains. The first consists of proteins in higher eukaryotes related to human SMARCAL1 itself with centrally located helicase-like regions and one or more Harp motifs immediately N-terminal to this. Mutants in the helicase-like region of human SMARCAL1 have been linked to a genetic disorder Schimke immuno-osseous dysplasia (206) although the molecular function of the SMARCAL1 protein is unknown. The bovine homologue ADAAD is stimulated by DNA single-double strand boundaries (207) |
| The second subtype is found in animal, plant and some protist genomes and contains SMARCAL1 subfamily members related in overall domain organization to the human ZRANB3 protein (Zinc finger, RAN-Binding domain containing 3). The helicase region is located at the N-terminus of the polypeptide, followed by an unusual zinc finger structure related to those found in Ran protein binding proteins (208), and a putative HNH type endonuclease domain at the C-terminus (209). No functional information about any proteins in this subtype is available | ||
| rapA group | The rapA group includes some 220 eubacterial and archaeal members with significantly more sequence variation than other subfamilies. Subsets of sequences are qualitatively visible within multiple alignments of the rapA group, but initial attempts to distinguish them have been unreliable due to the variability of microbial sequences and non-homogeneous sampling in sequenced organisms (e.g. half of all complete bacterial genomes are for a limited range of firmicute and gamma proteobacterial genera) | |
| Although the rapA group contains the conserved sequence patterns of the Snf2 family for the classical helicase-like motifs, other conserved blocks cannot be easily identified (Supplementary Figure S4). The characteristic extended span of at least 160 residues between helicase motifs III and IV (44) is maintained in the rapA group, but the central part of this region diverges markedly from the other subfamilies and lacks the highly conserved features characteristic of the Snf2 family. The specific difficulty of aligning this region has been remarked previously (20) | ||
| The rapA group also includes a number of polypeptides for which the helicase-like region comprises effectively the entire polypeptide, in contrast with other Snf2 family members which almost universally contain sequences outside the helicase-like region that are likely to form accessory domains or interaction surfaces | ||
| The only member of this group for which biological function has been investigated is E.coli rapA, also known as HepA (210), which influences polymerase recycling under high salt conditions, possibly by aiding the release of stalled polymerases (211) |
Summaries of known biochemical, biological and distinctive sequence of each subfamily. Background colouring of subfamilies for groupings as shown in Figure 1.
Since the subfamily assignments are based only on the common helicase-like region, this suggests that the ‘motor’ at the core of even large multiprotein remodeller complexes is tuned to the mechanistic requirements of its function. Such properties are not unprecedented for motor protein subfamilies. The ubiquitous kinesin and myosin proteins are divided into at least 14 and 17 subfamilies, respectively (39,40), and those subfamilies are recognized to reflect tuning of the motors for enzymatic properties linked to particular functional roles. As this also appears to be true for Snf2 family proteins we can anticipate that mechanistic features of the motors will be shared within subfamilies and groupings. This may be useful in helping to predict function of poorly characterized proteins. For example, owing to the recent observation that Swr1 functions in histone exchange (41–43), it is tempting to speculate that the Snf2 motors within other subfamilies in the Swr1-like grouping may be adapted for related purposes.
Owing to the remarkable diversity revealed by this classification and the occurrence of many subfamilies which have not been intensively investigated, we briefly summarize current functional and biochemical understanding and characteristic features of each subfamily in Table 3.
Defining the Snf2 family
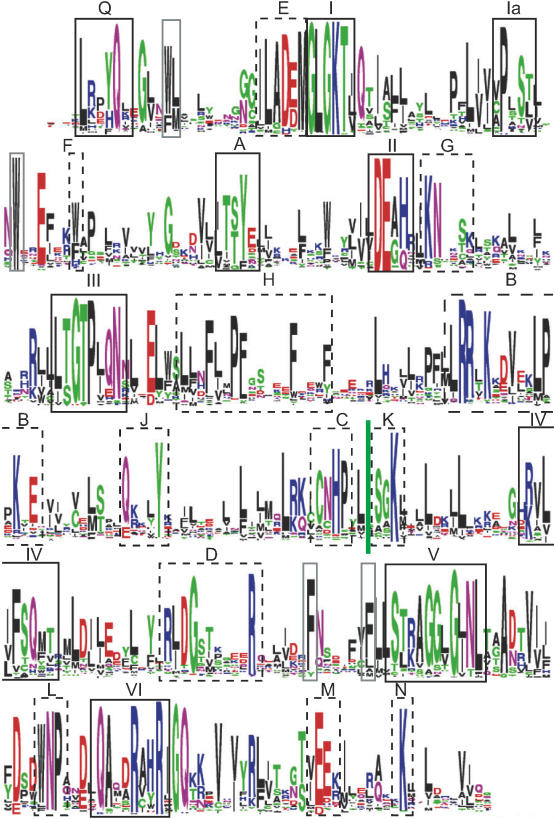
The survey of Snf2 family proteins enables detailed analysis of sequence conservation in the helicase-like region (Figure 2). This reveals a number of unique features distinguishing them from other helicase superfamily SF2 members. First, the conserved helicase motifs show a highly conserved character across the Snf2 family, and some motifs are extended by juxtaposed residues such as conserved blocks E and G (Figure 2 and Supplementary Figure S4). Second, the helicase-like region in the Snf2 family is significantly longer than for many other helicases, primarily due to an increased spacing between motifs III and IV of >160 residues compared to 38 and 78 for typical SF2 helicases NS3 and RecG, respectively (44). Third, a number of unique conserved blocks are found in Snf2 family proteins (Figure 2 and Supplementary Table S5). Several of these blocks have been noted previously (20,45–48), with conserved block B having been confused in a number of early manuscripts with motif IV. Conserved blocks B, C and K are of particular interest because they are located within the characteristic extended inter-motif III–IV region (Figure 3G).
Figure 2.
Conserved residues within Snf2 helicase-like region. Sequence logo of global multiple alignment of 1306 Snf2 helicase-like region for alignment positions with residues in >90% of proteins. Helicase motifs are indicated in solid black boxes with roman numerals I–IV, additional conserved blocks are indicated in dashed black boxes with uppercase letters A–N, and conserved hydrophobic residues packing in the core of the structure by grey solid boxes. Motif and box labels as in Thoma et al. (47) with extensions. A comparison to other nomenclatures is in Supplementary Table S5. See Table 4 for actual distances between conserved blocks.
Figure 3.
