Abstract
DNA polymerase lambda (polλ) is a recently identified DNA polymerase whose cellular function remains elusive. Here we show, that polλ participates at the molecular level in a chromosomal context, in the repair of DNA double strand breaks (DSB) via non-homologous end joining (NHEJ) in mammalian cells. The expression of a catalytically inactive form of polλ (polλDN) decreases the frequency of NHEJ events in response to I-Sce-I-induced DSB whereas inactivated forms of its homologues polβ and polμ do not. Only events requiring DNA end processing before ligation are affected; this defect is associated with large deletions arising in the vicinity of the induced DSB. Furthermore, polλDN-expressing cells exhibit increased sensitization and genomic instability in response to ionizing radiation similar to that of NHEJ-defective cells. Our data support a requirement for polλ in repairing a subset of DSB in genomic DNA, thereby contributing to the maintenance of genetic stability mediated by the NHEJ pathway.
INTRODUCTION
DNA polymerase lambda (polλ) is a recently identified DNA polymerase (1) belonging to the X family that includes the well-known polβ and the recently discovered polμ and polσ (2,3). Previous work suggested that polλ could be involved in the repair of DNA double strand breaks (DSB) by non-homologous end joining (NHEJ), based on its homology with the yeast DNA polymerase IV (4,5). In addition to identified major players of NHEJ in mammalian cells, i.e. DNA-PK, Ku, XRCC4 and DNA ligase IV, further candidates may be required to process the DNA breaks and to create 5′ and 3′ ends available for ligation (6). Though the participation of a DNA polymerase in the NHEJ process is still a matter of debate, recent in vitro biochemical studies suggest that polλ and/or polμ contribute to NHEJ function at incompatible DNA ends (7–9). However, the cellular function of polλ remains largely unknown; indeed, cells lacking polλ were apparently unimpaired in response to DNA damaging agents that produce DSB, methylation or interstrand cross links (10,11). The close homology among some of the 14 identified mammalian DNA polymerases raises the possibility of functional redundancy among at least some DNA polymerases. Such redundancy might compensate the absence of polλ and result in the lack of a defective response to DNA damaging agents observed in polλ-null cells (2).
In order to study the cellular role of polλ in the repair of DNA DSB we expressed a catalytically inactive mutant of polλ (polλDN) in cells bearing an integrated NHEJ or homologous recombination (HR) substrate. The overexpression of the polλDN acted as a competitive inhibitor of the endogenous polλ. No differences were observed either in HR efficiency or in NHEJ of fully complementary ends. In contrast, the expression of the enzymatically inactive form of polλ strongly affected the joining efficiency for mismatches, nucleotide gaps and flap structures generated by incompletely complementary ends. The molecular analysis of the junctions revealed the presence of large deletions in all the repaired junctions of the polλDN expressing cells. We further show that this NHEJ defect appear as increased sensitivity and chromosomal aberrations in cells in response to DNA DSB. Thus, it can be argued that polλ activity was specific of some DNA broken ends and was a determinant of the quality of the repair.
MATERIALS AND METHODS
Cells
The CHO-DRA10 cell line was cultured in α-MEM medium (GIBCO BRL) (12), and the C′10, A′7H, XR-1 cells (xrcc4 mutant cell line) and the complemented cell line (X4V) in DMEM medium (GIBCO BRL) as previously described (13,14). The DRA10-R2, DRA10-R15, C′10-R5, A′7H-RC clones, the DRA10-RD10, DRA10-RD14, C′10-RD13, A′7H-RD12 clones, the C′10-RDT2, A′7H-RDT3 clones, the C′10-BD6 and A′7H-BD5 clones and the C′10-muDB and A′7H-muD5 clones were obtained after transfection with the pIRESpuro2 vector (Clontech) containing the cDNA coding for the human wild-type DNA polymerase λ (polλ), the inactive form of DNA polymerase λ (polλDN), the truncated form (aa 245–575) of polλDN, the polβDN and the polμDN, respectively. Individual clones were obtained after transfection with jetPEI (Qbiogen, Illkirch, France) and selection with puromycin (5 μg/mL).
