Abstract
The addition of poly(A)-tails to RNA is a process common to almost all organisms. In eukaryotes, stable poly(A)-tails, important for mRNA stability and translation initiation, are added to the 3′ ends of most nuclear-encoded mRNAs, but not to rRNAs. Contrarily, in prokaryotes and organelles, polyadenylation stimulates RNA degradation. Recently, polyadenylation of nuclear-encoded transcripts in yeast was reported to promote RNA degradation, demonstrating that polyadenylation can play a double-edged role for RNA of nuclear origin. Here we asked whether in human cells ribosomal RNA can undergo polyadenylation. Using both molecular and bioinformatic approaches, we detected non-abundant polyadenylated transcripts of the 18S and 28S rRNAs. Interestingly, many of the post-transcriptionally added tails were composed of heteropolymeric poly(A)-rich sequences containing the other nucleotides in addition to adenosine. These polyadenylated RNA fragments are most likely degradation intermediates, as primer extension (PE) analysis revealed the presence of distal fragmented molecules, some of which matched the polyadenylation sites of the proximal cleavage products revealed by oligo(dT) and circled RT–PCR. These results suggest the presence of a mechanism to degrade ribosomal RNAs in human cells, that possibly initiates with endonucleolytic cleavages and involves the addition of poly(A) or poly(A)-rich tails to truncated transcripts, similar to that which operates in prokaryotes and organelles.
INTRODUCTION
Polyadenylation is an important post-transcriptional modification of prokaryotic, eukaryotic and organellar RNA. In bacteria, archaea and organelles, such as plant mitochondria and chloroplasts, polyadenylation is transient and occurs mainly on fragmented molecules as part of the RNA decay pathway (1–3). In general, this process consists sequentially of endonucleolytic cleavage, addition of degradation-stimulating poly(A) or poly(A)-rich sequences to the proximal cleavage products, and exonucleolytic degradation. In contrast to this form of degradation-stimulating polyadenylation, stable poly(A)-tails are added to the mature 3′ ends of most nuclear-encoded mRNAs and are important for proper translation initiation, mRNA stability and, at least in some cases, nuclear export (4–7). However, the categorization described above is not definitive since in animal mitochondria, RNA molecules are characterized with stable poly(A)-tails that are post-transcritionally added to their mature 3′ ends, akin to the case described for nuclear-encoded RNA, although their complete function remains unknown (8). Moreover, recent data have revealed that in addition to the full-length, stably polyadenylated transcripts described above, non-abundant, polyadenylated RNA fragments are also present in human mitochondria (9). The coexistence of non-abundant truncated polyadenylated transcripts together with those characterized with stable poly(A)-tails was also described for trypanosome mitochondria (10). Furthermore, a quality control mechanism, including transient polyadenylation which targets nuclear-encoded yeast RNA for degradation by the exosome complex, has recently been described (11–14). In these studies, in yeast cells in which the exosome activity was abolished, fragmented polyadenylated transcripts from intergenic regions of the nuclear genome accumulated. Transient polyadenylation of improperly folded tRNA molecules was witnessed as well. These accumulated observations suggest that polyadenylation-stimulated RNA degradation is a common process which occurs in most of the life kingdoms including bacteria, some archaea, chloroplasts, plant and animal mitochondria and nuclear-encoded transcripts (3). Whether or not the somewhat paradoxical coexistence of stable and unstable polyadenylation in the same system, as described above, occurs for nuclear-encoded RNA of higher eukaryotes, such as homo sapiens, is yet unknown. Recently, however, the accumulation of β-globin pre-mRNAs containing short A tails in the absence of the exosome has been reported in human cells (15).
Here we demonstrate, using molecular and bioinformatic tools, the presence of non-abundant polyadenylated transcripts polyadenylated at sites corresponding to both fragmented and full-length 18S and 28S rRNA in human cells. Surprisingly, a significant number of the post-transcriptionally added tails were not exclusively composed of adenosines but included the other nucleotides as well, similar to the degradation-stimulating tails previously observed in bacteria, organelles and archaea. Together, our results suggest the presence of a polyadenylation-stimulated degradation mechanism for human ribosomal RNA and reveal, for the first time, post-transcriptionally added extensions of heteropolymeric nature in human cells.
MATERIALS AND METHODS
Cells
The cancer cell lines used in this work were CCRF-CEM T-cell leukemia wt, MCF-7 epidermal breast cancer and CCRF-MTA-C3 cells (16). Primary human dermal fibroblasts were isolated from adult skin and cultivated as described previously (17).
Accession numbers
Numbering of the nucleotides of the 28S rRNA is according to the sequence which appears in the NCBI GenBank accession no, M11167. For 18S, the sequence that was used was that which appears in the ‘Human ribosomal DNA complete repeating unit’, sequence U13369.
RNA purification, oligo(dT) RT–PCR and circularized RNA RT–PCR
RNA isolation was performed using the Invisorb Spin cell-RNA Mini kit (Invitek Inc.). Oligo(dT) primed RT–PCR was performed as described previously (9). Briefly, an adaptor-dT16 oligonucleotide was used to prime the RT reaction which was applied to total RNA and the resulting cDNA was PCR amplified using the adaptor as a reverse primer and one of the gene specific forward primers shown in Supplementary Table S6. PCR products were then cloned and sequenced. Circularized RNA RT–PCR (cRT–PCR) was performed as described using the primers shown in Supplementary Table S6 (18).
