Abstract
The rDNA cluster in Saccharomyces cerevisiae is located 450 kb from the left end and 610 kb from the right end of chromosome XII and consists of ∼150 tandemly repeated copies of a 9.1 kb rDNA unit. To explore the biological significance of this specific chromosomal context, chromosome XII was split at both sides of the rDNA cluster and strains harboring deleted variants of chromosome XII consisting of 450 kb, 1500 kb (rDNA cluster only) and 610 kb were created. In the strain harboring the 1500 kb variant of chromosome XII consisting solely of rDNA, the size of the rDNA cluster was found to decrease as a result of a decrease in rDNA copy number. The frequency of silencing of URA3 inserted within the rDNA locus was found to be greater than in a wild-type strain. The localization and morphology of the nucleolus was also affected such that a single and occasionally (6–12% frequency) two foci for Nop1p and a rounded nucleolus were observed, whereas a typical crescent-shaped nucleolar structure was seen in the wild-type strain. Notably, strains harboring the 450 kb chromosome XII variant and/or the 1500 kb variant consisting solely of rDNA had shorter life spans than wild type and also accumulated extrachromosomal rDNA circles. These observations suggest that the context of chromosome XII plays an important role in maintaining a constant rDNA copy number and in physiological processes related to rDNA function in S.cerevisiae.
INTRODUCTION
The genomic structure and organization of the repeated ribosomal DNA cluster (rRNA genes) have been studied extensively in many organisms, from Escherichia coli to humans. With respect to the number of rDNA genes, E.coli has seven, four of which are located fairly close to the origin of replication but are not continuous, whereas the rDNAs of most eukaryotic organisms are tandemly repeated at one or a few chromosomal loci and are present in many copies. In contrast, certain organisms routinely harbor extrachromosomal rRNA genes (1).
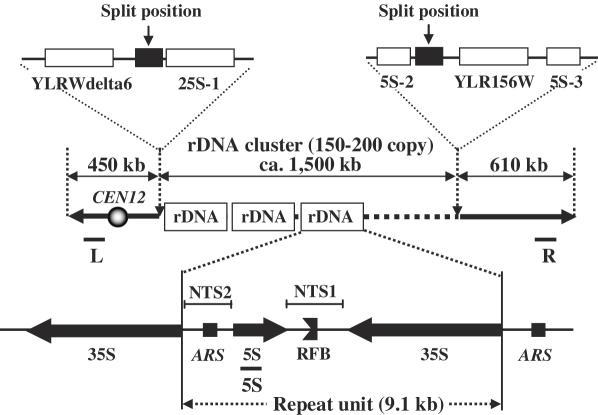
The rRNA of Saccharomyces cerevisiae is encoded by the ribosomal RNA genes called RDN1, an ∼1.5 Mb region consisting of ∼150 tandem repeated copies of a 9.1 kb unit, on chromosome XII (2). A single 9.1 kb unit consists of two transcribed regions, the 35S precursor rRNA and 5S rRNA, and two non-transcribed regions, NTS1 and NTS2 (Figure 1). The DNA encoding the 35S rRNA and 5S rRNA genes is transcribed by RNA polymerase I (Pol I) and III (Pol III), respectively. Each rDNA unit has a replication fork barrier (RFB) and an autonomous replicating sequence (ARS) in the NTS1 and NTS2 regions, respectively (3–5). It has been reported that only 50% of the rRNA genes are transcribed even in rapidly growing cells and that half of the rRNA genes are inactive (6).
Figure 1.
Structure of rDNA repeats in S.cerevisiae. The rDNA repeats, present in 150∼200 copies on chromosome XII, are indicated. Each repeat contains the coding sequence for 35S rRNA and 5S rRNA, transcribed in the direction of the arrows, and NTS1 and NTS2 regions that are not transcribed. NTS1 and NTS2 contain the RFB and ARS, respectively. The locations of the hybridization probes L, 5S and R are also shown.
The copy number of the rDNA unit varies and its regulation requires some special genes, including SIR2 and FOB1. The Sir2 protein, an NAD-dependent histone deacetylase (7–9), suppresses meiotic and mitotic recombination within the rDNA (10–12). The Fob1 protein is required for replication fork blocking activity at the RFB site in rDNA, which appears to stimulate recombination directly within rDNA repeats and to play an essential role in rDNA expansion/contraction (13,14). Accumulation of extrachromosomal rDNA circles (ERCs) (15) caused by recombination of the repeated sequence is promoted by FOB1 (16) and suppressed by SIR2 (11,17). Sinclair and Guarente (15) have shown that ERCs accumulate in aging yeast cells and that these circles can cause aging. rDNA transcription by Pol I is associated with a distinct crescent-shaped subnuclear structure, the nucleolus, which also appears to be the major site of ribosomal assembly. In a strain carrying a plasmid with a single rDNA unit transcribed by Pol I, the nucleolus was fragmented and distributed throughout the nucleus (18). Moreover, when a gene transcribed by Pol II is integrated into the tandem array of RDN1, transcription of the gene is silenced (19–21). This transcriptional silencing of marker genes inserted into rDNA is abolished by loss of SIR2 function (22). All of these observations suggest that rDNA is a suitable model for studying genomic stability as it relates to copy number fluctuation of repeated genes, but also the important processes of aging, nucleolus morphology and gene silencing.
In this study, we addressed the question of how chromosome XII context in S.cerevisiae affects rDNA copy number control and how it relates to various physiological endpoints relevant to rDNA function. To this end, we constructed deletion variants of chromosome XII, among them, a chromosome consisting solely of the rDNA cluster. A strain harboring this chromosome was found to have a decreased number of rDNA copies, a reduced life span, and an increased frequency of gene silencing, relative to the parent strain. We suggest that the entire chromosome XII context, including the non-rDNA-containing regions, plays an important role in maintaining normal rDNA function and constant rDNA copy number in S.cerevisiae.
MATERIALS AND METHODS
Strains, plasmids, media and transformation
The yeast strains and plasmids used in this study are described in Table 1. All strains were derived from FY833 (MATa ura3-52 his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63) (23). FY833 fob1::HIS3 (ID-10) was constructed by transforming FY833 with p2453 (also known as pTAK024) linearized with EcoRI (gift of T. Kobayashi) (13). Plasmids pUG6-CEN4, pUG6-CgHIS3, pUG6-CgTRP1 and pUG6-CgLEU2 (24) were used as templates for preparing splitting fragments. p3018 (also known as pSK-U) was used as a template plasmid for amplification of URA3 gene in the silencing analysis. p3151 (also known as YCp-FOB1, gift of T. Kobayashi) (14) and p3150 (also known as YCp-Nop1::GFP, gift of Y. Kikuchi) (25) were used to introduce the FOB1 gene and for visualization of the nucleolus, respectively. Strains were propagated in YPAD nutrient medium and synthetic complete (SC) medium as described (26) and were cultured at 30°C. The SC-Ura and 5-fluoroorotic acid (5-FOA) media were prepared as described (27). Yeast transformation was performed according to the high efficiency transformation protocol described by Gietz and Schiestl (28). For transformation of E.coli, a Z-Competent E.coli transformation kit system was used (Zymo Research, USA).