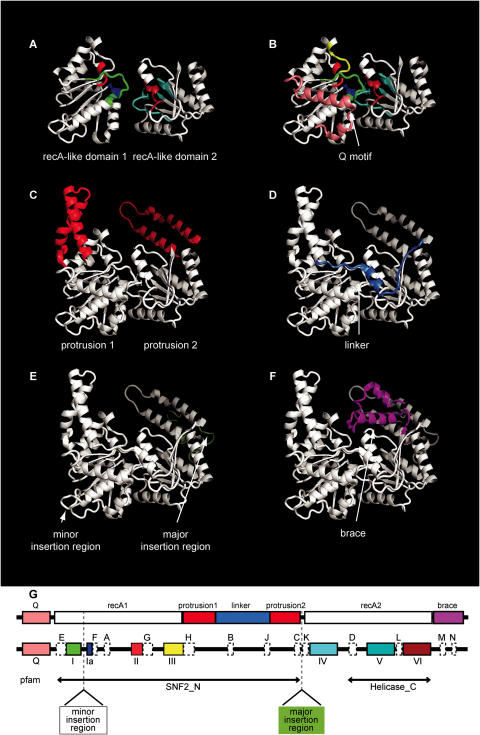
Conserved blocks contribute to distinctive structural features of Snf2 family proteins. Structural components of Snf2 family proteins relevant to the conservation are illustrated on the zebrafish Rad54A structure [pdb 1Z3I (153)]. (A) core recA-like domains 1 and 2 including colouring of helicase motifs (I in green, Ia in blue, II in bright red, III in yellow, IV in cyan, V in teal and VI in dark red). (B) Q motif (pink). (C) antiparallel alpha helical protrusions 1 and 2 (red) projecting from recA-like domains 1 and 2, respectively. (D) Linker spanning from protrusion 1 to protrusion 2 (middle blue). (E) Major insertion region behind protrusion 2 (light green). (F) triangular brace (magenta). (G) Schematic diagram showing location of structural elements and helicase motifs coloured as in A–F, with conserved blocks from Figure 2 shown as white boxes. Spans identified by Pfam profiles SNF2_N and Helicase_C are shown flanking the major insertion site.
The SMARCAL1 subfamily contains classical helicase motifs which are highly similar to the other subfamilies. It also has an extended motif III–IV spacing, but it nevertheless lack conserved blocks within the motif III–IV region (Supplementary Figure S4). The rapA group has similar properties but is more diverse in overall sequence and retains less similarity in the classical motifs. It is unclear whether the SMARCAL1 subfamily and particularly the rapA group will maintain the structural features of the Snf2 family and they are therefore at the limit of the definition of the Snf2 family. We have also noticed further protein groupings with extended spacing between motifs III and IV and detectable similarity to the classical helicase-like motifs of the Snf2 family sequences (Supplementary Figure S4). These include poxvirus NPH-I related proteins involved in transcription termination (49) and the FANCM/MPH1/Hef group of helicases encompassing yeast Mph1p, archaeal Hef and human FANCM proteins involved in DNA repair (50–52). However, those proteins show low similarity to the Snf2 family between motifs III and IV and appear to lack the characteristic conserved blocks C, J and K of the Snf2 family. Interestingly, comparison of the recently determined Pyrococcus furiosus archaeal Hef helicase structure reveals that the MPH1/Hef group has a related structural organization to Zebrafish Rad54, but contains only a single compact alpha-helical domain encoded between motifs III and IV (Supplementary Figure S6). It has been noted that this extra alpha-helical domain has some similarities with the thumb domain of Taq DNA polymerase which grips the DNA minor groove (53). It is therefore likely that the SMARCAL1 subfamily, rapA group, NPH-I and MPH1/Hef proteins reflect a continuum of diversity while sharing core features with the other Snf2 subfamilies.
Evolution of Snf2 family diversity
None of the 293 scanned archaeal or bacterial genomes contains a protein classified in any of the eukaryotic subfamilies (Supplementary Tables S1B and S1C). All identified archaeal and bacterial proteins belong to the SSO1653 subfamily and rapA group. Conversely, the SSO1653 subfamily and rapA group are likely to be specific to microbial organisms because the only two members of these families identified in eukaryotes (Supplementary Table S1A) appear to be false positives (data not shown). Over two-thirds of complete microbial genomes contain members of the SSO1653 subfamily and/or rapA group. This broad yet incomplete distribution suggests they perform non-essential functions that are sufficiently advantageous to maintain their prevalence.
Although rapA group proteins are distinguished by the lack of several features characteristic of eukaryotic Snf2 family members (see above), the SSO1653 subfamily carries all the Snf2 family sequence and structural hallmarks (Supplementary Figure S4). SSO1653 subfamily members are present in both bacteria and archaea, but they are not ubiquitous in archaeal genomes despite the presence of transcription, replication and repair mechanisms with significant similarity to those of eukaryotes (54,55). There is also no obvious linkage between the presence of histone-like proteins and SSO1653 subfamily members in archaeal genomes (Supplementary Table S1B). Furthermore, the SSO1653 subfamily falls in a grouping (Figure 1C) with the eukaryotic ERCC6 and Mot1 subfamilies whose biochemical role appears not to involve chromatin directly. In contrast to the limited archaeal and bacterial distribution of Snf2 family proteins, all eukaryote genomes contain multiple Snf2 family proteins. The early branching Giardia lamblia and the minimal Encephalozooan cuniculi genomes both encode six different Snf2 family genes falling into subfamilies represented across eukarya (Supplementary Table S1A), several of which have clear linkage to chromatin transactions. It is therefore possible that the microbial SSO1653 subfamily represents an ancestral Snf2-like form from which the eukaryotic subfamilies radiated. Such expansion of the Snf2 family early in eukaryote evolution (20) could have been coincident with the development of high-density nucleosomal packaging (56).
Distribution of Snf2 family members in complete genomes
The linkage between the primary sequence-based definitions of the subfamilies and distinct biological function is strongly supported by the presence of one or more subfamily members in each eukaryotic genome across large evolutionary ranges (Table 2 and Supplementary Table S1A). For example, a common set of subfamilies are found in almost all fungi, plant and animal genomes comprising Snf2, Iswi, Chd1, Swr1, Etl1, Mot1, Ino80, Rad5/16, ERCC6, Rad54 and SHPRH. Increased genomic complexity is also paralleled by increasing numbers of subfamilies and members: E.cuniculi with a genome encoding some 2000 gene products has 6 Snf2 family members from 6 subfamilies, whereas the S.cerevisiae genome encoding some 6000 genes has 17 Snf2 family members from 13 subfamilies, and the human genome encoding some 25 000 genes has 32 Snf2 family genes from 20 subfamilies (Table 2, part B).
The functional linkage across large evolutionary ranges suggests that each subfamily may have distinctive properties of their ATPase motors tuned to their function. This is supported by recent biochemical results demonstrating that helicase-like regions can be swapped within but not between subfamilies (57). However, a counterpoint is that functional redundancy can occur between subfamilies. For example, synthetic deletion of all three of the S.cerevisiae ISW1, ISW2 and CHD1 genes together is required to generate a strong phenotype (58,59). Redundancy also provides an explanation why some genomes lack certain members: the small genome of Schizosaccharomyces pombe lacks an Iswi subfamily member but maintains two Chd1 subfamily members. In addition to the 11 subfamilies represented broadly across eukaryotes are a number of others restricted to specific taxonomic ranges. For example, CHD7 members are found almost exclusively in animals, and ATRX members are found only in animals and plants.