Primer extension (PE) analysis
RNA (15 µg) was annealed to the primer, 28Sp1, shown in Supplementary Table S6. The primers were initially labeled with [32P] using polynucleotide kinase. PE reactions were performed as described previously (19). For the DNA sequencing ladder template, a region of 28S rRNA was PCR amplified using genomic DNA as a template and the product was then cloned into the pGEM-T plasmid (Promega Inc.). DNA sequencing was performed with the Sequenase kit (Amersham Inc.) using the same primer as for the PE. Both the sequencing and PE products were resolved by 7% denaturing PAGE and autoradiography. For quantification of the truncated 28S rRNA molecules relative to the amount of full-length 28S transcripts, an additional PE reaction was performed using the primer, 28Sp5, (shown in Supplementary Table S6) located 99 nt downstream from the mature 5′ end. A serial dilution of the resulting product was analyzed on the same gel.
The PolyAfinder tool
Originally, we established the PolyAfinder tool, (http://bioinfo.cs.technion.ac.il/polyafinder/), in order to search the human EST database for ESTs originating from polyadenylated human mitochondrial RNA (9). For the present work, an improved version of the tool, using a completely different algorithm, was employed. Briefly, PolyAfinder is supplied with an EST database, in this case, the non-redundant NCBI human EST database, depleted of any ESTs which do not contain a poly(A) region of minimal length (initially determined by the user). The analyzed gene sequence is aligned with the established data base using the BLAST algorithm. Within each retrieved EST, PolyAfinder searches for a poly(A) tail near the end of the alignment and ESTs positive for this screening stage are collected. This pool then undergoes further filtering criteria to ensure that only ESTs corresponding to polyadenylated RNA of the analyzed gene remain. One of the advantages of the new version of PolyAfinder is that ESTs containing heteropolymeric poly(A)-rich tails, and not only those harboring tails comprised exclusively of adenosines, can be detected as well. Although the recorded ESTs pass many stages of automatic screening, for this work, each of the detected ESTs was verified by manual examination, to ensure stringent homology to the rRNA genes. Furthermore, all heteropolymeric tail sequences were manually analyzed by BLAST to determine that they indeed result from random post-transcriptional nucleotide addition and do not originate from known genomic sequences, by lack of any such corresponding accessions in the non-redundant NCBI database. A detailed description of the new version, as well as the differences between it and the previous version, are listed in the website and described in the Supplementary Data.
RESULTS
Detection of non-abundant, internally polyadenylated rRNA transcripts by oligo(dT) primed RT–PCR
Following the observation that polyadenylation-stimulated RNA degradation pathways are present in almost every organism that has been analyzed, including bacteria, organelles and the yeast nucleus, we asked whether such a mechanism takes part in the degradation of nuclear-encoded RNA in human cells. Detection of truncated polyadenylated RNA molecules, alternatively known as internal polyadenylation [as opposed to mature 3′ end poly(A)] has served as a preliminary tell-tale sign of the presence of such mechanisms in past studies and therefore was our initial goal in this work. Ribosomal RNA was chosen for this analysis since we assumed that, as in bacteria, organelles and yeast nuclei, the partially digested polyadenylated transcripts are rapidly degraded, leaving only a minute amount of these molecules present in the cell at any point in time. Therefore, detection of such non-abundant molecules requires extensive PCR screening stages and the comparatively high level of rRNA molecules, relative to that of mRNAs, would increase the probability of detecting these truncated polyadenylated transcripts. Another reason for choosing to analyze ribosomal RNA is that mature 3′ end poly(A)-tails, a quality nuclear-encoded rRNA is known to be lacking of, have been shown to hamper isolation of the much less abundant internally polyadenylated transcripts by competing with the oligo(dT) primer (9).
Several sets of oligonucleotide primers were designed for oligo(dT) primed RT–PCR analysis of the 28S and 18S rRNAs, purified from either human cancer cell lines or primary fibroblasts. This approach was indeed successful in the isolation and cloning of tens of truncated polyadenylated rRNA molecules from the 18S and 28S rRNAs (Figure 1 and Supplementary Tables S3 and S4). It should be noted that total cellular rRNA, including both the precursor transcripts located in the nucleus and the mature rRNA located in the cytoplasm, was analyzed here. Therefore, the identified truncated polyadenylated transcripts could have initiated from either the precursor, mature transcript or both types of rRNAs. Despite the dominant amount of rRNA full-length transcripts in the cell, the detection of these truncated polyadenylated molecules required more PCR amplification cycles and selection/screening stages compared to similar analysis of prokaryotes and organelles in past studies. This indicates that the truncated and polyadenylated molecules are present at very low levels in the cell.
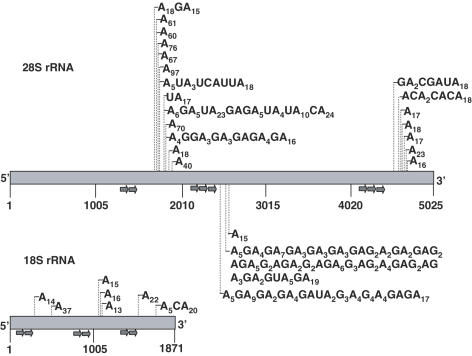
Figure 1.
Detection of 28S and 18S rRNA transcripts containing homopolymeric and heteropolymeric poly(A)-tails using oligo(dT) primed RT–PCR. The 28S and 18S rRNAs are schematically presented. The gene specific forward primers used for the PCR amplification and screening of the oligo(dT)-primed cDNAs are shown as arrows below the gene. Thin vertical lines indicate the positions of the added poly(A)-tail of each clone and the tail compositions are shown. The exact positions of the polyadenylated nucleotides, according to the gene sequence, are presented in the Supplementary Tables S3 and S4.