Table 1.
Yeast strains and plasmids used in this study
| Designation | Description |
|---|---|
| Strains | |
| FY833 | MATa ura3-52 his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 |
| FY834 | MATα ura3-52 his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 |
| W303-1B | MATα ura3-1 leu2-3,112 trp1 his3-11,15 ade2-1 |
| S288C | MATα SUC2 mal mel gal2 CUP1 flo1 flo8-1 |
| ID-1 | FY833 [CFXII/TRP1+rDNA-R proximal:1900 kb] [CFXII/rDNA-R distal:610 kb] |
| ID-2 | FY833 [CFXII/HIS3+rDNA-L proximal:450 kb] [CFXII/rDNA-L distal:2050 kb] |
| ID-8 | FY833 [CFXII/HIS3+rDNA-L proximal:450 kb][rDNA cluster+TRP1] [CFXII/rDNA-R distal:610 kb] |
| ID-9 | FY833 [CFXII/HIS3+rDNA-L proximal:450 kb][rDNA cluster+TRP1] [CFXII/rDNA-R distal:610 kb] |
| ID-10 | FY833 fob1::HIS3 |
| ID-11 | FY833 fob1::HIS3[CFXII/TRP1+rDNA-R proximal:1900 kb] [CFXII/rDNA-R distal:610 kb] |
| ID-12 | FY833 fob1::HIS3[CFXII/LEU2+rDNA-L proximal:450 kb] [rDNA cluster+TRP1] [CFXII/rDNA-R distal:610 kb] |
| ID-13 | FY833 fob1::HIS3[CFXII/LEU2+rDNA-L proximal:450 kb] [rDNA cluster+TRP1] [CFXII/rDNA-R distal:610 kb] [p3151(=YCp+FOB1)] |
| ID-16 | FY833 rdn18::URA3 |
| ID-18 | ID-1 rdn18::URA3 |
| ID-19 | ID-2 rdn18::URA3 |
| ID-20 | ID-8 rdn18::URA3 |
| ID-24 | ID-12 rdn18::URA3 |
| ID-29 | FY833[p3150(=YCp+Nop1::GFP)] |
| ID-30 | ID-1 [p3150(=YCp+Nop1::GFP)] |
| ID-31 | ID-2 [p3150(=YCp+Nop1::GFP)] |
| ID-32 | ID-8 [p3150(=YCp+Nop1::GFP)] |
| Plasmids | |
| p3121 | A derivative of pUG6 carrying CEN4 (= pUG6-CEN4) (24) |
| p3008 | A derivative of pUG6 carrying loxP-CgLEU2-loxP (= pUG6-CgLEU2) (24) |
| p3009 | A derivative of pUG6 carrying loxP-CgHIS3-loxP (= pUG6-CgHIS3) (24) |
| p3010 | A derivative of pUG6 carrying loxP-CgTRP1-loxP (= pUG6-CgTRP1) (24) |
| p2453 | A derivative of pUC18 carrying fob1Δ::HIS3 (= pTAK024) (13) |
| p3018 | A derivative of pBluscript II SK+ carrying URA3 gene (= pSK-U) |
| p3151 | A derivative of YCplac33 carrying FOB1 gene (= YCp-FOB1) (14) |
| p3150 | A derivative of YCp carrying Nop1::GFP (= YCp-Nop1::GFP) (25) |
Oligonucleotides, PCR and other methods
The oligonucleotides used in this study are described in Table 2. Saccharomyces Genome Database (http://yeastgenome.org/) was used to design the splitting points and the oligonucleotides. PCR was performed as described elsewhere (29). To split 610 kb from the right end of chromosome XII, two splitting fragments were prepared using the following strategy: one splitting fragment (R-I) consisted of CgTRP1, a telomeric 5′-(C4A2)6-3′ repeat sequence and a target sequence. The 1.2 kb CgTRP1 gene used to select yeast transformants was amplified by PCR using p3010 as template and loxP-F1 and Tr6-4 as primers. Independently, a 300 bp target sequence which corresponds to nucleotide position 468 931–469 225 of chromosome XII was amplified by PCR using genomic DNA from strain FY833 as template and rDNAr-1 and rDNAr-2 as primers. The 1.2 kb and the 300 bp PCR fragments were used as templates, and Tr6-4 and rDNAr-1 as primers for the second PCR to generate the 1.5 kb splitting fragment R-I. The other splitting fragment (R-II) containing CEN4, a telomeric repeat sequence and the target sequence was also amplified by two rounds of PCR as follows: a 0.8 kb PCR product containing CEN4, and telomeric repeat sequence was amplified by a first PCR using p3121 as template and loxP-F1 and Tr6-4 as primers. Separately, a 300 bp target sequence which corresponds to nucleotide position 469 226–469 530 of chromosome XII was amplified using genomic DNA from FY833 as template and rDNAr-3 and rDNAr-4 as primers. Then, a second PCR was performed using the 0.8 kb and the 300 bp PCR fragments as templates and Tr6-4 and rDNAr-4 as primers to generate the second 1.1 kb splitting fragment R-II. Next, two splitting fragments (L-I and L-II) for splitting 450 kb from the left end of chromosome XII were also prepared as described above. In this case, p3121, p3009 or p3008 (for splitting at the left end in the fob1 mutants) were used as template plasmids and rDNAl-1, rDNAl-2, rDNAl-3, rDNAl-4, loxP-F1 and Tr6-4 were used as primers. The target sequence corresponds to nucleotide position 451 070–451 786 of chromosome XII. rDNA5S-1, rDNA5S-2, HaY12P3F, HaY12P3R, HaY12P5F and HaY12P5R were used as primers for the amplification of probe-5S (5S), probe-R (R) and probe-L (L), respectively, for Southern hybridization. To introduce URA3 into the 18S rDNA region for silencing analysis, a DNA fragment containing URA3 was amplified by PCR using p3018 as template and ura3-f and ura3-r as primers. Si-69 and Si-72 were used as primers for the amplification of a probe to detect a single copy gene for rDNA copy number determination. CHEF gel electrophoresis and Southern hybridization were performed as described elsewhere (26).
Table 2.