Subfamily-specific properties
A number of specific features contribute to the distinction between subfamilies. First, the spacing between motifs III and IV is extended significantly beyond the minimal ∼160 residues for a number of subfamilies (Table 4). For the Rad5/16, Ris1 and SHPRH subfamilies, the additional sequences all include RING fingers, whereas for the Swr1 and EP400 subfamilies they comprise highly proline and serine/threonine-rich spans. Ino80 and ATRX subfamilies also contain large, novel and distinct spans. Remarkably, all these large extra insertions occur at the same location in the primary sequence, between conserved blocks C and K which we term the ‘major insertion site’ (Figure 3G and Supplementary Figure S7A). Even for the subfamilies without large insertions there is variation in the length of sequence in the major insertion site (Table 4). For example, the Zebrafish Rad54 structure contains some 25 more residues forming two additional small alpha helices compared to the Sulfolobus solfataricus SSO1653 structure. When Snf2 family members from different subfamilies are aligned, the variability of the major insertion region strongly disturbs the alignment such that a contiguous pattern becomes difficult to define. This has led to some of the Snf2 family proteins being described as having ‘split’ helicase-like ATPase regions. The discontinuity is also the cause of protein motif databases such as SMART and Pfam defining Snf2 family members as matching a bipartite combination of SNF2_N and Helicase_C profiles (Figure 3G). The C-terminal end of the SNF2_N profile corresponds to conserved block C.
Table 4.
Spacings between helicase motifs and major conserved blocks by subfamily
Means and standard deviations (in parentheses) by subfamily for the number of amino acids between helicase motifs and the conserved blocks B, C and K, calculated for 1306 Snf2 family members from Uniref assigned to subfamilies (Table 1). Spacings are calculated from the edges of the core conserved residues of motifs as marked in Supplementary Figure S4.
Second, subfamilies have characteristic small insertions at other sites (Table 4). Two such sites, also in the motif III–IV region, are located between conserved blocks H and B and between J and C (Figure 2). These are likely to influence the length of the long alpha helical protrusions 1 and 2, respectively (see below, Figure 3C), and there is a difference of some 40 residues between the shortest and longest subfamily lengths for each (Table 4). A ‘minor insertion site’ located between motifs I and Ia on the back of recA-like domain 1 is also occupied by recognizable domains in a few subfamilies from the Rad5/16-like grouping such as SHPRH (Supplementary Figure S3B). A number of other small insertions map to loops between various secondary structural elements (data not shown).
Third, although adhering to a general Snf2 family-specific pattern, individual subfamilies show characteristic patterns in the helicase motifs and in other conserved blocks (Supplementary Figure S4). For example, the well-known helicase motif II with typical DEAH pattern favours DEGH in the Snf2, Mot1 and Rad54 subfamilies, DEAQ in the Swr1, EP400, Ino80 and SSO1653 subfamilies or DESH in the SMARCAL1 subfamily. Likewise, for the typical conserved block E—motif I combined pattern ILADEMGLGKT all ATRX subfamily members have histidine instead of aspartate (i.e. ILAHEMGLGKT) and most Mot1 subfamily members have cysteine replacing alanine (i.e. ILCDEMGLGKT). It is also possible to identify other residues correlating with groups of subfamilies. For example, members of the Snf2, Iswi, Chd1, Mi-2, CHD7, ALC1, Rad54, ATRX and Arip4 subfamilies have an arginine immediately following the motif II DEAH. In the zebrafish Rad54 structure this residue R294 interacts with the sulphate which is suggested to mimic the ATP gamma phosphate.
Conserved blocks encode the unique structural features of the Snf2 family
Two structural determinations of the helicase-like regions of Snf2 family members have been presented recently: zebrafish Rad54 (pdb code 1Z3I) (47) and S.solfataricus SSO1653 (pdb codes 1Z6A, 1Z63, 1Z5Z) (46). As expected for members of the Snf2 family, the fold of each core recA-like domain in the Rad54 and SSO1653 structures is substantially similar and related to those of other known SF1 and SF2 helicases. In the zebrafish Rad54 structure the two recA-like domains are oriented equivalently to those of other known helicase structures (Figure 3A and B), whereas in the S.solfataricus SSO1653 structures recA-like domain 2 is flipped by 180° to an arrangement never previously observed for a helicase (Supplementary Figure S7B). This unusual orientation in SSO1653 is observed for both the DNA free and DNA-bound forms (46).
The most striking feature of the Snf2 family structures is the presence of several additional structural elements grafted onto the core helicase structure. These comprise antiparallel alpha helical protrusions from both recA-like domains 1 and 2 (Figure 3C), a structured linker between the recA-like domains (Figure 3D), the major insertion region at the back side of the domain 2 alpha helical protrusion (Figure 3E) and a triangular brace packed against the domain 2 alpha helical protrusion (Figure 3F). The two alpha-helical protrusions and linker are all encoded within the enlarged span between motifs III and IV. The triangular brace is encoded immediately downstream of motif VI.
Remarkably, the primary sequence features of the Snf2 family correspond directly to the additional structural elements (Figure 3G). First, the bases of the protrusions from recA-like domains 1 and 2 are both fixed by conserved blocks. For protrusion 1, this involves conserved block H composed of a repeating pattern of aromatic residues, with additional involvement of aromatics from conserved block A. For protrusion 2 this involves the arrangement of conserved blocks C, J and K. Second, the protrusions themselves are relatively conserved in sequence and length within subfamilies but not across the whole Snf2 family. Although there is no obvious correlation between the lengths of the protrusions 1 and 2, the distribution of protrusion lengths adheres to multiples of the alpha helical repeat (Supplementary Figure S8), suggesting that protrusions retain structure while varying in extension. Third, the Q motif structure found in many SF2 proteins utilizes a different arrangement of residues to DEAD box helicases such as eIF4A, where an aromatic residue orients the adenine base ring for contacts with a downstream glutamine (4) (Figures 3B). In the Snf2 family, the aromatic residue is contributed by conserved block F downstream of the glutamine. The Q motif affects ATP hydrolysis in DEAD box helicases and mutation of the core glutamine in yeast Snf2 subfamily member Sth1p causes slow growth (4). Fourth, the linker connecting protrusions 1 and 2 contains highly conserved dual arginines in conserved block B. Their central location between the ATP-associating and DNA-associating structural elements suggests that they may play an important role in the mechanism of Snf2 family enzymes. Consistent with this, mutation of the second arginine of the pair in Snf2p leads to effectively complete loss of function of the protein in vivo (48). Finally, the brace is composed of a principal alpha helix anchored by conserved block M into the junction at the base of protrusion 2 composed of conserved blocks C, J and K.