Some of the post-transcriptionally added tails contain heteropolymeric poly(A)-rich sequences
For the 18S rRNA, six of the seven obtained clones had homopolymeric poly(A) extensions and one clone contained a single C in the poly(A) tail (Figure 1). However, for the 28S rRNA, nine of the 23 clones harbored tails containing the other nucleotides in addition to adenosine (Figure 1). Using the BLAST algorithm, we manually searched the non-redundant NCBI database for any accessions containing these particular heteropolymeric sequences in order to ensure that they are indeed the result of random post-transcriptional addition. No such entries were disclosed. Such heteropolymeric extensions resemble the post-transcriptionally added tails produced by the enzyme, PNPase, in bacteria and organelles, which is known to be a major player in the poly(A)-stimulated RNA degradation pathway (19–22). In addition, it was recently found that heteropolymeric tails are produced by the archaeal exosome complex in hyperthermophilic archaea (18).
ESTs of polyadenylated rRNA transcripts
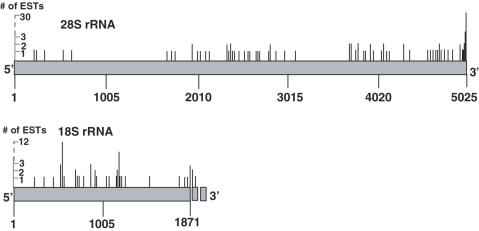
The human EST database includes approximately six million entries (November 2005), many of which were produced by oligo(dT) priming. We assumed that if fragmented polyadenylated rRNAs exist, their corresponding ESTs could be found among these accessions. We have previously described the ‘PolyAfinder’ tool that was developed and successfully applied in the detection of ESTs related to fragmented and polyadenylated transcripts of human mitochondrial RNA (9). Here, an improved and modified version of this bioinformatic tool, as described in the Materials and Methods and Supplementary Data, was applied. Aside from the significant improvement in calculation time and other computational parameters, the tool was modified to enable detection of truncated or full-length RNA with tails of heteropolymeric nature, as such molecules were revealed by the oligo(dT) RT–PCR analysis. Indeed, PolyAfinder succeeded in finding numerous ESTs originating from both the 18S and 28S rRNAs. For the 28S rRNA alone, 112 polyadenylated ESTs divided among 38 poly(A) sites along the gene sequence were found (Figure 2). Of these ESTs, 64 contained heteropolymeric poly(A)-rich tails while the rest had extensions made completely of adenosine residues (Tables 1 and 2 and Supplementary Tables S1 and S2). As with the oligo(dT) RT–PCR analysis, not only were the resulting EST sequences manually BLAST-analyzed to determine their positive homology to the original rRNA gene sequence, but each heteropolymeric tail was also checked to ensure negative homology to any accessions existing in the non-redundant NCBI database.
Figure 2.
Detection of ESTs related to polyadenylated 28S and 18S rRNA transcripts. ESTs containing the 28S or 18S rRNAs with poly(A) or poly(A)-rich tails were identified by the bioinformatic tool, PolyAfinder. The points of addition of the poly(A) or poly(A)-rich tails are presented as thin vertical lines, while the length of the line indicates the number of ESTs which were polyadenylated at this location. Examples of the sequences of several heteropolymeric tails are presented in Table 1 and a full list, including the polyadenylation positions according to the gene sequence as well, is presented in Supplementary Tables S1 and S2 of the supplementary Data.
Table 1.
28S rRNA ESTs containing heteropolymeric poly(A)-rich tails
| EST # | EST name | Poly(A) site | Poly(A)-rich tail (5′→3′) |
|---|---|---|---|
| 17 | BF436401 | 2350 | A6GA9GA3GA10GA6UA19 |
| 30 | CA309659 | 2788 | A6G3A7GACAGA2GA15 |
| 49 | CB048936 | 4026 | U2A17UA16UA22 |
| 52 | AA568606 | 4054 | A16GA2GA6GA2GAGA11 |
| 65 | BE874641 | 4715 | UCG2A3GA2CG3UC3AGACAUG2CA4GA11CACA3G2A3CA3GA3GA3GAGA4CA3 GA4UGA4GACGACGAGACGA5CA2G2A5CAG2A2CAGA5CA7GCAG2A6GA3 |
| 70 | CA309999 | 5017 | A12GAGAGA5GA3GA3GA10CA16GA3GA2GAG2AGA2GA2G2A10GA7 |
| 107 | AA737783 | 5024 | A4GA3UAG2A9GA7U2A18 |
Several ESTs corresponding to the 28S rRNA, harboring long heteropolymeric post-transcriptionally added tails, revealed by PolyAfinder are presented. Each of these sequences was manually analyzed to ensure that they are indeed randomly added nucleotides and therefore are not present as a sequence in the non-redundant genomic DNA or cDNA NCBI database. The full EST list is presented in Supplementary Table S1 and the numbering of the ETSs presented here is according to this table.
Table 2.
Fraction of homopolymeric and heteropolymeric tails detected in this work
| Gene | Method | Homopolymeric | Heteropolymeric | No tails |
|---|---|---|---|---|
| 28S rRNA | dT-RT–PCR | 16 | 7 | NA |
| 28S rRNA | ESTs | 66 | 46 | NA |
| 28S rRNA | cRT–PCR | 1 | 3 | 27 |
| 18S rRNA | dT-RT–PCR | 7 | 0 | NA |
| 18S rRNA | ESTs | 44 | 11 | NA |
The number of homopolymeric and heteropolymeric post-transcriptionally added tails, as revealed by the oligo(dT)-RT–PCR (dT-RT–PCR), the poly(A)-finder bioinformatic tool (ESTs) and cyclic RT–PCR (cRT–PCR) are shown. Tails containing only a single non-A nucleotide were counted as homopolymeric and not heteropolymeric because of the possibility of sequencing artifacts. The detailed sequences of the tails are presented in the Supplementary Data. NA: not applicable.