Generation of cells overexpressing polλ, polμ and polβ isoforms
Polλ expressing plasmids pIRES-polλ, pIRES-polλDN and pIRES-polλDNΔN (aa 245–575) were constructed by PCR amplification from a pRSETB plasmid carrying the cDNA sequence of the DNA polymerase λ gene. The polλ, polμ and polβ catalytically inactive mutants were constructed from pRSET plasmid, using the Quick Change mutagenesis kit (Stratagene) according to the manufacturer's instructions. Residues D427 and D429 in polλ, D330 and D332 in polμ, D190 and D192 in polβ (15) were replaced by Ala using primers. The resulting constructs were sequenced and error-free vectors were used to transfect cells.
Western blotting
Total cellular protein (50 µg) was separated by electrophoresis in a 10% SDS–PAGE and transferred to PVDF membrane. Polβ was detected using monoclonal antibody (18S) (Interchim) followed by incubation with horseradish peroxidase (HRP)-conjugated anti-mouse IgG, polλ was detected using polyclonal antibody (1/1000) (provided by Dr L Blanco, Madrid, Spain) and polμ was detected using polyclonal antibody (1/500) (AbCam) followed by incubation with HRP conjugated anti-rabbit IgG, and revealed using the ECL system (Amersham). Topoisomerase I western blotting was performed with the monoclonal anti-human C21 top1 antibody (Becton Dickinson). Equal loading was determined using monoclonal antibody to actin (1/5000) (AC10, Sigma).
Measurement of spontaneous homologous recombination
Determination of homologous recombination was achieved as previously described (12).
Measurement of NHEJ by FACS after induction of DSBs
Cells (5 × 105) were plated and allowed to attach overnight. Expression of the meganuclease I-Sce-I in cells was achieved by transient transfection of the expression plasmid pCMV–I-Sce-I (16) using Jet-PEI (Q-BIOgen, Illkrich, France). Seventy-two hours after transfection, cells were dissociated with PBS/EDTA (50 mM), washed in PBS and fixed in PBS/2% PAF for 20 min at room temperature. Cells were then stained with anti-H2Kd (1/30 mouse isotype, SF1-1.1, Pharmigen), or anti-CD4 (1/30 rat isotype, H129.19, Pharmigen) or anti-CD8 (1/30 rat isotype, 53-6.7, Pharmigen) for 30 min at room temperature in PBS/0.5% BSA. Cells were then incubated with anti-mouse-FITC (1/530 mouse isotype, F-2761, Molecular probe) for 30 min at room temperature. The frequency of H2K events allowed us to estimate the efficiency of I-Sce-I transfection and activity. Scoring of the NHEJ events affecting CD4 and CD8 was performed by flow cytometry using FACScan (Becton Dickinson).
Enrichment of CD4+ -expressing cells by MACS
Cells were dissociated with PBS/EDTA (50 mM) and washed with PBS/0.5% BSA/2mM EDTA. A total of 106 cells were stained with anti-CD4 for 15 min at room temperature, then incubated with beads coated with goat anti-rat IgG (Miltenyi Biotec) in PBS/0.5% BSA/2mM EDTA for 15 min at room temperature. After washing, the positively stained cells were separated onto miniMACS columns, and enriched ∼40%.
Junction sequence analysis
Genomic DNA was prepared from population of CD4 enriched cells (Puregene, Gentra Systems) and junctions of deletion events (CD4+) were amplified by PCR with the following primers (CMV1: 5′-TGGCCCGCCTGGCATTATGCCCAG-3′ and CD4int: 5′-GCTGCCCCAGAATCTTCCTCT-3′). PCR products were cloned in Topo-TA (Invitrogen), which allows isolation of individual clones, and sequenced.
Cytotoxicity studies
Cytotoxicities were determined by clonogenic assay (17). For ionizing radiation cells were exposed to a Co-source irradiator (IBL 437C type H, Oris industries SA). For campthotecin and mitomycin C (Sigma) cytotoxicities, cells were treated with increasing concentrations of drug for 24 h or 1 h, respectively. Survival was expressed as the plating efficiency of treated cells relative to the untreated control cells.
Karyotype analyses
Cells were either untreated or irradiated at 2 Gy, and metaphases spreads were prepared as described (18). Chromosomal aberrations were analysed using a Nikon microscope (TE300) (objective 100×).
Statistical analysis
Student' t paired analysis was used to examine differences between the two sets of results.