Oligonucleotides used in this study
| Oligonucleotides | Sequences |
|---|---|
| rDNAr-1 | 5′-GTTGTTTTTTTTTTCGCGCA-3′ |
| rDNAr-2 | 5′-CTGCAGCGTACGAAGCTTCAGCTGGCGGCCATGTGCCAGTTAAGCTATTT-3′ |
| rDNAr-3 | 5′-CTGCAGCGTACGAAGCTTCAGCTGGCGGCCCATGAAAAGGATGTAGAAAT-3′ |
| rDNAr-4 | 5′-AACGAACAACTTTATGAAGA-3′ |
| rDNAl-1 | 5′-TGTATATCACGTAATACACA-3′ |
| rDNAl-2 | 5′-CTGCAGCGTACGAAGCTTCAGCTGGCGGCCGCGTTCTATAGCAAACATGAGGAAATATCC-3′ |
| rDNAl-3 | 5′-CTGCAGCGTACGAAGCTTCAGCTGGCGGCCGCGTTCTATACGCTCTGATGGTGCGGAAAA-3′ |
| rDNAl-4 | 5′-TTTTTATTTCTTTCTAAGTG-3′ |
| Tr6-4 | 5′-(CCCCAA)6AGGCCACTAGTGGATCTGAT-3′ |
| loxP-F1 | 5′-GGCCGCCAGCTGAAGCTTCG-3′ |
| rDNA5S-1 | 5′-GCGGCCATATCTACCAGAAA-3′ |
| rDNA5S-2 | 5′-TGCGGAGTTGTAAGATGTAC-3′ |
| HaY12P3F | 5′-CGGAATTCGAACATGTTTGCGCCTT-3′ |
| HaY12P3R | 5′-ATAGCGTCATTTGGGCCGCTAGTAT-3′ |
| HaY12P5F | 5′-CGGTAGTGCTGTTGTTTGTCAGG-3′ |
| HaY12P5R | 5′-TTTCTCTTCTTCCAACCAGGC-3′ |
| ura3-f | 5′-TCCATGCTAATATATTCGAGCAATACGCCTGCTTTGAACACTCTAATTTTGGGGATCCACTAGTTCTAGA-3′ |
| ura3-r | 5′-CTAACCTTGAGTCCTTGTGGCTCTTGGCGAACCAGGACTTTTACTTTGAAAATTGGAGCTCCACCGCGGT-3′ |
| Si-69 | 5′-GGTTTACACCCCACCGTGAG-3′ |
| Si-72 | 5′-AGGGAACTGTTGGGATATAC-3′ |
| 18S-F | 5′-CCTGAGAAACGGCTACCACATC-3′ |
| 18S-R | 5′-ATTGTCACTACCTCCCTGAATTAGGA-3′ |
| ACT1-F | 5′-CGCTCCTCGTGCTGTCTTC-3′ |
| ACT1-R | 5′-TTGACCCATACCGACCATGATA-3′ |
Estimation of rDNA copy number and rRNA quantification
Genomic DNA digested with at a unique KpnI site in the rDNA region was subjected to 0.8% agarose gel electrophoresis and analyzed by Southern hybridization using 5S rDNA as probe (500 bp fragment prepared by PCR), which hybridizes to chromosomal rDNA. To determine rDNA copy number, a single copy gene, YDL183C on a chromosome IV fragment (130 872–131 834) prepared by PCR, was used as an internal control. Intensity of hybridization signal was determined by Scion image (http://www.scioncorp.com/) and the copy number was calculated by comparing the signal strength of the hybridizing bands.
To quantify the rRNA, real-time quantitative RT–PCR using an ABI Prism 7300 sequence detector (TaqMan PCR; Applied Biosystems, CA, USA) was performed. The TaqMan RT–PCRs were carried out by two-step RT–PCR using the High capacity cDNA Archive kit (Applied Biosystems) and TaqMan Universal PCR Master Mix (Applied Biosystems). The probe and primers used for detection of the 18S rRNA were 5′-CAGCAGGCGCGCAAATTACCCA-3′ (TaqMan probe), 18S-F and 18S-R (Table 2). To detect the ACT1 as endogenous control, 5′-TCTATCGTCGGTAGACCA AGACACCA-3′ (TaqMan probe), ACT1-F and ACT1-R (Table 2) were used as probe and primers, respectively. The TaqMan probe was labeled in the 5′ end with FAM (fluoresceine) and in the 3′ end with MGB (quencher).
Silencing analysis and localization of the nucleolus
An analysis of silencing was performed as described by Huang and Moazed (30). A fragment containing URA3 flanked by 50 bp of homologous region of 18S rDNA (nucleotide positions 456 940–456 980 and 456 981–457 030 on chromosome XII) was prepared and introduced into FY833, ID-1, ID-2, ID-8 and ID-12, to generate ID-16, ID-18, ID-19, ID-20 and ID-24, respectively. Integration of URA3 into the 18S rDNA locus was confirmed by PCR (data not shown). Strains were grown in YPAD to an OD600 of 1.5 and 3 µl each of 10-fold serial dilutions were spotted on YPAD, SC-Ura and 5-FOA media. Plates were incubated at 30°C for 2–4 days. Silencing of URA3 expression was evaluated qualitatively based on relative growth of the strains on SC-Ura (expression required for growth) and 5-FOA (silencing required for growth).
For determination of nucleolar localization, FY833, ID-1, ID-2 and ID-8 harboring p3150 were grown in YPAD for 1 day. An appropriate amount of cell suspension was mixed in Vectashield® Mounting Medium with DAPI (Vector Laboratories, Inc., Burlingame, USA), and held for 10 min in the dark. DAPI and GFP fluorescence were observed by fluorescence microscopy (BX61 Olympus, Japan) and photographed using a CCD camera (CCD-EX1; Universal Imaging Corporation, USA). DAPI and GFP images were merged using Photoshop (Adobe system). The shape and localization of the nucleolus were determined from an analysis of these images for at least 100 cells in each strain.
Life span analysis and detection of ERC
The methods used for life span analysis are similar to those described by Kennedy et al. (31). Life spans were determined by counting the number of daughter cells generated by a single mother cell. A micromanipulator (SINGER MSN system series 200, Singer instruments, UK) was used to tease away and remove each daughter cell that emerged as a bud from its mother cell. Mother cells are larger than daughter cells and are easily distinguishable. Virgin cells to be used as mother cells for life span analysis were generated by randomly picking cells from a log-phase liquid culture that had been streaked onto a solid YPAD plate. At least 40 mother cells were used from each strain to generate life span curves and to determine average longevity.
In order to detect ERCs, genomic DNA was first prepared by the yeast DNA miniprep method (32) and then ERCs were resolved by loading the same volume of DNA on a 0.6% agarose gel without ethidium bromide run at 1 V/cm for 30 h followed by Southern hybridization using 5S rDNA as a probe.