The major insertion region is immediately behind protrusion 2, almost diametrically opposite the ATP-binding site in the zebrafish Rad54 structure (Figure 3E). The nearest residues of the major insertion region in Rad54 are some 15 Å from DNA phosphates for docked DNA (Supplementary Figure S7A). However, an appropriately oriented alpha helix of some 20 residues would be sufficient to reach into the major groove, so large insertions at the major insertion site could potentially interact with DNA or other DNA-binding proteins bound in the groove. In the flipped conformation of domain 2 observed in the SSO1653 structure, the major insertion region is juxtaposed immediately adjacent to the DNA such that two non-conserved arginines from the major insertion region make direct DNA phosphate contacts.
As the distinctive structural features are defined by unique and highly conserved blocks, they are likely to confer properties to the ATPase motor that adapts the action of the core recA-like domains for a unique mechanism. We anticipate that while some features of the Snf2 family mechanism will be common to SF2 translocases, other aspects will be distinctive. Knowledge of the conserved residues and their structural location provides important information for understanding these distinctions.
Other levels of Snf2 family identity
We have demonstrated that the common helicase-like region is sufficient to enable classification of Snf2 family members. However, almost all Snf2 family polypeptides contain significant additional sequences likely to harbour accessory domains. For some subfamilies there is good correlation with the presence of particular accessory domain combinations (Supplementary Table S9). For example, almost all Snf2 subfamily members contain a bromodomain, ISWI members contain a SANT domain, and Chd1, Mi-2 and CHD7 members contain a chromodomain. However, many domain profiles in resources used for domain analysis have unidentified function or are unreliable in the context of Snf2 proteins. For example, Pfam lacks a SANT-specific profile and detects <10% of SANT domains with a more generic general ‘Myb_DNA-binding’ profile. We are currently undertaking further analysis to improve the relevant profiles and analyse the linkage of Snf2 family accessory domains in detail.
Finally, many Snf2 family proteins are part of larger multi-protein complexes. Accessory motifs within these complexes are also likely to adapt the function of Snf2 motors for different purposes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We gratefully acknowledge Chris Stockdale, Charlie Bond and Helder Ferreira for comments on this manuscript and Diego Miranda-Saavedra and Jim Procter for helpful discussions regarding the analyses. We are also indebted to Andrew Waterhouse for providing essential enhancements to Jalview, and to Jonathan Monk for excellent systems support. We also thank Prof. U. Baumann for coordinates of eIF4A with ADP, Dr Kathleen Sandman and Prof. John Reeve for providing a table of histone-like proteins in archaeal genomes, and Dr Malcolm White for communicating unpublished observations about the SSO1653 protein. AF and TOH are funded by a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science to TOH. Funding to pay the Open Access publication charges for this article was provided by a Wellcome Trust VIP award to the School of Life Sciences in Dundee.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure–function relationship. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 2.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 5.Subramanya H.S., Bird L.E., Brannigan J.A., Wigley D.B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 6.Singleton M.R., Wigley D.B. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye J., Osborne A.R., Groll M., Rapoport T.A. RecA-like motor ATPases—lessons from structures. Biochim. Biophys. Acta. 2004;1659:1–18. doi: 10.1016/j.bbabio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Becker P.B., Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 9.Saha A., Wittmeyer J., Cairns B.R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehouse I., Stockdale C., Flaus A., Szczelkun M.D., Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaskelioff M., Van Komen S., Krebs J.E., Sung P., Peterson C.L. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 12.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T.L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl Acad. Sci. USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Komen S., Petukhova G., Sigurdsson S., Stratton S., Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 14.Havas K., Flaus A., Phelan M., Kingston R., Wade P.A., Lilley D.M., Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 15.Liu R., Liu H., Chen X., Kirby M., Brown P.O., Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 16.Rouleau N., Domans'kyi A., Reeben M., Moilanen A.M., Havas K., Kang Z., Owen-Hughes T., Palvimo J.J., Janne O.A. Novel ATPase of SNF2-like protein family interacts with androgen receptor and modulates androgen-dependent transcription. Mol. Biol. Cell. 2002;13:2106–2119. doi: 10.1091/mbc.01-10-0484.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu S., Owen-Hughes T., Friedberg E.C., Waters R., Reed S.H. The yeast Rad7/Rad16/Abf1 complex generates superhelical torsion in DNA that is required for nucleotide excision repair. DNA Repair (Amst) 2004;3:277–287. doi: 10.1016/j.dnarep.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Lia G., Praly E., Ferreira H., Stockdale C., Tse-Dinh Y.C., Dunlap D., Croquette V., Bensimon D., Owen-Hughes T. Direct observation of DNA distortion by the RSC chromatin remodeling complex. Mol. Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auble D.T., Hansen K.E., Mueller C.G., Lane W.S., Thorner J., Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 20.Eisen J.A., Sweder K.S., Hanawalt P.C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindback T., Kolsto A.B. A Bacillus cereus member of the SNF2 family. Microbiology. 1997;143:171–174. doi: 10.1099/00221287-143-1-171. [DOI] [PubMed] [Google Scholar]
- 22.Coleman M.A., Eisen J.A., Mohrenweiser H.W. Cloning and characterization of HARP/SMARCAL1: a prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics. 2000;65:274–282. doi: 10.1006/geno.2000.6174. [DOI] [PubMed] [Google Scholar]
- 23.Linder B., Cabot R.A., Schwickert T., Rupp R.A. The SNF2 domain protein family in higher vertebrates displays dynamic expression patterns in Xenopus laevis embryos. Gene. 2004;326:59–66. doi: 10.1016/j.gene.2003.09.053. [DOI] [PubMed] [Google Scholar]
- 24.Bakshi R., Prakash T., Dash D., Brahmachari V. In silico characterization of the INO80 subfamily of SWI2/SNF2 chromatin remodeling proteins. Biochem. Biophys. Res. Commun. 2004;320:197–204. doi: 10.1016/j.bbrc.2004.05.147. [DOI] [PubMed] [Google Scholar]
- 25.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 32.Zmasek C.M., Eddy S.R. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 33.Bingham J., Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 2000;16:660–661. doi: 10.1093/bioinformatics/16.7.660. [DOI] [PubMed] [Google Scholar]
- 34.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 35.Madera M. 2005. Hidden Markov models for detection of remote homology. Cambridge University PhD thesis. [Google Scholar]