RESULTS AND DISCUSSION
The expression of polλDN decreases the frequency of I-Sce-I induced NHEJ events
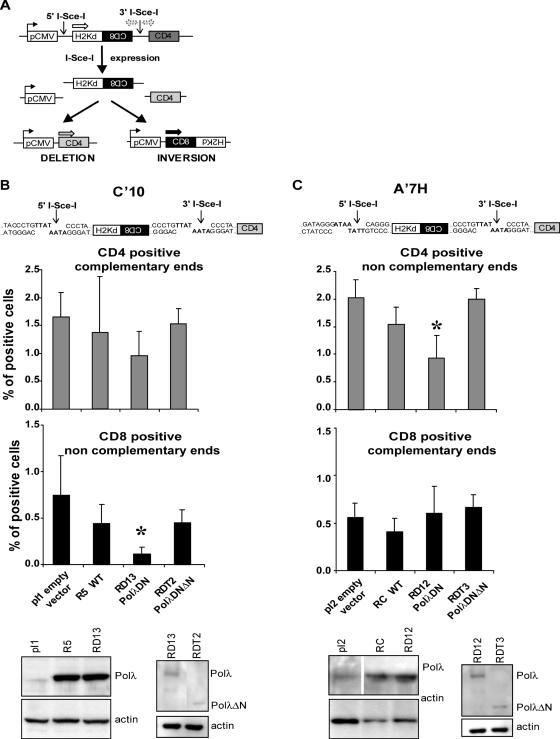
We generated a polymerase-deficient form of polλ (polλDN) by directed mutagenesis, changing two amino acids of the catalytic triad, Asp 427 and Asp 429 to Ala. Following expression and purification of the protein, the absence of DNA polymerase activity was confirmed by an in vitro primer extension assay (see Supplementary Data). We then expressed the WT, the polλDN and a truncated form of the polλDN (polλDNΔN) in CHO cells containing an integrated chromosomal NHEJ substrate (Figure 1A and B) designed to allow the measurement of NHEJ events at the molecular level by targeting breaks to this substrate (13). The polλ WT and polλDN forms were approximately overexpressed 10-fold compared with the parental cell lines (Figure 1). In the first cell line (C′10) the two I-Sce-I sites are in direct orientation creating fully complementary ends for deletion events visualized by joining of the CMV promoter to CD4; in contrast, inversion events that require partial pairing of 3′ overhangs and gap filling are recognized by juxtaposition of CD8 sequences to the CMV promoter (Figure 1A and B). Overexpression of the WT form of polλ did not alter the frequency of deletion (CD4; P = 0.62) or inversion events (CD8; P = 0.27). In contrast, whereas the frequency of deletion (CD4) events involving fully complementary ends was not significantly affected (P = 0.12; Figure 1B) expression of the polλDN form (RD13) resulted in a 7-fold decrease (P = 0.04) in the frequency of inversion (CD8) events involving incompletely complementary ends. Therefore, the absence of an efficient polλ appears to be prejudicial to the repair of events that potentially require DNA synthesis. This bias was confirmed by an additional test making use of an independent cell line, A′7H, in which (contrary to the C′10 system) the inverted orientation of the two I-Sce-I sites creates incompletely complementary ends for deletion events (CD4) but fully complementary ends for inversion events (CD8) (Figure 1A and C). No effect was observed for the frequencies of deletion (CD4) (P = 0.11) versus inversion (CD8) events (P = 0.42) in A′7H cells overexpressing the WT form of polλ. In contrast, in polλDN-expressing A′7H cells, deletion events (CD4) decreased by 2.3-fold (P = 0.02) while the frequency of inversion events (CD8) was unchanged (P = 0.83) (Figure 1C). To exclude the trivial possibility that the observed differences might be the simple result of varying efficiencies in I-Sce-I transfection efficiency and/or activity, we examined total H2K expression in treated cells. Deletion and inversion events that constitute total recombination activity concomitantly both reduce the number of remaining H2K-expressing gene copies (Figure 1). We find that H2K expression diminished similarly for all the cell lines tested (data not shown). We conclude that the differing results reflect the choice of recombination mode, and not simply access to cleaved DNA. Both in A′7H and C′10 cell lines, the affected events are those involving incompletely complementary ends that potentially require gap filling. Expressing the polλDN form led to a stronger reduction of inversion (CD8) events in the C′10 cells than for molecularly equivalent deletion (CD4) events in the A′7H cells. One possible explanation is that generating an inversion in C′10 cells requires two ligation events involving incompletely complementary ends whereas only one ligation event is necessary to produce a deletion event (CD4) in the A′7H cells (Figure 1B and C). These data support the need for a functional polλ for the maturation of some DNA ends during the repair of DSB by NHEJ. In addition, we generated a truncated form of the polλDN protein that lacks the N-terminal region comprising the BRCT and the proline rich domains that mediate protein interaction and control polλ activity (7,19). The expression of this truncated protein did not change the frequency of NHEJ events detected compared with the empty vector transfected (pI1 and pI2) control cell lines (Figure 1). These results show that polλ needs to interact with functional partners to perform its repair activity, as suggested by in vitro studies (7,9,20).