RESULTS
Construction of an artificial chromosome consisting solely of the rDNA cluster
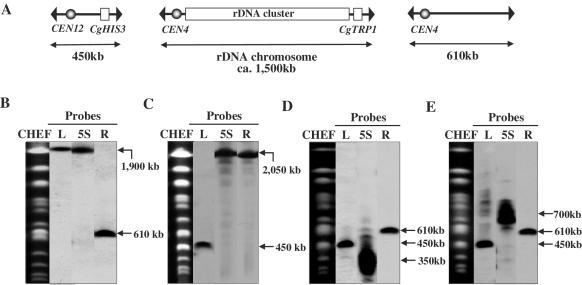
The structure and organization of the yeast rDNA repeating unit located 450 kb from the left end and 610 kb from the right end of chromosome XII are shown in Figure 1. The size of chromosome XII is ∼2–3 Mb depending upon the rDNA copy number. To explore the biological significance of the context and organization of chromosome XII, we constructed a chromosome consisting of only the rDNA cluster (Figure 2A) by using the PCR-mediated chromosome splitting method (24). To split 610 kb from the right end of chromosome XII, two splitting fragments, R-I and R-II, were prepared as described in Materials and Methods. These 2 splitting fragments were introduced into FY833 and 19 Trp+ transformants were analyzed for karyotype by CHEF gel electrophoresis and Southern hybridization. Fifteen transformants (79%) showed that the 2.5 Mb chromosome XII was successfully split into 1900 and 610 kb chromosomes (Figure 2B). One such transformant was called ID-1.
Figure 2.
(A) Structure of split chromosomes and rDNA chromosome and analysis of split-chromosomes by CHEF gel electrophoresis and Southern hybridization. Chromosomal DNA was isolated from the following strains: (B) ID-1, (C) ID-2, (D) ID-8 and (E) ID-9. For Southern hybridization analysis, L, R and 5S containing 1 kb of chromosome XII 50 kb from the left or right end, or 500 bp of 5S rDNA, respectively, were used as probes. The positions of the probes are indicated in Figure 1.
We also split chromosome XII 450 kb from its left end. Two splitting fragments, L-I and L-II as described in Materials and Methods, were introduced into FY833 and three His+ transformants were analyzed for karyotype by CHEF gel electrophoresis and Southern hybridization. All transformants (100%) showed that the 2.5 Mb chromosome XII was successfully split into 450 and 2050 kb variants (Figure 2C). One such transformant was designated ID-2.
Because strain ID-1 was viable, we performed subsequent splitting 450 kb from the left end of the 1900 kb split chromosome. Two splitting fragments, L-I and L-II, were prepared by PCR and introduced into strain ID-1. If the 1900 kb split chromosome was split as expected at the target site, 450 kb and ∼1500 kb chromosomes should have been generated, the latter consisting solely of the rDNA cluster. Of 18 transformants tested 2 (ID-8 and ID-9) were found to harbor 450 kb bands in addition to the pre-existing 610 kb split chromosome (Figure 2D and E). However, when Southern hybridization analysis was conducted using the 5S rDNA as a probe, broad bands centered at about 350 kb (ID-8) (Figure 2D) and 700 kb (ID-9) (Figure 2E) in size were detected, although the expected size of the rDNA cluster was ∼1500 kb. Henceforth, a chromosome consisting solely of the rDNA cluster will be referred to as the ‘rDNA chromosome.’ To confirm variation of size of rDNA chromosome, we isolated larger number of single colonies from the strains ID-8 and ID-9. The results revealed that shortened and variable-sized rDNA chromosomes were also observed. Furthermore, when fluctuation of size of rDNA chromosome was examined in diploids made by crossing ID-8 with FY834 (harboring no split rDNA chromosome) and ID-9 with FY834, variable-sized rDNA chromosomes were observed. Theses results suggest that while it is possible to create an rDNA chromosome, such a structure is unstable and undergoes spontaneous rearrangement leading to smaller size variants.
rDNA copy number on the rDNA chromosome is less than on the chromosome XII but quantity of rRNA was indistinguishable from that in wild-type strain
To examine whether the faster migration of the rDNA chromosome on the CHEF gel was indeed caused by size reduction, rDNA copy number was estimated in the transformants carrying the rDNA chromosome. rDNA copy number is controlled by the RFB which allows progression of the DNA replication fork in the direction of 35S rRNA transcription, but not in the opposite direction (13). Fob1p is a trans-acting factor required for RFB to function and therefore, in a fob1 mutant, the necessary homologous recombination does not occur, resulting in a fixed number of rDNA copies (33).
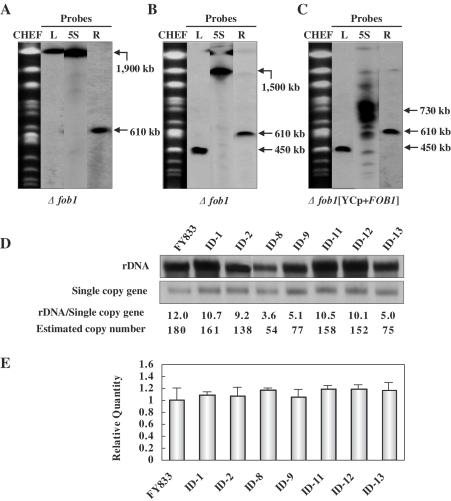
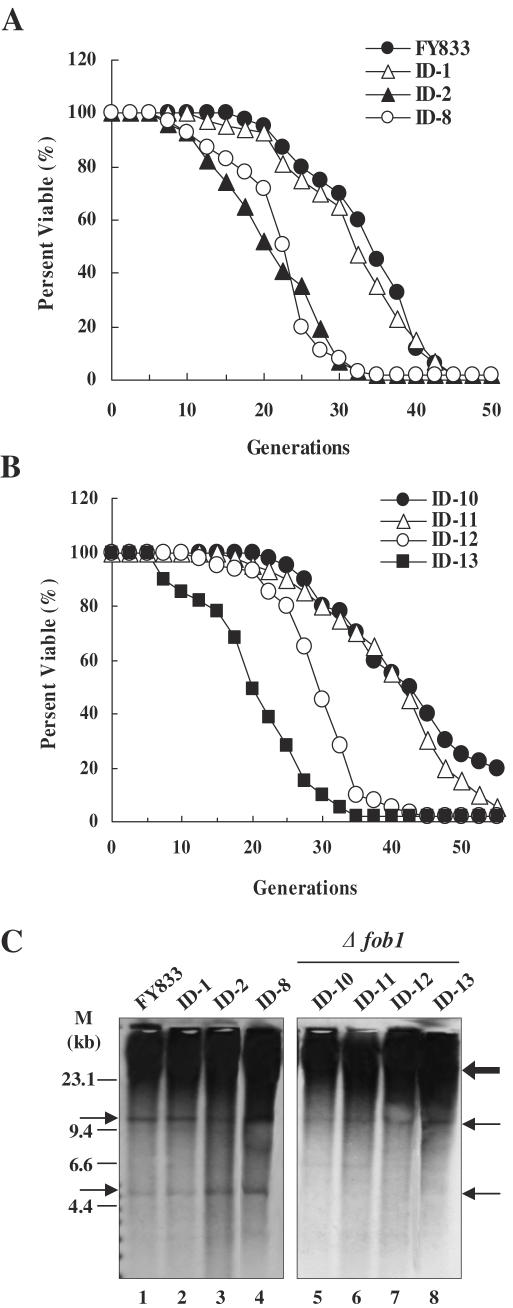