- 36.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuster-Bockler B., Bateman A. Visualizing profile–profile alignment: pairwise HMM logos. Bioinformatics. 2005;21:2912–2913. doi: 10.1093/bioinformatics/bti434. [DOI] [PubMed] [Google Scholar]
- 38.DeLano W.L. The PyMOL User's Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 39.Miki H., Okada Y., Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Hodge T., Cope M.J. A myosin family tree. J. Cell Sci. 2000;113:3353–3354. doi: 10.1242/jcs.113.19.3353. [DOI] [PubMed] [Google Scholar]
- 41.Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 42.Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 44.Flaus A., Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 45.Bork P., Koonin E.V. An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res. 1993;21:751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durr H., Korner C., Muller M., Hickmann V., Hopfner K.P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 47.Thoma N.H., Czyzewski B.K., Alexeev A.A., Mazin A.V., Kowalczykowski S.C., Pavletich N.P. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature Struct. Mol. Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 48.Richmond E., Peterson C.L. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henikoff S. Transcriptional activator components and poxvirus DNA-dependent ATPases comprise a single family. Trends Biochem. Sci. 1993;18:291–292. doi: 10.1016/0968-0004(93)90037-n. [DOI] [PubMed] [Google Scholar]
- 50.Meetei A.R., Medhurst A.L., Ling C., Xue Y., Singh T.R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C.G., et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nature Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J.B., Johnson M., Langevin F., Pace P., Patel K.J. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nature Struct. Mol. Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 52.Prakash R., Krejci L., Van Komen S., Anke Schurer K., Kramer W., Sung P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- 53.Nishino T., Komori K., Tsuchiya D., Ishino Y., Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Bartlett M.S. Determinants of transcription initiation by archaeal RNA polymerase. Curr. Opin. Microbiol. 2005;8:677–684. doi: 10.1016/j.mib.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 55.Kelman Z., White M.F. Archaeal DNA replication and repair. Curr. Opin. Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Minsky A., Ghirlando R., Reich Z. Nucleosomes: a solution to a crowded intracellular environment? J. Theoret. Biol. 1997;188:379–385. doi: 10.1006/jtbi.1997.0525. [DOI] [PubMed] [Google Scholar]
- 57.Fan H.Y., Trotter K.W., Archer T.K., Kingston R.E. Swapping function of two chromatin remodeling complexes. Mol. Cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 58.Alen C., Kent N.A., Jones H.S., O'Sullivan J., Aranda A., Proudfoot N.J. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 59.Tsukiyama T., Palmer J., Landel C.C., Shiloach J., Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laurent B.C., Yang X., Carlson M. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol. Cell. Biol. 1992;12:1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cairns B.R., Lorch Y., Li Y., Zhang M., Lacomis L., Erdjument-Bromage H., Tempst P., Du J., Laurent B., Kornberg R.D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 62.Tamkun J.W., Deuring R., Scott M.P., Kissinger M., Pattatucci A.M., Kaufman T.C., Kennison J.A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 63.Muchardt C., Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khavari P.A., Peterson C.L., Tamkun J.W., Mendel D.B., Crabtree G.R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 65.Whitehouse I., Flaus A., Cairns B.R., White M.F., Workman J.L., Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 66.Kassabov S.R., Zhang B., Persinger J., Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 67.Narlikar G.J., Phelan M.L., Kingston R.E. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 68.Lorch Y., Zhang M., Kornberg R.D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 69.Bruno M., Flaus A., Stockdale C., Rencurel C., Ferreira H., Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xue Y., Canman J.C., Lee C.S., Nie Z., Yang D., Moreno G.T., Young M.K., Salmon E.D., Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl Acad. Sci. USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sif S., Saurin A.J., Imbalzano A.N., Kingston R.E. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harikrishnan K.N., Chow M.Z., Baker E.K., Pal S., Bassal S., Brasacchio D., Wang L., Craig J.M., Jones P.L., Sif S., et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nature Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 73.Xu W., Cho H., Kadam S., Banayo E.M., Anderson S., Yates J.R., III, Emerson B.M., Evans R.M. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunaief J.L., Strober B.E., Guha S., Khavari P.A., Alin K., Luban J., Begemann M., Crabtree G.R., Goff S.P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 75.Zhang H.S., Gavin M., Dahiya A., Postigo A.A., Ma D., Luo R.X., Harbour J.W., Dean D.C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 76.Moshkin Y.M., Armstrong J.A., Maeda R.K., Tamkun J.W., Verrijzer P., Kennison J.A., Karch F. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 2002;16:2621–2626. doi: 10.1101/gad.231202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson C.J., Chao D.M., Imbalzano A.N., Schnitzler G.R., Kingston R.E., Young R.A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 78.Metivier R., Penot G., Hubner M.R., Reid G., Brand H., Kos M., Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 79.Huang J., Hsu J.M., Laurent B.C. The RSC nucleosome-remodeling complex is required for Cohesin's association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 80.Peterson C.L., Workman J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 81.Hassan A.H., Neely K.E., Workman J.L. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 82.Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 83.Elfring L.K., Deuring R., McCallum C.M., Peterson C.L., Tamkun J.W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell. Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsukiyama T., Daniel C., Tamkun J., Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 85.Ito T., Bulger M., Pazin M.J., Kobayashi R., Kadonaga J.T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 86.Varga-Weisz P.D., Wilm M., Bonte E., Dumas K., Mann M., Becker P.B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 87.Hamiche A., Kang J.G., Dennis C., Xiao H., Wu C. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl Acad. Sci. USA. 2001;98:14316–14321. doi: 10.1073/pnas.251421398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clapier C.R., Nightingale K.P., Becker P.B. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–655. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fazzio T.G., Gelbart M.E., Tsukiyama T. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell. Biol. 2005;25:9165–9174. doi: 10.1128/MCB.25.21.9165-9174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dirscherl S.S., Krebs J.E. Functional diversity of ISWI complexes. Biochem. Cell Biol. 2004;82:482–489. doi: 10.1139/o04-044. [DOI] [PubMed] [Google Scholar]