The locations of the poly(A) and poly(A)-rich sites in the EST sequences were spread along the entire length of the 18S and 28S rRNA sequences. Interestingly, for the 28S rRNA, a relatively large number of ESTs with poly(A) sites in close proximity to the mature 3′ end were obtained. As for the mature 3′ end of the 18S rRNA, three ESTs polyadenylated at this position were found. In addition, three more ESTs with poly(A) sites a number of nucleotides downstream from the mature 3′ end (in the transcribed spacer region) were observed (Supplementary Table S2). These ESTs could be related to precursor molecules of the 18S rRNA.
In addition to the occurrence of endonucleolytic cleavage followed by polyadenylation of the proximal cleavage products as described above, we considered the possibility that, transcripts can undergo polyadenylation at points at which 3′→5′ exonucleolytic degradation stalls at strong secondary structures of the rRNA. As the tertiary structure of the human ribosomal complex is yet unknown, we used the predicted hypothesized secondary structure to check for correlation between strong secondary structures and polyadenylation sites detected either by oligo(dT) primed RT–PCR or the PolyAfinder. The result of this analysis is presented in Supplementary Figure S1. Although no significant correlation was found, such a re-adenylation process cannot be ruled out.
These results revealed the presence of truncated polyadenylated human rRNA molecules, therefore suggesting the presence of a poly(A)-stimulated RNA degradation mechanism in this system. Furthermore, together with the tails identified using oligo(dT) RT–PCR, we demonstrated that unlike the poly(A)-tails witnessed in human mitochondria, yeast nuclei and the mature 3′ end of most nuclear-encoded mRNAs, which are comprised exclusively by adenosines, human 28S and 18S rRNA can undergo heteropolymeric polyadenylation (Table 2). To our knowledge, this is the first report of heteropolymeric extensions in human cells.
PE analysis revealed the corresponding 28S rRNA distal fragments
The oligo(dT)-primed RT–PCR and the PolyAfinder bioinformatic analyses identified numerous 28S rRNA molecules of which their polyadenylated 3′ ends were dispersed along the full sequence. In the general scheme of poly(A)-stimulated RNA degradation, the transcript can undergo endonucleolytic cleavage, after which the proximal product is polyadenylated and degraded. Identification of distal cleavage products with 5′ ends adjacent to the 3′ ends of the proximal polyadenylated molecules, mapped by the oligo(dT) RT–PCR technique, could suggest that endonucleolytic cleavage is responsible for their formation. PE is a technique which can demonstrate such complementarity and has been applied for this purpose in previous studies, for instance, to show that fragmented polyadenylated chloroplast RNA molecules isolated from spinach chloroplasts resulted from endonucleolytic cleavage (19). In addition, using this technique, the relative amounts of the degradation intermediates compared to the full-length mature RNA, can be estimated. Here we applied this type of analysis for the region of 1670–1800 nt of the 28S rRNA.
Total RNA purified from several different cancer cell lines, as well as from primary fibroblasts, was annealed to a 32P-labeled oligonucleotide primer located downstream to the region in which the first group of oligo(dT) RT–PCR clones terminated (1670–1800 nt). Following reverse transcription, the products were analyzed alongside a DNA sequencing ladder produced with the same primer (Figure 3). The resulting PE signals showed a similar profile for RNA obtained from each of the four cell types and repeated itself when applied to different RNA preparations. The general grouping of the PE signals corresponded with the 3′ ends of the clones identified by the oligo(dT) RT–PCR assay, while two PE signals perfectly complemented two of the truncated transcripts detected by oligo(dT) RT–PCR (Figure 4).
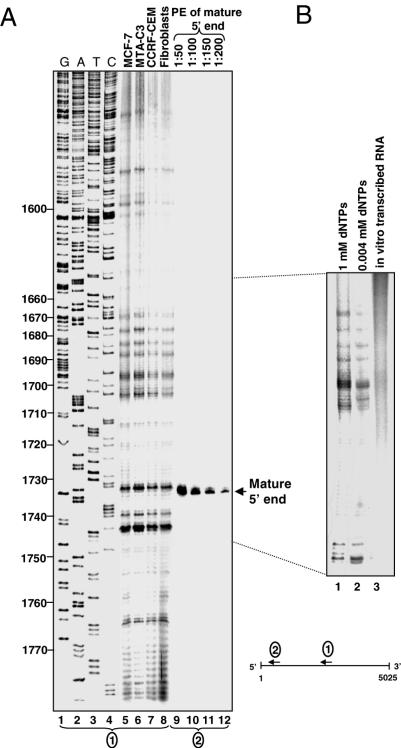
Figure 3.
PE analysis of the 28S rRNA. (A) In an attempt to detect distal cleavage products, total RNA isolated from the cell types indicated at the top of lanes 5–8 was annealed to [32P] primer 28Sp1 and extended with reverse transcriptase. The resulting cDNA was fractionated on 7% denaturing PAGE along with a DNA sequencing ladder generated using a 28S rRNA DNA template and the same primer (lanes 1–4). The nucleotide numbers of the sequence of 28S rRNA are indicated to the left. In order to estimate the amount of the putative truncated transcripts, the mature 5′ end of the molecule was detected by PE with primer 28Sp5, located 99 nt downstream to the mature 28S rRNA 5′ end, using RNA purified from CCRF-CEM cells. Appropriated dilutions of this reaction, as shown at the top of lanes 9–12 were fractionated on the same gel. (B) Control reactions, in which RNA purified from CCRF-CEM cells was analyzed by PE in the presence of dNTPs at concentrations of 1 or 0.004 mM (lanes 1 and 2, respectively) in order to distinguish whether the observed signals were caused by modified nucleotides, are shown. Note that the significant signal located at nucleotide 1733 was not detected in this experiment. In lane 3, in vitro transcribed RNA corresponding to the analyzed region of 28S rRNA, was analyzed by PE to ensure that the signals did not arise from secondary structures that inhibit the reverse transcriptase.
