Abstract
DNA strand exchange, the central step of homologous recombination, is considered to occur approximately independently of DNA sequence content. However, certain prokaryotic and eukaryotic genomic loci display either an enhanced or reduced frequency of genetic exchange. Here we show that the Homo sapiens DNA strand exchange protein, HsRad51, shows a preference for binding to single-stranded DNA sequences primarily rich in G-residues and poor in A- and C-residues, and that these DNA sequences manifest enhanced HsRad51 protein-dependent homologous pairing. Both of these properties are common to all DNA strand exchange proteins examined thus far. These preferred DNA pairing sequences resemble those found at genetic loci in human cells that cause genomic instability and lead to genetic diseases.
INTRODUCTION
Homologous recombination is an ubiquitous process important for DNA repair, generation of genetic diversity, and the proper segregation of chromosomes during meiosis. The reciprocal exchange of DNA strands promoted by DNA strand exchange proteins is a central step in homologous recombination (1). Although DNA strand exchange is considered to occur essentially independently of DNA sequence content, certain loci within prokaryotic and eukaryotic genomic DNA display either an enhanced or reduced frequency of genetic exchange.
Previously it was shown that both the prototypic bacterial DNA strand exchange protein, the RecA protein from Escherichia coli, and its eukaryotic counterpart, the Rad51 protein from Saccharomyces cerevisiae (ScRad51), show a preference for binding to certain single-stranded DNA (ssDNA) sequences that are over-represented in G and T mononucleotide residues (hereafter referred to as GT-rich sequences). The RecA protein selected a set of ssDNA sequences that contained, on average, 38.3% G-residues and 37.3% T-residues and the ScRad51 protein selected a set of ssDNA sequences that contained 44.6% G- and 30.5% T-residues (2,3). These GT-rich sequences also display an enhanced RecA or Rad51 protein-dependent pairing activity, suggesting that such sequences are potentially more active recombinationally than DNA sequences rich in A- and C-residues (2,3). In support of this notion, sequences that are similar to those selected are found in numerous unstable loci within the genomes of different eukaryotic organisms (4–6). This finding raises the possibility that the increased instability of those GT-rich loci in vivo is due to the increased ability of DNA strand exchange proteins to bind to these sequences and to promote DNA recombination.
Rad51 protein homologues have been identified throughout the Eucarya, from yeast to man (7,8). The ScRad51 protein is involved, in cooperation with other proteins from the Rad52 epistasis group, in homologous recombination that leads to the repair of dsDNA breaks in yeast (9–11). The mouse Rad51 protein was identified on the basis of its homology to the ScRad51 protein, and was shown to be particularly important in maintaining genome integrity and viability, since a knock-out allele of the mouse RAD51 gene resulted in early embryonic lethality (12,13). The human Rad51 (HsRad51) protein was also identified based on homology to the ScRad51 protein, and it possesses the same biochemical activities (8). Here we show that, similar to the ScRad51 and E.coli RecA proteins, HsRad51 protein has a preference for binding and homologously pairing ssDNA sequences that are over-represented for G-residues and under-represented for A- and C-residues; when the binding site size (3 nt) of a monomer is considered, a bias for GT-richness in the most frequent triplets is evident.
MATERIALS AND METHODS
In vitro selection assay
SKBT18 was constructed so that it contains a random internal region of 18 nt and two defined regions on the 5′ and 3′ ends that are 18 nt in length (2). The in vitro selection process began with about 6 × 1013 molecules of these 54mer oligodeoxyribonucleotides. Radiolabeled oligonucleotide (50 µM nt) was mixed with selection buffer [30 mM Tris-acetate (pH 7.5), 20 mM Mg-acetate, 1 mM DTT and 2.5 mM ATP]. Selection was initiated by the addition of HsRad51 protein, which was a generous gift from Patrick Sung (University of Texas, San Antonio; currently at Yale University); incubation was at 37°C for 1 h and the filter binding was performed as described previously (2,3).