- 91.Eberharter A., Becker P.B. ATP-dependent nucleosome remodelling: factors and functions. J. Cell Sci. 2004;117:3707–3711. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- 92.Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nature Rev. Mol. Cell Biol. 2002;3:422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- 93.Poot R.A., Bozhenok L., van den Berg D.L., Hawkes N., Varga-Weisz P.D. Chromatin remodeling by WSTF-ISWI at the replication site: opening a window of opportunity for epigenetic inheritance? Cell Cycle. 2005;4:543–546. doi: 10.4161/cc.4.4.1624. [DOI] [PubMed] [Google Scholar]
- 94.Jarvis C.D., Geiman T., Vila-Storm M.P., Osipovich O., Akella U., Candeias S., Nathan I., Durum S.K., Muegge K. A novel putative helicase produced in early murine lymphocytes. Gene. 1996;169:203–207. doi: 10.1016/0378-1119(95)00843-8. [DOI] [PubMed] [Google Scholar]
- 95.Sun L.Q., Arceci R.J. Altered epigenetic patterning leading to replicative senescence and reduced longevity. A role of a novel SNF2 factor, PASG. Cell Cycle. 2005;4:3–5. doi: 10.4161/cc.4.1.1341. [DOI] [PubMed] [Google Scholar]
- 96.Vongs A., Kakutani T., Martienssen R.A., Richards E.J. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 97.Dennis K., Fan T., Geiman T., Yan Q., Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Muegge K. Lsh, a guardian of heterochromatin at repeat elements. Biochem. Cell Biol. 2005;83:548–554. doi: 10.1139/o05-119. [DOI] [PubMed] [Google Scholar]
- 99.Brzeski J., Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- 100.Guan X.Y., Fang Y., Sham J.S., Kwong D.L., Zhang Y., Liang Q., Li H., Zhou H., Trent J.M. Recurrent chromosome alterations in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2000;29:110–116. [PubMed] [Google Scholar]
- 101.Mao M., Fu G., Wu J.S., Zhang Q.H., Zhou J., Kan L.X., Huang Q.H., He K.L., Gu B.W., Han Z.G., et al. Identification of genes expressed in human CD34(+) hematopoietic stem/progenitor cells by expressed sequence tags and efficient full-length cDNA cloning. Proc. Natl Acad. Sci. USA. 1998;95:8175–8180. doi: 10.1073/pnas.95.14.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yan L., Echenique V., Busso C., SanMiguel P., Ramakrishna W., Bennetzen J.L., Harrington S., Dubcovsky J. Cereal genes similar to Snf2 define a new subfamily that includes human and mouse genes. Mol. Genet. Genomics. 2002;268:488–499. doi: 10.1007/s00438-002-0765-3. [DOI] [PubMed] [Google Scholar]
- 103.Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., Bycroft M., Ladurner A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Delmas V., Stokes D.G., Perry R.P. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc. Natl Acad. Sci. USA. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brehm A., Tufteland K.R., Aasland R., Becker P.B. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 106.Tajul-Arifin K., Teasdale R., Ravasi T., Hume D.A., Mattick J.S. Identification and analysis of chromodomain-containing proteins encoded in the mouse transcriptome. Genome Res. 2003;13:1416–1429. doi: 10.1101/gr.1015703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Woodage T., Basrai M.A., Baxevanis A.D., Hieter P., Collins F.S. Characterization of the CHD family of proteins. Proc. Natl Acad. Sci. USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tran H.G., Steger D.J., Iyer V.R., Johnson A.D. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lusser A., Urwin D.L., Kadonaga J.T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nature Struct. Mol. Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 110.Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pray-Grant M.G., Daniel J.A., Schieltz D., Yates J.R., III, Grant P.A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 112.Krogan N.J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D.P., Beattie B.K., Emili A., Boone C., et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kelley D.E., Stokes D.G., Perry R.P. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma. 1999;108:10–25. doi: 10.1007/s004120050347. [DOI] [PubMed] [Google Scholar]
- 114.Walfridsson J., Bjerling P., Thalen M., Yoo E.J., Park S.D., Ekwall K. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jae Yoo E., Kyu Jang Y., Ae Lee M., Bjerling P., Bum Kim J., Ekwall K., Hyun Seong R., Dai Park S. Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem. Biophys. Res. Commun. 2002;295:970–974. doi: 10.1016/s0006-291x(02)00797-0. [DOI] [PubMed] [Google Scholar]
- 116.Seelig H.P., Moosbrugger I., Ehrfeld H., Fink T., Renz M., Genth E. The major dermatomyositis-specific Mi-2 autoantigen is a presumed helicase involved in transcriptional activation. Arthritis Rheum. 1995;38:1389–1399. doi: 10.1002/art.1780381006. [DOI] [PubMed] [Google Scholar]
- 117.Bowen N.J., Fujita N., Kajita M., Wade P.A. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 118.Bouazoune K., Mitterweger A., Langst G., Imhof A., Akhtar A., Becker P.B., Brehm A. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schultz D.C., Friedman J.R., Rauscher F.J., III Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Korenjak M., Taylor-Harding B., Binne U.K., Satterlee J.S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 121.White P.S., Thompson P.M., Gotoh T., Okawa E.R., Igarashi J., Kok M., Winter C., Gregory S.G., Hogarty M.D., Maris J.M., et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 122.Thompson P.M., Gotoh T., Kok M., White P.S., Brodeur G.M. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 123.Ogas J., Kaufmann S., Henderson J., Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl Acad. Sci. USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C., et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nature Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 125.Jongmans M., Admiraal R., van der Donk K., Vissers L., Baas A., Kapusta L., van Hagen A., Donnai D., de Ravel T., Veltman J., et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J. Med. Genet. 2006;43:306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schuster E.F., Stoger R. CHD5 defines a new subfamily of chromodomain-SWI2/SNF2-like helicases. Mamm. Genome. 2002;13:117–119. doi: 10.1007/s00335-001-3042-6. [DOI] [PubMed] [Google Scholar]
- 127.Shur I., Benayahu D. Characterization and functional analysis of CReMM, a novel chromodomain helicase DNA-binding protein. J. Mol. Biol. 2005;352:646–655. doi: 10.1016/j.jmb.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 128.Daubresse G., Deuring R., Moore L., Papoulas O., Zakrajsek I., Waldrip W.R., Scott M.P., Kennison J.A., Tamkun J.W. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999;126:1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- 129.Srinivasan S., Armstrong J.A., Deuring R., Dahlsveen I.K., McNeill H., Tamkun J.W. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 130.Wu W.H., Alami S., Luk E., Wu C.H., Sen S., Mizuguchi G., Wei D., Wu C. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nature Struct. Mol. Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 131.Noh Y.S., Amasino R.M. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell. 2003;15:1671–1682. doi: 10.1105/tpc.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He Y., Amasino R.M. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 133.Ruhf M.L., Braun A., Papoulas O., Tamkun J.W., Randsholt N., Meister M. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development. 2001;128:1429–1441. doi: 10.1242/dev.128.8.1429. [DOI] [PubMed] [Google Scholar]