Figure 4.
cRT–PCR analysis detected additional 28S fragments which terminate in the region disclosed by oligo(dT) RT–PCR and PE. The combined results of the analysis of the 28S rRNA for truncated and polyadenylated fragments using cRT–PCR, oligo(dT) RT–PCR (dT-RT–PCR) and PE are schematically presented along the region of 1250–2100 nt. Horizontal lines represent the cDNAs obtained by each technique and are drawn to scale. The primers used for the PCR amplification and screening of the dT-RT–PCR and cRT–PCR cDNA, as well as the PE are shown by arrows. Details about the cRT–PCR clones are displayed in the Supplementary Table S5 and the clone # in the figure is related to that which appears in this table. The defined region of 1670 to 1780 that is enriched with 5′ and 3′ ends of detectable cleavage products is indicated with a gray background. Inset: A diagram explaining the experimental design. Following cleavage, the 28S rRNA is separated to proximal (upstream) and distal (downstream) fragments. PE discloses the 5′ end of the distal fragment while cRT–PCR and oligo(dT) RT–PCR analysis reveal the 3′ end of the proximal fragment, to which the poly(A) tail is added. Arrow heads indicate the primers used and dashed lines the cDNAs produced.
As stated, PE signals can be the result of termination of synthesis by the reverse transcriptase, as it reaches the 5′ end of a fragmented molecule which may be a distal endonucleolytic cleavage product (Figure 4 inset). Alternatively, these signals can result from stalling of the reverse transcriptase at modified nucleotides and/or RNA secondary structures. Indeed, the 28S rRNA is characterized with many modified nucleotides although only one 2′-O-methylated nucleotide was identified in the region disclosed by the PE of this work, at position 1739 (23). Also, aside from this methyl-group, there are no known snoRNAs which are predicted to guide nucleotide methylation in this region (23). In order to confirm that the observed PE signals did not result from RT stalling at methylated nucleotides, the PE analysis was repeated in the presence of reduced concentration of dNTPs. Under this condition, if a PE signal is the result of stalling at a modified nucleotide, its intensity should be enhanced compared to a parallel PE reaction containing a high concentration of dNTPs (23–26). As shown in Figure 3B lanes 1 and 2, no differences in the intensity of the PE signals were obtained, suggesting that these PE signals are not the result of modified nucleotides, rather termination of reverse transcription upon reaching the 5′ end of a fragmented transcript. This was further supported by the fact that the PE signals matched the 3′ end termination points of the cDNAs obtained by the oligo(dT) and circularized RT–PCR (cRT–PCR) methods (described below) (Figure 4).
In order to determine whether the formation of secondary structures inhibits reverse transcription during PE and is, therefore, the cause of the observed PE signals, an RNA corresponding to 1501–1861 nt of the mature 28S molecule was transcribed in vitro using T7 RNA polymerase and the corresponding DNA template. PE analysis of this RNA did not result in the typical signal pattern obtained from the RNA purified from the cells (Figure 3B lane 3). These negative controls suggested that the PE signals resulted mostly from the 5′ ends of fragmented 28S rRNA molecules.
The fragmented 28S rRNA molecules are non-abundant
Assuming that the PE signals indicated fragmented 28S rRNA molecules we intended to determine their relative abundance compared to that of the full-length transcript. To this end, we performed PE with a primer located 99 nt downstream from the 5′ end of the mature 28S molecule. Serial dilutions of the reaction product were electrophorased on the same gel as the other PE experiments (Figure 3A lanes 9–12). Quantification of the autoradiograph revealed that most of the PE signals were below that of 1:100 dilution of the mature 5′ end signal. This result suggested that the fragmented 28S rRNA molecules are indeed non-abundant compared to the mature full-length molecule, most likely because these partially degraded molecules are rapidly digested. This minute amount, compared to the mature full-length transcript, also explained why attempts to detect the fragmented molecules using the ribonuclease protection method failed (data not shown).
cRT–PCR further disclosed fragmented 28S rRNA molecules with 3′ ends located in the examined region
In order to further explore the possibility that the clones isolated by oligo(dT) RT–PCR and the signals disclosed by the PE were indeed distal and proximal endo-cleavage products of the 28S rRNA, respectively, we chose to apply a third method, the cRT–PCR technique. Purified total RNA molecules were circularized by ligation of their 5′ and 3′ ends with T4 RNA ligase, followed by RT–PCR, cloning, PCR screening using the corresponding pairs of primers and finally sequencing (Figure 4 and Supplementary Table S5). This analysis revealed fragmented 28S rRNA molecules with 5′ ends dispersed between 1275–1500 nt and 3′ ends located in the distinct region where the 3′ ends of the first group of oligo(dT) primed RT–PCR clones and the 5′ ends of the PE signals where localized (Figure 4). Most of the clones did not harbor post-transcriptionally added tails as opposed to those isolated by the poly(A)-biased technique of oligo(dT) RT–PCR (Table 2 and Supplementary Table S5). This could be explained by the short half-life of the post-transcriptionally added tails that once synthesized, undergo rapid degradation. Alternatively, these fragments may be a population of molecules caught between the sequential cleaving and polyadenylation stages. Previous work on hyperthermophilic Archaea, showed similar results of non-polyadenylated fragments detected using cRT–PCR, while oligo(dT) RT–PCR analysis revealed poly(A) extensions (18). The 3′ ends of seven of the obtained 28S rRNA clones were located at 1687 nt, a position which matches the 5′ end of a putative distal cleavage product mapped by the PE approach (Figure 4). The combined results of these three techniques, oligo(dT) RT–PCR, cRT–PCR and PE, support the proposition that endonucleolytic cleavage can occur in this region of the 28S rRNA, followed by polyadenylation of the proximal fragments.