To investigate the mechanism underlying the reduction in the size of the rDNA chromosome, an rDNA chromosome was created in a fob1 background (ID-10) by splitting chromosome XII in the right-hand region of the rDNA cluster, resulting in strain ID-11 (Figure 3A), and subsequently splitting the left-hand region of the rDNA cluster in ID-11, resulting in strain ID-12. CHEF gel analysis coupled with Southern hybridization indicated that a size reduction in the rDNA chromosome did not occur in strain ID-12 (Figure 3B). When a YCp-type plasmid harboring FOB1 (p3151) was introduced into the fob1 strain ID-12 (designated ID-13), the size of the rDNA cluster was found to decrease (Figure 3C), indicating that the decrease was caused by FOB1-dependent recombination within the rDNA cluster. To confirm this, genomic DNA was digested at a unique KpnI site in the rDNA unit and subjected to electrophoresis followed by Southern hybridization using 5S rDNA as a probe. As shown in Figure 3D, the copy number of the rDNA cluster in strains ID-8, ID-9 and ID-13 which exhibited a decrease in the size of the rDNA cluster is significantly reduced relative to that in other strains that did not undergo a change in rDNA size. These results indicate that the decrease in the size of rDNA cluster was indeed caused by the decrease in rDNA copy number. However, it should be noted that rDNA copy number differs among cells and during growth because the hybridization signal corresponding to the rDNA chromosome was broad.
Figure 3.
Analysis of split-chromosomes in the fob1 mutants by CHEF gel electrophoresis and Southern hybridization (A–C). Chromosomal DNA was isolated from the following strains: (A) ID-11, (B) ID-12 and (C) ID-13. (D) Analysis of rDNA copy numbers. Genomic DNA was digested at a unique KpnI site in the rDNA unit and subjected to electrophoresis followed by Southern hybridization using 5S rDNA as a probe. A single copy gene was used as an internal control for normalization. Because the normalization factor for the 5S rDNA probe was found to be 15 relative to the single copy gene, rDNA copy numbers were determined by multiplying by 15. (E) Relative quantification of 18S rRNA using real-time RT–PCR. The 18S rRNA amount is divided by the ACT1 amount to obtain a normalized 18S rRNA value and the normalized amount of 18S rRNA in FY833 (WT) was used to compare the relative amount of 18S rRNA in different strains.
We also determined the quantity of rRNA in the ID-8, ID-9 and ID-13 strains in which rDNA copy number was decreased by real-time quantitative PCR. Result revealed that quantity of rRNA was indistinguishable from that in the ID-1, ID-2 and wild-type strains in spite of decreased rDNA copy number (Figure 3E). We suppose that transcription of rDNA by Pol I may be increased according to decrease of rDNA copy number and consequently the synthesis of rRNA may be adjusted. In other words, artificially constructed rDNA chromosome was also normally regulated by Pol I-transcription machinery.
Growth rate decreases in the strain containing the rDNA chromosome
To determine the physiological effect of moving the rDNA cluster from its original position in chromosome XII, we next examined specific growth rate (µ) of strains having split-chromosomes. Growth of FY833 (wild-type strain), ID-1, ID-2, ID-8 and ID-9 was monitored by observing optical densities of the cultures at OD660, and specific growth rates were calculated (Table 3). The results reveal that the strains harboring the rDNA chromosome, ID-8 and ID-9, exhibited a significantly slower growth rate relative to the parental strain (FY833), and ID-1 and ID-2. Widianto et al. (34) reported that the growth rates of haploid strains having 17, 18, 19, 20 and 21 chromosomes were indistinguishable from that of a wild-type haploid having 16 chromosomes. It has also been reported that a hyper-recombination mutant which displays extremely high rates of intrachromosomal recombination has a reduced growth rate (35). Therefore, we presume that the recombination rate within the rDNA cluster may increase due to removal of the cluster from chromosome XII. We examined this possibility by determining specific growth rates in the fob1 mutants, ID-10 (fob1), ID-11 (fob1), ID-12 (fob1) and found that they were not significantly different from that of wild type. When FOB1 was introduced into strain ID-12, the specific growth rate of the resultant strain (ID-13) was found to decrease (Table 3). In addition, ∼10% marker loss (71 Ura− colonies/674 total colonies tested and 94 Ura− colonies/939 total colonies tested) within the rDNA cluster was observed in strains ID-19 (constructed by integrating URA3 into RND18 of ID-2) and ID-20 (constructed by integrating URA3 into RND18 of ID-8), respectively. This suggests that the decreased specific growth rate is at least partly caused by hyper-recombination within the rDNA cluster removed from its original position on chromosome XII, but not due to an increased number of chromosomes. However, we suggest that this is not the only mechanism because strain ID-2 displaying hyper-recombination exhibited similar growth rate to that of wild-type strain FY833. In addition sir2 mutants with hyper-recombination of rDNA cluster is known to grow normally (11). We presume that another possible mechanism is that rDNA chromosome might be lost at relatively high frequency during mitotic division which leads to cell death. This was revealed by measuring the frequency of loss of rDNA chromosome, 6% (58 Trp− colonies/970 total colonies tested), observed in diploid made by crossing ID-8 with W303-1B, whereas Trp− colonies was not observed in diploid constructed by crossing ID-1 with W303-1B as control strain.
Table 3.