- 134.Doyon Y., Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 135.Kusch T., Florens L., Macdonald W.H., Swanson S.K., Glaser R.L., Yates J.R., III, Abmayr S.M., Washburn M.P., Workman J.L. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 136.Johnston H., Kneer J., Chackalaparampil I., Yaciuk P., Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J. Biol. Chem. 1999;274:16370–16376. doi: 10.1074/jbc.274.23.16370. [DOI] [PubMed] [Google Scholar]
- 137.Cai Y., Jin J., Florens L., Swanson S.K., Kusch T., Li B., Workman J.L., Washburn M.P., Conaway R.C., Conaway J.W. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 138.Monroy M.A., Schott N.M., Cox L., Chen J.D., Ruh M., Chrivia J.C. SNF2-related CBP activator protein (SRCAP) functions as a coactivator of steroid receptor-mediated transcription through synergistic interactions with CARM-1 and GRIP-1. Mol. Endocrinol. 2003;17:2519–2528. doi: 10.1210/me.2003-0208. [DOI] [PubMed] [Google Scholar]
- 139.Eissenberg J.C., Wong M., Chrivia J.C. Human SRCAP and Drosophila melanogaster DOM are homologs that function in the notch signaling pathway. Mol. Cell. Biol. 2005;25:6559–6569. doi: 10.1128/MCB.25.15.6559-6569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Samuelson A.V., Narita M., Chan H.M., Jin J., de Stanchina E., McCurrach M.E., Fuchs M., Livingston D.M., Lowe S.W. p400 is required for E1A to promote apoptosis. J. Biol. Chem. 2005;280:21915–21923. doi: 10.1074/jbc.M414564200. [DOI] [PubMed] [Google Scholar]
- 141.Chan H.M., Narita M., Lowe S.W., Livingston D.M. The p400 E1A-associated protein is a novel component of the p53 → p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fuchs M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 143.Underhill C., Qutob M.S., Yee S.P., Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 144.Cai Y., Jin J., Tomomori-Sato C., Sato S., Sorokina I., Parmely T.J., Conaway R.C., Conaway J.W. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 145.Doyon Y., Selleck W., Lane W.S., Tan S., Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bachhawat N., Ouyang Q., Henry S.A. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- 147.Ebbert R., Birkmann A., Schuller H.J. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 1999;32:741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- 148.Shen X., Mizuguchi G., Hamiche A., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 149.Jin J., Cai Y., Yao T., Gottschalk A.J., Florens L., Swanson S.K., Gutierrez J.L., Coleman M.K., Workman J.L., Mushegian A., et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 150.Morrison A.J., Highland J., Krogan N.J., Arbel-Eden A., Greenblatt J.F., Haber J.E., Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 151.van Attikum H., Fritsch O., Hohn B., Gasser S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 152.Tsukuda T., Fleming A.B., Nickoloff J.A., Osley M.A. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Downs J.A., Cote J. Dynamics of chromatin during the repair of DNA double-strand breaks. Cell Cycle. 2005;4:1373–1376. doi: 10.4161/cc.4.10.2108. [DOI] [PubMed] [Google Scholar]
- 154.Soininen R., Schoor M., Henseling U., Tepe C., Kisters-Woike B., Rossant J., Gossler A. The mouse Enhancer trap locus 1 (Etl-1): a novel mammalian gene related to Drosophila and yeast transcriptional regulator genes. Mech. Dev. 1992;39:111–123. doi: 10.1016/0925-4773(92)90030-n. [DOI] [PubMed] [Google Scholar]
- 155.Adra C.N., Donato J.L., Badovinac R., Syed F., Kheraj R., Cai H., Moran C., Kolker M.T., Turner H., Weremowicz S., et al. SMARCAD1, a novel human helicase family-defining member associated with genetic instability: cloning, expression, and mapping to 4q22–q23, a band rich in breakpoints and deletion mutants involved in several human diseases. Genomics. 2000;69:162–173. doi: 10.1006/geno.2000.6281. [DOI] [PubMed] [Google Scholar]
- 156.Clark M.W., Zhong W.W., Keng T., Storms R.K., Barton A., Kaback D.B., Bussey H. Identification of a Saccharomyces cerevisiae homolog of the SNF2 transcriptional regulator in the DNA sequence of an 8.6 kb region in the LTE1-CYS1 interval on the left arm of chromosome I. Yeast. 1992;8:133–145. doi: 10.1002/yea.320080208. [DOI] [PubMed] [Google Scholar]
- 157.Schoor M., Schuster-Gossler K., Roopenian D., Gossler A. Skeletal dysplasias, growth retardation, reduced postnatal survival, and impaired fertility in mice lacking the SNF2/SWI2 family member ETL1. Mech. Dev. 1999;85:73–83. doi: 10.1016/s0925-4773(99)00090-8. [DOI] [PubMed] [Google Scholar]
- 158.Ouspenski I.I., Elledge S.J., Brinkley B.R. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 1999;27:3001–3008. doi: 10.1093/nar/27.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barton A.B., Kaback D.B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the genes in the FUN38-MAK16-SPO7 region. J. Bacteriol. 1994;176:1872–1880. doi: 10.1128/jb.176.7.1872-1880.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tan T.L., Kanaar R., Wyman C. Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amst) 2003;2:787–794. doi: 10.1016/s1568-7864(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 161.Krogh B.O., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 162.Klein H.L. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tanaka K., Kagawa W., Kinebuchi T., Kurumizaka H., Miyagawa K. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 2002;30:1346–1353. doi: 10.1093/nar/30.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tan T.L., Essers J., Citterio E., Swagemakers S.M., de Wit J., Benson F.E., Hoeijmakers J.H., Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 165.Petukhova G., Van Komen S., Vergano S., Klein H., Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 166.Alexeev A., Mazin A., Kowalczykowski S.C. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nature Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 167.Alexiadis V., Kadonaga J.T. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gibbons R.J., Picketts D.J., Villard L., Higgs D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 169.McDowell T.L., Gibbons R.J., Sutherland H., O'Rourke D.M., Bickmore W.A., Pombo A., Turley H., Gatter K., Picketts D.J., Buckle V.J., et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl Acad. Sci. USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gibbons R.J., McDowell T.L., Raman S., O'Rourke D.M., Garrick D., Ayyub H., Higgs D.R. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nature Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 171.Janne O.A., Moilanen A.M., Poukka H., Rouleau N., Karvonen U., Kotaja N., Hakli M., Palvimo J.J. Androgen-receptor-interacting nuclear proteins. Biochem. Soc. Trans. 2000;28:401–405. [PubMed] [Google Scholar]