DISCUSSION
Polyadenylation-stimulated RNA degradation is broadly dispersed throughout the kingdoms of life
Following its initial discovery in Escherichia coli, the polyadenylation-stimulated RNA degradation pathway was described in other bacteria, as well as in archaea, chloroplasts and mitochondria (3,18,19,21,27–29). Interestingly, polyadenylation of mitochondrial RNA has a varying function or doesn't occur at all, depending on the organism (30). Recently, we found that in addition to stable mature 3′ end polyadenylation, the polyadenylation-stimulated degradation pathway must likely operates in human mitochondria as well, as was evident by the molecular and bioinformatic detection of fragmented and non-abundant polyadenylated transcripts related to rRNAs, mRNAs, tRNAs and intergenic regions of the human mitochondrial genome (9). A similar observation of coexistence of stable and unstable poly(A)-tails was also described for trypanosome mitochondria (10). In both systems the molecular mechanism that differentiates between the stable poly(A)-tails and those that play a role in the RNA degradation process is yet unknown.
In addition, a polyadenylation-stimulated degradation pathway was recently described for nuclear-encoded transcripts in yeast, revealing yet a third system that includes both types of polyadenylation (11–13). Furthermore, polyadenylation of rRNA in yeast has been reported (31,32), as well as in the polymorphic fungus, Candida albicans (33,34) and the trypanosome, Leishmania (35). Taking these observations into account, we decided to determine whether a polyadenylation-stimulated degradation pathway exists for nuclear-encoded RNA in human cells as well. Indeed, our detection of non-abundant polyadenylated rRNA fragments and the strict correlation between the presence of such molecules and that of a polyadenylation-stimulated degradation pathway, in every system investigated as of today, strongly suggest that as in bacteria, archaea, organelles and yeast nuclei, a similar mechanism is involved in the decay of RNA which is encoded in the nuclear genome in human cells. While this work was being prepared for publication, the accumulation of human β-globin pre-mRNAs containing short oligo(A) tails in the absence of the exosome was reported (15). This observation indeed further supports our hypothesis.
The function of rRNA polyadenylation in human cells
There are several possible explanations for the function of the rRNA polyadenylation that we observed. The first one is that rRNA molecules, although having no recognizable poly(A) signal, are polyadenylated by the poly(A) polymerization complex at cryptic polyadenylation signals, to a certain level, similar to the case of non-stop RNA decay (36,37). However, this possibility seems unlikely because of the dispersed characteristic of the polyadenylation sites along the rRNA molecule. In addition, the heteropolymeric tails described in this work are probably not the product of the Pol II associated polyadenylation complex as it is specific to adenosine (see below). The second possibility is that the polyadenylation described here is evidence of a polyadenylation-stimulated degradation pathway which functions as a quality control system that rapidly removes unfolded, and improperly processed molecules, or those which are not embedded in the functional ribosome, while the functional rRNA molecules are degraded by a non polyadenylation-stimulated degradation pathway. A third possibility is that the polyadenylated transcript fragments are polyadenylated degradation intermediates originating from mature and functional rRNA, located in the cytoplasm. The second and third possibilities, which indicate a polyadenylation-stimulated degradation pathway, are supported by the observations that in yeast strains in which the exosome activity was inhibited or abolished, the amount of polyadenylated rRNA and other transcripts was significantly increased (11–13,31,32,38). Similar results were reported recently for the human β-globin pre-RNA (15). This significant increase witnessed in the yeast system indicated the existence of a central and essential poly(A)-stimulated degradation pathway and the truncated polyadenylated rRNA which we detected here appear to be intermediates of a parallel pathway which occurs in human cells.
Following transcription in the nucleolus, the pre-rRNA transcript is subjected to rapid and efficient processing, removing transcribed spacer regions, generating the mature molecules which may be incorporated into functional ribosomes and accumulate to a very large amount in the cytoplasm (39,40). While the half-life of precursor rRNA is very short (several minutes), that of the mature cytoplasmic molecules is among the longest observed (several days) (41–43). As stated, since the RNA analyzed here included both nuclear and cytoplasmic fractions, the identified polyadenylated molecules could be the result of a polyadenylation-stimulated degradation pathway which is only present in one of these compartments or contrarily in both. One possibility, for example, is that the homopolymeric tails occur in the nucleus, similar to the observation made in yeast, (11–13), while the heteropolymeric tails are produced in the cytoplasm (see below). The identification of three ESTs related to the 18S rRNA, in which polyadenylation occurred within a spacer region which exists in the precursor rRNA but not in the mature molecule supports the possibility that at least some of the detected molecules originate from precursor transcripts in a process that takes place in the nucleus. (Figure 2 and Supplementary Table S2). Experiments to explore these possibilities are underway.
Does the exosome produce the heteropolymeric poly(A)-rich tails?
In the yeast nucleus, the RNA polyadenylating enzyme was identified as Trf4p which is a component of the polyadenylation complex termed TRAMP. This enzyme showed specificity to adenosines when analyzed in vitro (11–13,44,45). In addition, a second protein of the β-like nucleotidyltransferase family, Trf5p, also displayed specific poly(A) polymerase activity when analyzed in vitro (44–46). Following polyadenylation by the TRAMP complex, the polyadenylated transcripts are degraded by the exosome. In light of the adenosine-specificity found for the two polymerases mentioned above, we were surprised to find polyadenylated rRNA transcripts in human cells in which some of the tails were comprised not only of adenosines but also of other nucleotides as well.