Specific growth rates (µ) of yeast strains having different split-chromosomes
| Strains | Split chromosomes (Chr comosome XII) | Specific growth rate (h−1) | |
|---|---|---|---|
| FOB1 | FY833 | 2500 kb | 0.46 ± 0.04 |
| ID-1 | 1900 and 610 kb | 0.44 ± 0.02 | |
| ID-2 | 450 and 2050 kb | 0.49 ± 0.05 | |
| ID-8 | 450, 350 kb (solely rDNA cluster) and 610 kb | 0.34 ± 0.02 | |
| ID-9 | 450 kb, 700 kb (solely rDNA cluster) and 610 kb | 0.36 ± 0.04 | |
| Δfob1 | ID-10 | 2500 kb | 0.47 ± 0.05 |
| ID-11 | 1900 and 610 kb | 0.48 ± 0.03 | |
| ID-12 | 450 kb, 1500 kb (solely rDNA cluster) and 610 kb | 0.42 ± 0.03 | |
| ID-13 (YCp-FOB1) | 450 kb, 730 kb (solely rDNA cluster) and 610 kb | 0.37 ± 0.02 |
Silencing and localization of the nucleolus is affected by structural alteration of the rDNA cluster
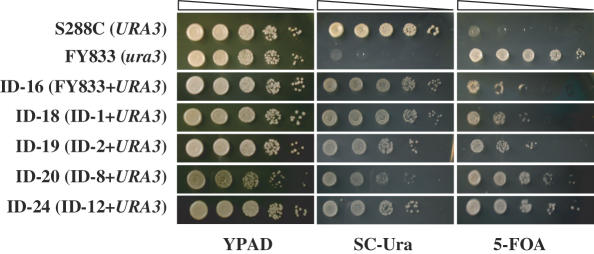
It is known that in S.cerevisiae, gene silencing is observed at telomeres, at HML and HMR loci, and in the rDNA cluster region (19,36,37). As an example of the latter, transcription of Pol II-dependent reporter genes is silenced if inserted within the rDNA locus. To examine the effect of structural alterations of chromosome XII on silencing, the URA3 gene was integrated into the 18S rDNA region of strains FY833 (WT), ID-1 (harboring 610 and 1900 kb split chromosomes), ID-2 (harboring 450 and 2050 kb split chromosomes), ID-8 (harboring 450, 610 and 350 kb split chromosomes, the latter consisting solely of the rDNA cluster) and ID-12 (fob1 mutant harboring 450, 610 and 1500 kb split chromosomes, the latter consisting solely of the rDNA cluster), generating ID-16, ID-18, ID-19, ID-20 and ID-24, respectively. Silencing of URA3 was evaluated qualitatively as a decrease in survival on medium lacking uracil and an increase in survival on 5-FOA medium (Figure 4). In SC-Ura, the growth of strains ID-16, ID-18 and ID-19 was reduced compared with strain S288C (URA3+). Growth in the strain containing the rDNA chromosome, ID-20, was reduced to an even greater extent. In addition, strains ID-20 and ID-24 exhibited better growth than strain ID-16, ID-18 and ID-19 on 5-FOA medium. These results indicate that silencing of URA3 inserted within the rDNA locus was enhanced in the strains containing the rDNA chromosome compared with wild type (ID-16) and strains ID-18 and ID-19.
Figure 4.
Silencing level in various strains containing split-chromosomes. Each strain was grown in YPAD to an OD600 = 1.5 and 3 µl each of 10-fold serial dilutions were spotted on YPAD, SC-Ura and 5-FOA media to determine silencing of URA3 expression at the rDNA locus.
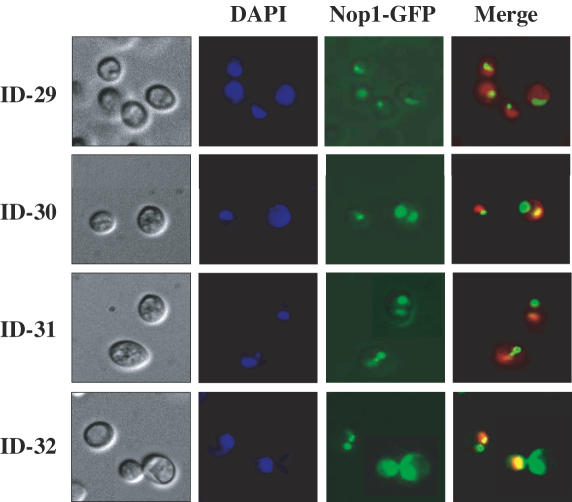
The nucleolus is the site of rDNA transcription by Pol I, processing of rRNA transcripts, and assembly of ribosomes and is a crescent-shaped structure that makes extensive contact with the nuclear envelope. It was reported that the localization and morphology of the nucleolus are affected by structural modification of the rDNA cluster (38). A strain carrying a plasmid with a single rDNA unit transcribed by Pol I was observed to contain a fragmented nucleolus distributed throughout the nucleus (18). To examine the morphology and localization of the nucleolus in strains FY833, ID-1, ID-2 and ID-8 carrying split-chromosomes, plasmid p3150 expressing a GFP-tagged Nop1p, a nucleolar marker protein, was introduced into each, resulting in strains ID-29, ID-30, ID-31 and ID-32, respectively. The results of this analysis are shown in Figure 5. Nop1p was localized at the nuclear periphery in the form of a typical crescent-shaped nucleolar structure in the control strain ID-29. In contrast, a single and occasionally (6∼12% frequency) two foci for Nop1p and a rounded nucleolus were seen in strains ID-30, ID-31 and ID-32, while only one focus for Nop1p observed in strain ID-29, suggesting that morphology and localization of the nucleolus are affected by splitting chromosome XII itself or by release of the rDNA cluster from chromosome XII.
Figure 5.
Analysis of morphology and localization of the nucleolus in various strains in which chromosome XII was split. Strains ID-29, ID-30, ID-31 and ID-32 were analyzed for localization of the nucleolar protein Nop1p. The frequency in which Nop1p appears in two foci in strains ID-30, ID-31 and ID-32 was 6.5, 6.4 and 12.6%, respectively.
Life span is reduced by splitting of the 450 kb region from left end of chromosome XII
In S.cerevisiae, hyper-recombination in the rDNA cluster has been demonstrated to be a cause of aging in mother cells. In old mother cells which display a shortened life span, DNA circles containing a variable number of rDNA repeats (ERC) arise, suggesting that the ERCs are a cause of aging (15). A fob1 mutation was reported to result in a decreased rDNA recombination rate leading to a decreased amount of ERCs, and consequently an increased life span (16). The average life span of strains FY833 and ID-1 (harboring 610 and 1900 kb split chromosomes) was 34.1 and 31.9 generations, respectively, while that of strains ID-2 (harboring 450 and 2050 kb split chromosomes) and ID-8 (harboring 450, 610 and 350 kb split chromosomes, the latter consisting solely of the rDNA cluster) was 20.6 and 22.7 generations, respectively (Figure 6A). On the other hand, average life span of the fob1 mutants (ID-10, ID-11 and ID-12), in which the rDNA copy number was fixed, was 42.5, 41.2 and 30.1 generations, respectively. However, it is noted that deletion of fob1 does not completely rescue the life span effects, suggesting that this is a fob1-independent life span shortening effect of splitting the chromosomes. When FOB1 was introduced into strain ID-12, the life span of the resultant strain (ID-13) was reduced again (19.9 generation) (Figure 6B), indicating that the strains harboring split chromosomes (450 kb) consisting of the left region of chromosome XII or/and only of the rDNA cluster (1500 kb) have a shorter life span than the other strains including wild-type. To explore whether splitting of the 450 kb region from the left end of chromosome XII was responsible for the decreased life span, we analyzed the life span of strains in which chromosome XII was split 193 kb (nucleotide position 193 850) or 330 kb (nucleotide position 329 560) from the left end, respectively. The life span of these strains was indistinguishable from that of the wild-type strain (data not shown).