- 172.Domanskyi A., Virtanen K.T., Palvimo J.J., Janne O.A. Biochemical characterization of androgen receptor-interacting protein 4. Biochem. J. 2006;393:789–795. doi: 10.1042/BJ20050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kanno T., Mette M.F., Kreil D.P., Aufsatz W., Matzke M., Matzke A.J. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 174.Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nature Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 175.Kanno T., Aufsatz W., Jaligot E., Mette M.F., Matzke M., Matzke A.J. A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO Rep. 2005;6:649–655. doi: 10.1038/sj.embor.7400446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.DiPaolo C., Kieft R., Cross M., Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol. Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 177.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 178.Chen S., Davies A.A., Sagan D., Ulrich H.D. The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res. 2005;33:5878–5886. doi: 10.1093/nar/gki902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Guzder S.N., Sung P., Prakash L., Prakash S. The DNA-dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J. Biol. Chem. 1998;273:6292–6296. doi: 10.1074/jbc.273.11.6292. [DOI] [PubMed] [Google Scholar]
- 180.Ramsey K.L., Smith J.J., Dasgupta A., Maqani N., Grant P., Auble D.T. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol. Cell. Biol. 2004;24:6362–6378. doi: 10.1128/MCB.24.14.6362-6378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hewetson A., Hendrix E.C., Mansharamani M., Lee V.H., Chilton B.S. Identification of the RUSH consensus-binding site by cyclic amplification and selection of targets: demonstration that RUSH mediates the ability of prolactin to augment progesterone-dependent gene expression. Mol. Endocrinol. 2002;16:2101–2112. doi: 10.1210/me.2002-0064. [DOI] [PubMed] [Google Scholar]
- 182.Moinova H.R., Chen W.D., Shen L., Smiraglia D., Olechnowicz J., Ravi L., Kasturi L., Myeroff L., Plass C., Parsons R., et al. HLTF gene silencing in human colon cancer. Proc. Natl Acad. Sci. USA. 2002;99:4562–4567. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang Z., Buchman A.R. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Girdham C.H., Glover D.M. Chromosome tangling and breakage at anaphase result from mutations in lodestar, a Drosophila gene encoding a putative nucleoside triphosphate-binding protein. Genes Dev. 1991;5:1786–1799. doi: 10.1101/gad.5.10.1786. [DOI] [PubMed] [Google Scholar]
- 185.Jiang Y., Liu M., Spencer C.A., Price D.H. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol. Cell. 2004;14:375–385. doi: 10.1016/s1097-2765(04)00234-5. [DOI] [PubMed] [Google Scholar]
- 186.Hara R., Selby C.P., Liu M., Price D.H., Sancar A. Human transcription release factor 2 dissociates RNA polymerases I and II stalled at a cyclobutane thymine dimer. J. Biol. Chem. 1999;274:24779–24786. doi: 10.1074/jbc.274.35.24779. [DOI] [PubMed] [Google Scholar]
- 187.Sood R., Makalowska I., Galdzicki M., Hu P., Eddings E., Robbins C.M., Moses T., Namkoong J., Chen S., Trent J.M. Cloning and characterization of a novel gene, SHPRH, encoding a conserved putative protein with SNF2/helicase and PHD-finger domains from the 6q24 region. Genomics. 2003;82:153–161. doi: 10.1016/s0888-7543(03)00121-6. [DOI] [PubMed] [Google Scholar]
- 188.Ramakrishnan V., Finch J.T., Graziano V., Lee P.L., Sweet R.M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 189.Clark K.L., Halay E.D., Lai E., Burley S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 190.Burley S.K., Xie X., Clark K.L., Shu F. Histone-like transcription factors in eukaryotes. Curr. Opin. Struct. Biol. 1997;7:94–102. doi: 10.1016/s0959-440x(97)80012-7. [DOI] [PubMed] [Google Scholar]
- 191.Aasland R., Gibson T.J., Stewart A.F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 192.Ragvin A., Valvatne H., Erdal S., Arskog V., Tufteland K.R., Breen K., AM O.Y., Eberharter A., Gibson T.J., Becker P.B., et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J. Mol. Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 193.Davis J.L., Kunisawa R., Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chicca J.J., II, Auble D.T., Pugh B.F. Cloning and biochemical characterization of TAF-172, a human homolog of yeast Mot1. Mol. Cell. Biol. 1998;18:1701–1710. doi: 10.1128/mcb.18.3.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Auble D.T., Wang D., Post K.W., Hahn S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol. Cell. Biol. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dasgupta A., Juedes S.A., Sprouse R.O., Auble D.T. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Topalidou I., Papamichos-Chronakis M., Thireos G., Tzamarias D. Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J. 2004;23:1943–1948. doi: 10.1038/sj.emboj.7600199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J.H. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 199.van Gool A.J., Verhage R., Swagemakers S.M., van de Putte P., Brouwer J., Troelstra C., Bootsma D., Hoeijmakers J.H. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Svejstrup J.Q. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 2003;116:447–451. doi: 10.1242/jcs.00271. [DOI] [PubMed] [Google Scholar]
- 201.Park J.S., Marr M.T., Roberts J.W. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 202.Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R.E., Hoeijmakers J.H., Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Beerens N., Hoeijmakers J.H., Kanaar R., Vermeulen W., Wyman C. The CSB protein actively wraps DNA. J. Biol. Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 204.Makarova K.S., Aravind L., Koonin E.V. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 2002;27:384–386. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- 205.Liu J., Gagnon Y., Gauthier J., Furenlid L., L'Heureux P.J., Auger M., Nureki O., Yokoyama S., Lapointe J. The zinc-binding site of Escherichia coli glutamyl-tRNA synthetase is located in the acceptor-binding domain. Studies by extended x-ray absorption fine structure, molecular modeling, and site-directed mutagenesis. J. Biol. Chem. 1995;270:15162–15169. doi: 10.1074/jbc.270.25.15162. [DOI] [PubMed] [Google Scholar]
- 206.Boerkoel C.F., Takashima H., John J., Yan J., Stankiewicz P., Rosenbarker L., Andre J.L., Bogdanovic R., Burguet A., Cockfield S., et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nature Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 207.Muthuswami R., Truman P.A., Mesner L.D., Hockensmith J.W. A eukaryotic SWI2/SNF2 domain, an exquisite detector of double-stranded to single-stranded DNA transition elements. J. Biol. Chem. 2000;275:7648–7655. doi: 10.1074/jbc.275.11.7648. [DOI] [PubMed] [Google Scholar]
- 208.Plambeck C.A., Kwan A.H., Adams D.J., Westman B.J., van der Weyden L., Medcalf R.L., Morris B.J., Mackay J.P. The structure of the zinc finger domain from human splicing factor ZNF265 fold. J. Biol. Chem. 2003;278:22805–22811. doi: 10.1074/jbc.M301896200. [DOI] [PubMed] [Google Scholar]
- 209.Hsia K.C., Chak K.F., Liang P.H., Cheng Y.S., Ku W.Y., Yuan H.S. DNA binding and degradation by the HNH protein ColE7. Structure (Camb) 2004;12:205–214. doi: 10.1016/j.str.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 210.Muzzin O., Campbell E.A., Xia L., Severinova E., Darst S.A., Severinov K. Disruption of Escherichia coli hepA, an RNA polymerase-associated protein, causes UV sensitivity. J. Biol. Chem. 1998;273:15157–15161. doi: 10.1074/jbc.273.24.15157. [DOI] [PubMed] [Google Scholar]
- 211.Sukhodolets M.V., Cabrera J.E., Zhi H., Jin D.J. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes Dev. 2001;15:3330–3341. doi: 10.1101/gad.936701. [DOI] [PMC free article] [PubMed] [Google Scholar]