Figure 1.
Impact of the polλDN form expression on intrachromosomal NHEJ. (A) Substrate used to measure NHEJ. The only expressed gene is H2-Kd, which is under control of the pCMV promoter. CD8 is not expressed because it is in inverted orientation. CD4 is not expressed because it is too far from the pCMV promoter. Two I-Sce-I sites are present in non-coding sequences. After cleavage by I-Sce-I, the internal H2-Kd/CD8 fragment is excised. Re-joining of the DNA ends can lead to two different measurable events, deletion which leads to expression of the CD4 gene and inversion which leads to expression of the CD8 gene. (B and C) Representation of the sequences of the I-Sce-I restriction sites in the C′10 and A′7H cell lines, respectively. In the C′10 cells the two I-Sce-I sites are in direct orientation whereas in the A′7H cells the two I-Sce-I sites are in inverted orientation (upper panel). Evaluation of the frequency of the deletion (CD4) and inversion (CD8) events in the different cell lines (median panel). Results are the mean +/− SD of three independent experiments. Asterisks represent significant statistical difference (P < 0.05). Western blot for polλ expression in the different cell lines (lower panel).
Expression of an inactive form of the closest homologues of polλ, polβ and polμ does not affect I-Sce-I induced NHEJ events
To confirm the specificity of the effect observed with the polλDN form, we expressed equivalent catalytically inactive forms of polβ and polμ, the two closest homologues of polλ (11). Polβ was two-fold overexpressed in transfected cells compared with their corresponding control cells (Figure 2A and B). Although the expression of the inactive form of polβ was relatively weak compared with that of polλDN, a sensitization to methyl methane sulfonate treatment was observed, meaning that the inhibition of the endogenous polβ for base excision repair activity was effective (data not shown). Expression of the inactive form of polμ was also achieved in the same cell lines and the level of overexpression was similar to that obtained with polλ (Figure 2A and B). The absence of polμ activity was checked by an in vitro assay with the purified protein (see Supplementary Data). The expression of the inactive polβ (polβDN) and polμ (polμDN) in the C′10 cell line did not significantly alter the frequency of NHEJ events (P = 0.82 and P = 0.98 for CD4, P = 0.49 and P = 0.15 for CD8 for polμDN and polβDN expressing cells, respectively) compared with the control cell lines (Figure 2A). Identical results were obtained for the A′7H cell line (P = 0.47 and 0.25 for CD4, P = 0.82 and 0.92 for CD8, for polμ and polβ expressing cells, respectively) (Figure 2B). Therefore, these data underline the high specificity of polλ to repair the incompletely complementary DNA ends generated in our cellular system by I-Sce-I cleavage. Previous results indicated that polμ could participate in NHEJ in vitro (7,9). However, as in the case of polλ no cellular phenotype was observed after exposure to DNA damaging agents in mutant cells harbouring an invalidated polμ gene (21). This might indicate that polμ and polλ have overlapping or redundant function(s). The absence of effect of catalytically inactive polμ on deletion and inversion frequencies seen through CD4 and CD8 markers might reflect the DNA substrate specificity of this DNA polymerase. Owing to its similarity to TdT and its terminal transferase activity, it appears plausible that polμ is preferentially involved in the repair of unpaired DNA ends (9), eschewing DNA ends presenting partial annealing with gap and/or mismatch when normal polλ is present.
Figure 2.
Effect of the expression of the polβ and polμ inactive form on NHEJ events. (A and B) Representation of the sequences of the I-Sce-I restriction sites in the C′10 and A′7H cell lines, respectively. In the C′10 cells the two I-Sce-I sites are in direct orientation whereas in the A′7H cells the two I-Sce-I sites are in inverted orientation (upper panel). Evaluation of the frequency of the deletion (CD4) and inversion (CD8) events in the different cell lines (median panel). Results are the mean +/− SD of three independent experiments. Western blots for polβ and polμ expression in the different cell lines (lower panel).