Figure 6.
Life span of various strains containing spilt-chromosomes. (A) Average life spans for FY833 (WT), ID-1, ID-2 and ID-8 strains were 34.1, 31.9, 20.6 and 22.7 generations, respectively. (B) Average life spans for the fob1 mutants, ID-10, ID-11, ID-12 and ID-13 were 42.5, 41.2, 30.1 and 19.9 generations, respectively. (C) Detection of the ERCs. DNA isolated from various strains was subjected to electrophoresis (0.6% agarose gel for 30 h at 1 V/cm) followed by Southern hybridization using 5S rDNA as a probe. Thin arrows represent ERCs and thick arrow indicates hybridization signal of the genomic rDNA copies. M; Lambda DNA/HindIII marker.
Because strains ID-2 and ID-8, but not strain ID-1, displayed a decrease in life span, we examined whether this phenotype could be attributed to an abundance of ERCs by estimating ERC accumulation (Figure 6C). In strains ID-2 (lane 3) and ID-8 (lane 4), more ERCs accumulated compared with wild-type (FY833, lane 1) and ID-1 (lane 2). In the fob1 mutants, ERC accumulation was not observed, as expected (Figure 6C, lanes 5–8) as deletion of FOB1 suppresses blocking of the replication fork and rDNA recombination. All of these observations suggest that splitting of the 450 kb region from the left end of chromosome XII promotes hyper-recombination within the rDNA cluster and that this hyper-recombination affects life span through accumulation of ERCs.
DISCUSSION
While rRNA genes are transcribed with high efficiency in order to keep pace with metabolic activity (39), the rate of rRNA synthesis is regulated in response to environmental conditions. During exponential growth, about half of the rRNA genes are transcribed by Pol I (40). French et al. (41) reported that rRNA synthesis rates were approximately the same in a strain with a typical number of rRNA genes (143 copies in this case) and in a strain with a reduced number (42 copies), but which grew as well as the control strains. However, the average number of Pol I molecules per gene was found to be higher in the strain with the reduced number of rRNA genes. In the present study, we demonstrate that rDNA copy number fluctuated and decreased when the rDNA cluster consisting of ∼150 tandem repeats was removed from its original position on chromosome XII (Figure 3D). Nonetheless, the quantity of rRNA in the ID-8 and ID-9 strains in which rDNA copy number was reduced was indistinguishable from that in the ID-1, ID-2 and wild-type strains as determined by real-time RT-PCR (Figure 3E). Therefore, we suggest that flanking upstream and/or downstream regions of the rDNA cluster may be necessary for maintenance of normal rDNA copy number, ∼150, and that fluctuation of copy number may occur if an unusual chromosome structure such as an rDNA-only chromosome arises. It is also possible that yeast regulates the rate of rRNA synthesis from the rDNA genes even on an rDNA-only chromosome in response to copy number fluctuation.
In the strain harboring an unusual chromosome consisting solely of the rDNA cluster, silencing of URA3 inserted within the rDNA locus was found to increase relative to the wild-type strain and strains harboring a split chromosome consisting of the upstream or downstream region of chromosome XII (Figure 4). Transcriptional silencing in S.cerevisiae occurs at the silent mating-type loci HML and HMR, and at telomeres mediated by the SIR complex consisting of Sir2p, Sir3p and Sir4p (42). Sir2p, but not Sir3p or Sir4p, is required for silencing in the rDNA cluster region (21). Smith et al. (43) have shown that alterations in SIR2 gene dosage dramatically affect the level of rDNA silencing, suggesting that a limiting pool of Sir2p is distributed between telomeres, HM loci and rDNA. Therefore, we presume that the increase in silencing may have resulted from a greater localized concentration of Sir2p at the rDNA cluster. One possible mechanism for this is that more telomere-associated Sir2p moved to the rDNA. In this regard, a telomeric repeat sequence, 5′-(C4A2)6-3′ was artificially added to the split rDNA chromosome. Therefore, we presume that telomere length could be shortened due to this artificial telomere seeding. If such is the case, some telomere-associated Sir2p may have translocated to the rDNA cluster, increasing the amount of Sir2p available for rDNA silencing. We examined this possibility by determining telomere length in the FY833, ID-1, ID-2, ID-8 and ID-12 strains. However, telomere length was found to be the same in all strains (data not shown), suggesting that the increased silencing in the rDNA locus was not caused by an increased amount of Sir2p in the rDNA cluster region through shortened telomere length. The fact that both silencing and rDNA recombination increased in strains ID-8 and ID-12 suggests that these are Sir2-independent events because increased Sir2 activity at the rDNA locus is expected to increase silencing, but decrease recombination. Another possibility is that an unusual rDNA structure arose as a result of the splitting, and this unusual structure affects silencing. Because the rRNA content in the strain harboring the rDNA chromosome did not change significantly, in spite of the decrease in rDNA copy number, we presume that Pol I in the strain harboring the rDNA chromosome was able to transcribe rDNA genes more efficiently in order to meet metabolic demand. If so, increased transcription of rDNA by Pol I may prevent access of Pol II machinery, resulting in an increase in silencing. Cioci et al. (44) reported that silencing of reporters is much stronger in a mutant with ∼25 rDNA copies, all of which are transcriptionally active, than in a control strain with ∼190 copies, and that rDNA chromatin structure that favors Pol I transcription of rRNA genes decreases or silences expression of a Pol II reporter. Therefore, we presume that increased silencing may be a consequence of decreased copy number of rDNA generated by release of the rDNA cluster from chromosome XII, but not by removal per se of the rDNA cluster.