Heteropolymeric poly(A)-rich tails were previously identified in E.coli strains deleted of the poly(A)-polymerase gene or the RNA-binding protein Hfq, as well as in other bacteria such as Bacillus subtilis, Streptomycis coelicolor and cyanobacteria (21,22,27,28). In addition, such tails were described in spinach chloroplasts as part of the poly(A)-stimulated RNA degradation process (19). In all of these systems, it was found that the enzyme which produces these heteropolymeric tails is polynucleotide phosphorylase (PNPase) (3,47). This enzyme can reversibly degrade or polymerize RNA in vitro using all 4 nt, producing heteropolymeric poly(A)-rich tails and is one of the major exoribonucleases present in prokaryotes and organelles. The PNPase structure is a homo-trimeric, doughnut shape complex which is believed to be similar to the structure of the eukaryotic exosome, a multi protein complex containing several exoribonucleases that are homologues of the phosphorylase domains of PNPase (48). Indeed, the crystallographic structure of the archaeal exosome of the hyperthermophile, Solfolobus solfactaricus, was recently described, revealing a doughnut shape structure very similar to that of PNPase (49). Moreover, when polyadenylation was analyzed in this archaea, non-abundant and fragmented transcripts harboring heteropolymeric poly(A)-rich tails, synthesized by the archaeal exosome, were obtained (18). This suggested a similar polyadenylation functionality of the prokaryotic/organellar PNPase and the archaeal exosome, in addition to their structural likeness. On this line, it is tempting to suggest that the heteropolymeric poly(A)-rich tails of rRNAs, which we observed here in human cells, are generated by the human exosome. This would mean that not only does the human exosome play a central role in exonucleolytically degrading nuclear-encoded RNA, as is well established, but also maintains heteropolymeric polyadenylation activity as do PNPase and the archaeal exosome. However, approximately half of the poly(A) extensions which we observed were homopolymeric in nature. Therefore, the exosome maybe responsible for the heteropolymeric poly(A)-rich tails, while poly(A) polymerases, such as the human homologous of Trf4p and Trf5p could be the source of the homopolymeric poly(A)-tails. We must note that in vitro polymerization activity has not yet been described for the eukaryotic exosome and genetic evidences in yeast showed that the association of the TRAMP complex seems to be required for the polyadenylation of the transcripts prior to their exosomal degradation. In addition, RT–PCR analysis of the poly(A)-tails of yeast rRNA is still lacking and may reveal heteropolymeric extensions. Clearly, more experimental data are required to determine whether the heteropolymeric poly(A)-rich tails are indeed produced by the exosome in human cells.
The mechanism of degradation
The rate-limiting step in the general scheme of polyadenylation-stimulated degradation in bacteria is believed to be endonucleolytic cleavage. Then, the proximal cleavage product is polyadenylated, followed by exoribonucleolytic digestion. The PE, oligo(dT) RT–PCR and cRT–PCR analyses performed in this work, identified a region (perhaps one of several) of the 28S rRNA that appears to undergo endonucleolytic cleavage, since both the proximal and distal products were detected. Moreover, the PE analysis disclosed similar signal profiles for all RNA preparations isolated from several cancer cell lines as well as from primary fibroblasts. Although we cannot completely exclude the possibility that some of these signals resulted from PE stalling at post-transcriptionally modified nucleotides or RNA secondary structures, we suggest that the majority of signals resulted from reverse transcription termination at the 5′ ends of transcript fragments (Figure 4 inset). Furthermore, our negative controls as well as previous extensive methyl-group mapping of human rRNA show that the all but one of the signals cannot be explained by this type of PE stalling. Additionally, the agreement between several different independent methods applied, further suggests the occurrence of endonucleolytic cleavage. As described, most of the degradation intermediates detected by PE are at least 100 times lower in quantity compared to the mature full-length transcript, as disclosed by the PE assay performed adjacent to the mature 5′ end of the 28S rRNA.
In summary, our results suggest that a molecular mechanism of polyadenylation-stimulated RNA decay, as witnessed in bacteria, archaea, organelles and yeast nuclei, is involved in the degradation of rRNA in human cells. The identification of the major components which govern this process is required, as the next step towards characterizing the molecular properties and biological function of this pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The author thank Eran Bram, Yehuda G Assaraf, Simona Zisman and Dina Ron for the cancer cells and primary fibroblasts. This work was supported by grants from the Israel Science Foundation (ISF) to G.S. and D.G., and Binational Science Foundation (BSF) and Binational Agriculture Science and Development Foundation (BARD) Foundations to G.S. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dreyfus M., Regnier P. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- 2.Kushner S.R. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- 3.Slomovic S., Portnoy V., Liveanu V., Schuster G. RNA polyadenylation in prokaryotes and organelles; different tails tell different tales. Crit. Rev. Plant Sci. 2006;25:65–77. [Google Scholar]
- 4.Edmonds M. A history of poly(A) sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 5.Shatkin A.J., Manley J.L. The ends of the affair: capping and polyadenylation. Nature Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 6.de Moor C.H., Richter J.D. Translational control in vertebrate development. Int. Rev. Cytol. 2001;203:567–608. doi: 10.1016/s0074-7696(01)03017-0. [DOI] [PubMed] [Google Scholar]