The repair junction is altered in the presence of the polλDN form
In order to determine the molecular impact of polλ on the repair of I-Sce-I-induced DSB in a chromosomal context, we compared the resealing patterns of the partially complementary ends associated with a decreased frequency of deletion events (CD4+; Figure 1). We investigated the repair junction in the A′7H cell lines expressing the WT and the polλDN form, after enrichment by cell sorting of the CD4-positive cell population that results from excision and deletion of the internal I-Sce-I fragment following the joining of the two incompletely complementary ends (Figure 1A). Figure 3A shows examples of junctions from A′7H cells overexpressing the WT or the catalytically inactive form of polλ. In the cell line overexpressing the WT form of polλ, most of the junctions revealed processing of the four protruding nucleotides (8+3+4/18; 83%). The three classes of junctions observed could result from partial pairing of the four protruding nucleotides, leading to mismatches, gaps and flap structures (Figure 3B). The other clones showed deletions (3/18; 17%) of the sequenced junctions. Overall, our results closely resemble those previously described by Guirouilh-Barbat et al. in the same untransfected cell line (13). These results indicate that the overexpression of polλ does not significantly affect the repair junction of two partially complementary ends, either quantitatively or qualitatively. In contrast, expression of the polλDN was associated with large deletions in all the junctions sequenced (Figure 3A). The same 235 bp deletion from the I-Sce-I site to the CMV sequence was observed at the repair junctions for 19 out of 22 independent clones (Figure 3A). This deletion is, in practice, the largest we can observe, since larger deletions which affect the CMV promoter activity would eliminate the expression of the CD4 marker used to select mutant cells. Accordingly, the decrease in observed CD4 frequency could reflect such extensive deletions that affect the CMV promoter driving CD4 expression. These results show that a functional polλ is necessary for the maturation of partially complementary ends, and that the presence of an inactive polλ leads to the degradation in only one way until the finding of microhomology to allow resealing. Consequently the absence of an efficient polλ to process the DNA ends appears both to induce large deletions at the DNA break sites, and to prevent processing of the DNA ends by an alternative pathway (Figure 3C).
Figure 3.
Sequencing of the repair junctions. (A) Examples of sequences of the repair junctions of the A′7H cell line expressing the different forms of polλ. The structures of the implicated DNA ends are shown at the top of the figure. Black squares, location of the microhomologies. (B) Models for the different classes of end-joining events involving the four protruding nucleotides of the I-Sce-I cleavage site, using microhomology. (C) Percentages of the different types of NHEJ junction in the polλ WT and polλDN expressing cells.
The expression of the inactive form of polλ affects cell sensitivity to ionizing radiation and camptothecin without affecting homologous recombination
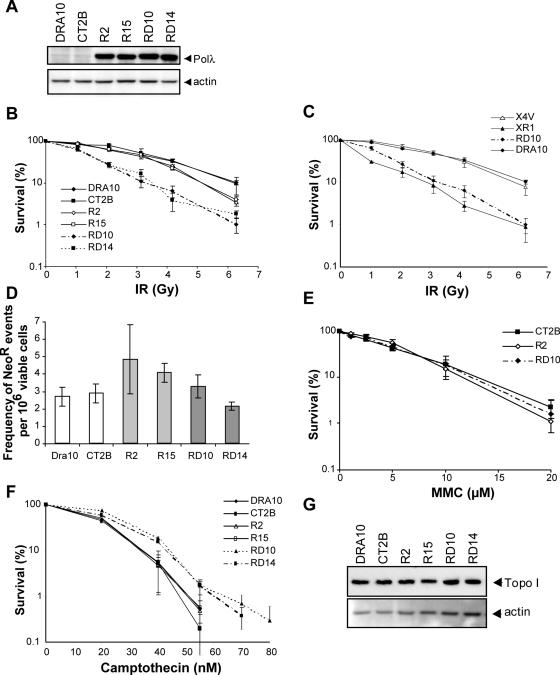
We next evaluated the cellular effects of polλDN in response to DNA damaging agents, in independent clones expressing the different forms of polλ at a similar level (Figure 4A). The cellular hallmark of a defect in the NHEJ function is hypersensitivity to ionizing radiation (IR) (22–24). We therefore tested the possible involvement of polλ in this pathway by examining cell survival after IR exposure. Cell lines expressing the polλDN form (RD10, RD14) showed high sensitivity to IR while cells overexpressing the WT form of polλ (R2, R15) were only slightly more sensitive than control cells (Figure 4B). In parallel, the cellular sensitivity to IR was compared with cell lines defective or complemented for the NHEJ protein XRCC4. Cells expressing the polλDN showed the same cellular sensitivity to IR as the XRCC4 defective cells (XR1), and the parental