Strains ID-2 and ID-8 were found to have a shorter life span than the other strains and ERCs were found to accumulate in them (Figure 6A and B). As a result of splitting chromosome XII, the total number of chromosomes increased from 16 in wild type to 17 in ID-2 and 18 in ID-8. An increase in chromosome number may result in a shorter life span. Thus, we examined the effect of an increased number of chromosomes on life span in strain YMH471 (34) harboring 21 chromosomes. The life span of this strain was found to be no different than that of wild type (data not shown). It has been established that trans-acting factors such as Fob1p and Sir2p are important for regulating copy number of rDNA, silencing and aging (16,17,22,30).
Michel et al. (45) reported that short rDNA clones that arose from a single-step deletion event in a chromosome with normal rDNA length caused an increase in telomeric and mating-type gene silencing. They also reported that short rDNA strains (∼135 copies) display normal rDNA silencing and a life span indistinguishable from that of wild type. In contrast, in the present study, when the rDNA cluster consisting of ∼150 tandem repeats was removed from its original position on chromosome XII, rDNA copy number decreased and the resulting short rDNA strains (54∼77 copies) displayed a shortened life span and an increase in transcriptional silencing in the rDNA cluster region. These contradictory results may be due to significant differences related to the context of the chromosome harboring the shortened rDNA cluster.
Our results suggest that the context of chromosome XII itself also influences a variety of rDNA-related phenotypes. The unique chromosomes described in this study in which the rDNA cluster region has been precisely manipulated should be useful reagents for evaluating rDNA-dependent cellular processes.
Acknowledgments
This study was partially supported by the Ministry of Education, Culture, Sports, Science and Technology, a Grant-in-Aid for Scientific Research B, 13580064, 2003 to 2005, and was carried out as a part of The Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers by the Ministry of Economy, Trade & Industry (METI), and entrusted by the New Energy and Industrial Technology Development Organization (NEDO).
Conflict of interest statement. None declared.
REFERENCES
- 1.Sucgang R., Chen G., Liu W., Lindsay R., Lu J., Muzny D., Shaulsky G., Loomis W., Gibbs R., Kuspa A. Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res. 2003;31:2361–2368. doi: 10.1093/nar/gkg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petes T.D. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl Acad. Sci. USA. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer B.J., Fangman W.L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 4.Campbell J.L., Newlon C.S. Chromosomal DNA replication. In: Broach J.R., Pringle R., Jones E.W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 41–146. [Google Scholar]
- 5.Linskens M.H., Huberman J.A. Organization of replication of ribosomal DNA in Saccharomyces. Mol. Cell. Biol. 1988;11:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warner J.R. The economics of ribosomal biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 7.Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 8.Landry J., Sutton A., Tafrov S.T., Heller R.C., Stebbins J., Pillus L., Sternglanz R. The silencing protein Sir2 and its homologs are NAD-dependent deacetylases. Proc. Natl Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith J.S., Brachmann C.B., Celic I., Kenna M.A., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl Acad. Sci. USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benguria A., Hernandez P., Krimer D.B., Schvartzman J.B. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:893–898. doi: 10.1093/nar/gkg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb S., Esposito R.E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T., Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi T., Heck D.J., Nomura M., Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinclair D.A., Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 16.Defossez P.A., Prusty R., Kaeberlein M., Lin S.J., Ferrigno P., Silver P.A., Keil R.L., Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 17.Kaeberlein M., McVey M., Guarente M. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oakes M., Aris J.P., Brockenbrough J.S., Wai H., Vu L., Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryk M., Banerjee M., Murphy M., Knudsen K.E., Garfinkel D.J., Curcio M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 20.Fritze C.E., Verschueren K., Strich R., Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith J.S., Boeke J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 22.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 23.Winston F., Dollard C., Ricupero-Hovasse S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 24.Sugiyama M., Ikushima S., Nakazawa T., Kaneko Y., Harashima S. PCR-mediated repeated chromosome splitting in Saccharomyces cerevisiae. BioTechniques. 2005;38:909–914. doi: 10.2144/05386RR01. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki T., Toh-e A., Kikuchi Y. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol. 2000;20:7971–7979. doi: 10.1128/mcb.20.21.7971-7979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y.H., Kaneko Y., Fukui K., Kobayashi A., Harashima S. A yeast artificial chromosome-splitting vector designed for precise manipulation of specific plant chromosome region. J. Biosc. Bioeng. 2005;99:55–60. doi: 10.1263/jbb.99.55. [DOI] [PubMed] [Google Scholar]
- 27.Burke D., Dawson D., Stearns T. Methods In Yeast Genetics. Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 174–180. [Google Scholar]
- 28.Gietz R.D., Schiestl R.H. Transforming yeast with DNA. Methods Mol. Cell. Biol. 1995;5:255–269. [Google Scholar]
- 29.Kim Y.H., Sugiyama M., Yamagishi K., Kaneko Y., Fukui K., Kobayashi A., Harashima S. A versatile and general splitting technology for generating targeted YAC subclones. Appl. Microbiol. Biotechnol. 2005;69:65–70. doi: 10.1007/s00253-005-1970-x. [DOI] [PubMed] [Google Scholar]
- 30.Huang J., Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy B.K., Austriaco N.R., Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke D., Dawson D., Stearns T. A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. Methods In Yeast Genetics; pp. 110–111. [Google Scholar]
- 33.Johzuka K., Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S.cerevisiae. Genes Cells. 2002;7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 34.Widianto D., Yamamoto E., Sugiyama M., Mukai Y., Kaneko Y., Oshima Y., Nishizawa M., Harashima S. Creating a Saccharomyces cerevisiae haploid strain having 21 chromosomes. J. Biosci. Bioeng. 2003;95:89–94. doi: 10.1016/S1389-1723(03)80154-8. [DOI] [PubMed] [Google Scholar]
- 35.Aguilera A., Klein H.L. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol. Cell. Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rine J., Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. Position effect at S.cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 38.Oakes M.L., Siddiqi I., Vu L., Aris J., Nomura M. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol. 1999;19:8559–8569. doi: 10.1128/mcb.19.12.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 40.Meier A., Thoma F. RNA polymerase i transcription factors in active yeast rRNA gene promoters enhance UV damage formation and inhibit repair. Mol. Cell. Biol. 2005;25:1586–1595. doi: 10.1128/MCB.25.5.1586-1595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.French S.L., Osheim Y.N., Cioci F., Nomura M., Beyer A.L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aparicio O.M., Billington B.L., Gottschling D.E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S.cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 43.Smith J.S., Brachmann C.B., Pillus L., Boeke J.D. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cioci F., Vu L., Eliason K., Oakes M., Siddiqi I.N., Nomura M. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell. 2003;12:135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- 45.Michel A.H., Kornmann B., Dubrana K., Shore D. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 2005;19:1199–1210. doi: 10.1101/gad.340205. [DOI] [PMC free article] [PubMed] [Google Scholar]