- 7.Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 9.Slomovic S., Laufer D., Geiger D., Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao C.Y., Read L.K. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell. Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 12.Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new Poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 14.Chanfreau G.F. CUTting genetic noise by polyadenylation-induced RNA degradation. Trends Cell Biol. 2005;15:635–637. doi: 10.1016/j.tcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 15.West S., Gromak N., Norbury C.J., Proudfoot N.J. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell. 2006;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Ifergan I., Shafran A., Jansen G., Hooijberg J.H., Scheffer G.L., Assaraf Y.G. Folate deprivation results in the loss of breast cancer resistance protein (BCRP/ABCG2) expression. A role for BCRP in cellular folate homeostasis. J. Biol. Chem. 2004;279:25527–25534. doi: 10.1074/jbc.M401725200. [DOI] [PubMed] [Google Scholar]
- 17.Stark H.J., Baur M., Breitkreutz D., Mirancea N., Fusenig N.E. Organotypic keratinocyte cocultures in defined medium with regular epidermal morphogenesis and differentiation. J. Invest. Dermatol. 1999;112:681–691. doi: 10.1046/j.1523-1747.1999.00573.x. [DOI] [PubMed] [Google Scholar]
- 18.Portnoy V., Evguenieva-Hackenberg E., Klein F., Walter P., Lorentzen E., Klug G., Schuster G. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polyadenylates RNA in Sulfolobus. EMBO Rep. 2005;6:1188–1193. doi: 10.1038/sj.embor.7400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisitsky I., Klaff P., Schuster G. Addition of poly(A)-rich sequences to endonucleolytic cleavage sites in the degradation of spinach chloroplast mRNA. Proc. Natl Acad. Sci. USA. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohanty B.K., Kushner S.R. Polynucleotide phosphorylase functions both as a 3′ to 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rott R., Zipor G., Portnoy V., Liveanu V., Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- 22.Mohanty B.K., Maples V.F., Kushner S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 23.Rebane A., Roomere H., Metspalu A. Locations of several novel 2′-O-methylated nucleotides in human 28S rRNA. BMC Mol. Biol. 2002;3:1–5. doi: 10.1186/1471-2199-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ofengand J., Bakin A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 1997;266:246–268. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- 25.Higa S., Maeda N., Kenmochi N., Tanaka T. Location of 2′-O-methyl nucleotides in 26S rRNA and methylation guide snoRNAs in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2002;297:1344–1349. doi: 10.1016/s0006-291x(02)02377-x. [DOI] [PubMed] [Google Scholar]
- 26.Maden B.E., Hughes J.M. Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem. Chromosoma. 1997;105:391–400. doi: 10.1007/BF02510475. [DOI] [PubMed] [Google Scholar]
- 27.Campos-Guillen J., Bralley P., Jones G.H., Bechhofer D.H., Olmedo-Alvarez G. Addition of Poly(A) and heteropolymeric 3′ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J. Bacteriol. 2005;187:4698–4706. doi: 10.1128/JB.187.14.4698-4706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bralley P., Jones G.H. cDNA cloning confirms the polyadenylation of RNA decay intermediates in Streptomyces coelicolor. Microbiology. 2002;148:1421–1425. doi: 10.1099/00221287-148-5-1421. [DOI] [PubMed] [Google Scholar]
- 29.Hayes R., Kudla J., Gruissem W. Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem. Sci. 1999;24:199–202. doi: 10.1016/s0968-0004(99)01388-2. [DOI] [PubMed] [Google Scholar]
- 30.Gagliardi D., Stepien P.P., Temperley R.J., Lightowlers R.N., Chrzanowska-Lightowlers Z.M. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Kuai L., Fang F., Butler J.S., Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang F., Hoskins J., Butler J.S. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell. Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleischmann J., Liu H., Wu C.P. Polyadenylation of ribosomal RNA by Candida albicans also involves the small subunit. BMC Mol. Biol. 2004;5:17. doi: 10.1186/1471-2199-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleischmann J., Liu H. Polyadenylation of ribosomal RNA by Candida albicans. Gene. 2001;265:71–76. doi: 10.1016/s0378-1119(01)00362-6. [DOI] [PubMed] [Google Scholar]
- 35.Decuypere S., Vandesompele J., Yardley V., De Donckeri S., Laurent T., Rijal S., Llanos-Cuentas A., Chappuis F., Arevalo J., Dujardin J.C. Differential polyadenylation of ribosomal RNA during post-transcriptional processing in Leishmania. Parasitology. 2005;131:321–329. doi: 10.1017/s0031182005007808. [DOI] [PubMed] [Google Scholar]
- 36.Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T.H. Quality control of mRNA 3′ end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 37.Frischmeyer P.A., van Hoof A., O'Donnell K., Guerrerio A.L., Parker R., Dietz H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 38.Milligan L., Torchet C., Allmang C., Shipman T., Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell. Biol. 2005;25:9996–10004. doi: 10.1128/MCB.25.22.9996-10004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fatica A., Tollervey D. Making ribosomes. Curr. Opin. Cell. Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 40.Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 41.Gillery P., Georges N., Wegrowski J., Randoux A., Borel J.P. Protein synthesis in collagen lattice-cultured fibroblasts is controlled at the ribosomal level. FEBS Lett. 1995;357:287–289. doi: 10.1016/0014-5793(94)01375-b. [DOI] [PubMed] [Google Scholar]
- 42.Halle J.P., Muller S., Simm A., Adam G. Copy number, epigenetic state and expression of the rRNA genes in young and senescent rat embryo fibroblasts. Eur. J. Cell Biol. 1997;74:281–288. [PubMed] [Google Scholar]
- 43.Lazdins I.B., Delannoy M., Sollner-Webb B. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma. 1997;105:481–495. doi: 10.1007/BF02510485. [DOI] [PubMed] [Google Scholar]
- 44.Haracska L., Johnson R.E., Prakash L., Prakash S. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol. Cell Biol. 2005;25:10183–10189. doi: 10.1128/MCB.25.22.10183-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egecioglu D.E., Henras A.K., Chanfreau G.F. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houseley J., Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2005;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bollenbach T.J., Schuster G., Stern D.B. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- 48.Symmons M.F., Williams M.G., Luisi B.F., Jones G.H., Carpousis A.J. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- 49.Lorentzen E., Walter P., Fribourg S., Evguenieva-Hackenberg E., Klug G., Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nature Struct. Mol. Biol. 2005;12:575–585. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.