DRA10 cells were as sensitive to IR as the XRCC4-complemented cells (X4V) (Figure 4C). Though these results are in accordance with a potential role of polλ in NHEJ, cells defective in the HR pathway are also known to display increased sensitivity to IR, albeit to a lesser extent than NHEJ defective cells (25). To rule out the possibility that a defect in HR may account for this phenotype, we evaluated the HR frequency in cell lines containing a HR substrate composed of two inactive copies of the neomycin resistance gene as direct repeats. The frequency of obtaining neomycin-resistant colonies reflects HR events between the two inactive copies that result in a functional neomycin gene (16). No significant differences were observed among the tested cell lines (Figure 4D), suggesting that the cellular phenotype of IR hypersensitivity was not related to a defect in HR in cells expressing the normal and the inactive forms of polλ. We also treated the cell lines with mitomycin C (MMC), since sensitivity to this drug is a hallmark of a defective HR pathway (26). No difference in MMC sensitivity was observed (Figure 4E). These results reinforce the interpretation that polλ does not act in the HR pathway. Recent work in chicken DT40 cells showed that a defect in the NHEJ pathway increases cellular resistance to camptothecin (CPT), an inhibitor of the topoisomerase I, while a defect in the HR pathway has an opposite effect (27). Our examination of cellular sensitivity to CPT showed that cells overexpressing polλ were not affected while cells expressing the polλDN form displayed a resistance to CPT compared to control cells (Figure 4F). This resistance was not due to a variation in topoisomerase I content (Figure 4G), nor could it be attributed to a difference in topoisomerase I activity since equivalent amounts of total cell extracts formed similar levels of topoisomerase I-DNA cleavage complexes in vitro (data not shown). We therefore conclude that the cellular resistance to CPT is likely related to a defect in NHEJ.
Figure 4.
Cellular sensitivity to DNA damaging agents of the polλDN expressing cells. (A) Western blot analysis of the expression of the different forms of polλ in the DRA10 cell line. DRA10 is the parental cell line, CT2B is a cell line expressing the empty vector, R2 and R15 are independent cell lines overexpressing the WT form of polλ, RD10 and RD14 are independent cell lines expressing the polλDN form. (B) Cell survival after ionizing radiation and (C) Comparison of cell survival after ionizing radiation of polλDN-transfected cells with the NHEJ-defective cells, XR-1 (xrcc4 mutant cell line) and the XRCC4-complemented cell line (X4V). (D) Homologous recombination activity. (E) Cell survival after treatment with mitomycin C. (F) Cell survival after camptothecin treatment. (G) Topoisomerase I expression in the different polλ expressing cell lines. All cell survival and homologous recombination results are the mean +/− SEM of 3 independent experiments.
The cellular expression of the polλDN form induces genetic instability
Defective NHEJ leads to the accumulation of spontaneous or IR-induced chromosomal aberrations (28,29). In light of the effects of expressing polλDN, we performed karyotypic analyses of the different cell lines without or after treatment by IR. Chromosomal aberrations were increased in non-irradiated cells expressing the polλDN form, and this genetic instability was further amplified after IR treatment (Table 1). A large part of the abnormalities were tri- and quadriradial chromosomes together with chromosome breaks (Figure 5), which are often observed in NHEJ defective cells (28,29). The intermediate effect observed in cells overexpressing the WT form of polλ can be correlated with the slight sensitization to IR (Figure 4). The overexpression of polλ might substitute for some cellular components affecting NHEJ activity. Identical chromosomal instability was observed with a human fibroblast cell line (MRC5) transfected with the polλDN form, allowing us to rule out a species-specific effect of the DNA polymerase (data not shown).
Table 1.
Increased genomic instability in polλDN expressing cells
| Clone | Number of metaphases | Cells with aberrations (%) | Aberration type dicentric/fusion (%) breaks (%) | ||
|---|---|---|---|---|---|
| UT | CT2B | 103 | 1.0 | 1.0 | 0.0 |
| R2 | 104 | 2.9 | 1.9 | 1.0 | |
| RD10 | 90 | 7.8 | 6.7 | 1.1 | |
| IR | CT2B | 107 | 8.4 | 6.5 | 1.9 |
| R2 | 122 | 19.7 | 16.4 | 3.3 | |
| RD10 | 94 | 59.5 | 57.1 | 24.6 | |
Metaphase spreads were analysed from untreated (UT) and 2 Gy treated cells (IR). CT2B, R2 and RD10 are independent cell lines transfected with the empty vector, the polλ WT and the polλDN forms, respectively.
Figure 5.
Chromosomal aberrations induced by IR in polλDN expressing cells. Photographs of metaphase spreads of polλDN expressing cells 24 h after treatment with IR at 2 Gy. (b, break; t, triradial).
Altogether, these results strongly support a role of polλ in the NHEJ-DSB repair process following genotoxic stress. This study demonstrates that polλ participates in the NHEJ pathway in mammalian cells. Importantly, it has a very specific role in the processing of DNA ends requiring some potential gap filling before ligation leading to the protection of DNA ends against extensive degradation (Figure 3C).
Recent studies suggest that another DNA polymerase of the X family, polμ, could be involved in the repair of DSB presenting non-compatible DNA ends (7,9). However, biochemical and cellular studies strongly suggest that polμ is more specialized in the processing of DNA end structures containing no homology and found in V(D)J recombination during the repair of Igκ recombination intermediates, in accordance with its specific ability to polymerise across a DSB with no pairing between the primer and the template (9). In addition, a study by Lee et al. (8) using cell extracts and DNA ends presenting a limited homology (2 nt and a 2 nt gap) similar to our intracellular NHEJ substrate, showed that immunodepletion of polλ, but not polμ, abolished the joining of these DNA ends in vitro.
Overall, these results show that the contribution of each polymerase in the repair of DSB depends on the sequence and specific overhang structure of the DNA ends. Whether another DNA polymerase can substitute for or act in concert with polλ to repair the different types of DSB induced by genotoxic stress during NHEJ remains to be determined.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
Acknowledgments
The authors thank Dr Luis Blanco for providing the cDNA of polλ, Dr Said Aoufouchi for providing the cDNA of polμ and Dr Maria Jasin for providing the CHO-DRA10 cell line and the I-Sce-I expression vector. The authors thank Dr David Cribbs for his critical reading of the manuscript. This work was supported by ‘Ligue Nationale contre le Cancer équipe labellisée’ (JSH), (BSL), the Cancéropôle Grand Sud-Ouest (CC) and Electricité de France (YC). Funding to pay the Open Access publication charges for this article was provided by Electricité de France (EDF).
Conflict of interest statement. None declared.
REFERENCES
- 1.Garcia-Diaz M., Dominguez O., Lopez-Fernandez L.A., de Lera L.T., Saniger M.L., Ruiz J.F., Parraga M., Garcia-Ortiz M.J., Kirchhoff T., del Mazo J., et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 2.Shcherbakova P.V., Bebenek K., Kunkel T.A. Functions of eukaryotic DNA polymerases. Sci. Aging Knowledge Environ. 2003;2003:RE3. doi: 10.1126/sageke.2003.8.re3. [DOI] [PubMed] [Google Scholar]
- 3.Ramadan K., Shevelev I., Hubscher U. The DNA-polymerase-X family: controllers of DNA quality? Nature Rev. Mol. Cell Biol. 2004;5:1038–1043. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- 4.Tseng H.M., Tomkinson A.E. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 2002;277:45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- 5.Wilson T.E., Lieber M.R. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 6.Budman J., Chu G. Processing of DNA for nonhomologous end-joining by cell-free extract. EMBO J. 2005;24:849–860. doi: 10.1038/sj.emboj.7600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y., Lu H., Tippin B., Goodman M.F., Shimazaki N., Koiwai O., Hsieh C.L., Schwarz K., Lieber M.R. A biochemically defined system for mammalian nonhomologous dna end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Lee J.W., Blanco L., Zhou T., Garcia-Diaz M., Bebenek K., Kunkel T.A., Wang Z., Povirk L.F. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 9.Nick McElhinny S.A., Havener J.M., Garcia-Diaz M., Juarez R., Bebenek K., Kee B.L., Blanco L., Kunkel T.A., Ramsden D.A. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi Y., Watanabe M., Okada Y., Sawa H., Takai H., Nakanishi M., Kawase Y., Suzuki H., Nagashima K., Ikeda K., et al. Hydrocephalus, situs inversus, chronic sinusitis, and male infertility in DNA polymerase lambda-deficient mice: possible implication for the pathogenesis of immotile cilia syndrome. Mol. Cell Biol. 2002;22:2769–2776. doi: 10.1128/MCB.22.8.2769-2776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trivedi R.N., Almeida K.H., Fornsaglio J.L., Schamus S., Sobol R.W. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 12.Canitrot Y., Capp J.P., Puget N., Bieth A., Lopez B., Hoffmann J.S., Cazaux C. DNA polymerase beta overexpression stimulates the Rad51-dependent homologous recombination in mammalian cells. Nucleic Acids Res. 2004;32:5104–5112. doi: 10.1093/nar/gkh848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guirouilh-Barbat J., Huck S., Bertrand P., Pirzio L., Desmaze C., Sabatier L., Lopez B.S. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Delacote F., Han M., Stamato T.D., Jasin M., Lopez B.S. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Date T., Yamamoto S., Tanihara K., Nishimoto Y., Matsukage A. Aspartic acid residues at positions 190 and 192 of rat DNA polymerase beta are involved in primer binding. Biochemistry. 1991;30:5286–5292. doi: 10.1021/bi00235a023. [DOI] [PubMed] [Google Scholar]
- 16.Liang F., Han M., Romanienko P., Jasin M. Homology-directed repair is a major double-strand-break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frechet M., Canitrot Y., Bieth A., Dogliotti E., Cazaux C., Hoffmann J.S. deregulated DNA polymerase beta strengthens ionizing radiation-induced nucleotidic and chromosomal instabilities. Oncogene. 2002;21:2320–2327. doi: 10.1038/sj.onc.1205295. [DOI] [PubMed] [Google Scholar]
- 18.Bergoglio V., Pillaire M.J., Lacroix-Triki M., Raynaud-Messina B., Canitrot Y., Bieth A., Gares M., Wright M., Delsol G., Loeb L.A., et al. Deregulated DNA polymerase beta induces chromosome instability and tumorigenesis. Cancer Res. 2002;62:3511–3514. [PubMed] [Google Scholar]
- 19.Shimazaki N., Yoshida K., Kobayashi T., Toji S., Tamai K., Koiwai O. Over-expression of human DNA polymerase lambda in E.coli and characterization of the recombinant enzyme. Genes Cells. 2002;7:639–651. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 20.Fan W., Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem. Biophys. Res. Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Bertocci B., De Smet A., Berek C., Weill J.C., Reynaud C.A. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003;19:203–211. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 22.Li Z., Otevrel T., Gao Y., Cheng H.L., Seed B., Stamato T.D., Taccioli G.E., Alt F.W. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 23.Riballo E., Critchlow S.E., Teo S.H., Doherty A.J., Priestley A., Broughton B., Kysela B., Beamish H., Plowman N., Arlett C.F., et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 24.Gu Y., Jin S., Gao Y., Weaver D.T., Alt F.W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl Acad. Sci. USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Gent D.C., Hoeijmakers J.H., Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 26.Hochegger H., Sonoda E., Takeda S. Post-replication repair in DT40 cells: translesion polymerases versus recombinases. Bioessays. 2004;26:151–158. doi: 10.1002/bies.10403. [DOI] [PubMed] [Google Scholar]
- 27.Adachi N., So S., Koyama H. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 2004;279:37343–37348. doi: 10.1074/jbc.M313910200. [DOI] [PubMed] [Google Scholar]
- 28.Karanjawala Z.E., Grawunder U., Hsieh C.L., Lieber M.R. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 1999;9:1501–1504. doi: 10.1016/s0960-9822(00)80123-2. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson D.O., Sekiguchi J.M., Chang S., Frank K.M., Gao Y., DePinho R.A., Alt F.W. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl Acad. Sci. USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]