The retained oligonucleotides (ranging from 5 to 50% depending on the cycle number) were eluted with elution buffer for use in asymmetric PCR (2,3). The eluted ssDNA was used in the following cycle of selection with HsRad51 protein and amplification by PCR. The DNA concentration at each cycle varied due to the amount of amplification, but the ratio of HsRad51 protein to DNA remained at the same 1:50 ratio (protein: nucleotides). A total of five cycles of selection and amplification were carried out. Cloning and sequencing of the selected sequences was performed as described previously (2,3).
Joint molecule formation assays
The oligodeoxyribonucleotides used were (2): SKBT16 [contains selected sequence 1: d(AGCTTGCATGCCTGCAGGGGCGTGTGTGGTGGTGTGCTAGGATCCCCGGGTACC)]; SKBT17 [contains the complement of SKBT16: d(GGTACCCGGGGATCCTAGCACACCACCACACACGCCCCTGCAGGCATGCAAGCT)]; SKBT19 [pairs on the opposite side of the plasmid: d(TGAACGAAATAGACAGATCGCTGAGATAGGTGCCTCACTGATTAAGCATTGGTA)]; SKBT20 [pairs adjacent to SKBT16: d(TCCCAGTCACGACGTTGTAAAACGACGGCCAGTGCCAAGCTTGCATGCCTGCAG)]; TELO [contains 3 tandem telomeric repeat sequences: d(AAGCTTGCATGCCTGCAGTTAGGGTTAGGGTTAGGGTAGGATCCCCGGGTACCG)]; and TELO complement [contains the complement of TELO: d(CGGTACCCGGGGATGCTACCCTAACCCTAACCCTAACTGCAGGCATGCAAGCTT)].
Oligodeoxyribonucleotides were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Joint molecule formation took place in a reaction mixture (260 µl) containing 2.5 mM Tris-acetate (pH 7.5), 20 mM Mg-acetate, 1 mM DTT, 25 mM ATP, 1 µM (nucleotides) 54mer, 0.33 µM HsRad51 protein, 18 µM nucleotides plasmid DNA [pBT54CN1 (2), a pUC19 derivative]. Reactions were initiated by the addition of plasmid after pre-incubation of HsRad51 protein and oligonucleotide for 5 min at 37°C. At various times, 40 µl aliquots were removed and added to 5 µl each of a 10% SDS/0.5 M EDTA mixture and DNA loading buffer. The reactions were subjected to agarose gel electrophoresis in 1% agarose gels run in TAE buffer for 180 V-hour. The gels were dried on DE-81 paper, imaged using a Storm PhosphorImager and quantified using ImageQuaNT 4.0 software. The percentage of joint molecule formation was determined relative to the limiting amount of plasmid DNA used in each reaction.
RESULTS
To determine whether the HsRad51 protein has a preference for DNA sequences with a particular composition, we performed in vitro selection experiments starting with a pool of 6 × 1013 54mers (SKBT18). These 54mers are composed of a random internal region of 18 nt flanked by defined regions of 18 nt (2). A limiting concentration of HsRad51 protein was used to guarantee equal competition between all of the 54mers. The resultant Rad51 protein–ssDNA complexes which comprised the highest affinity class were isolated by binding to nitrocellulose filters, and these selected ssDNA sequences were amplified by PCR. After five rounds of selection and amplification, the selected 54mers were cloned, and 41 randomly chosen clones were sequenced. Table 1 shows the sequences of the internal 18mer regions selected by the HsRad51 protein. The average base composition of these sequences is highly over-represented in G-residues (52%), while under-represented in A- and C-residues (7.2 and 16.7%, respectively). The average base composition for T-residues remains at almost a statistical average (24%). Interestingly, the HsRad51 protein selected a particular sequence five times (clone 1) that was also selected by two other DNA strand exchange proteins, the E.coli RecA and S.cerevisiae Rad51 proteins (2,3). The frequent selection of this sequence is not due to its over-representation in the initial pool as confirmed by previous sequencing experiments (2,3). The HsRad51 protein also selected a sequence (clone 9) four times, which differed from this sequence by only a single base.
Table 1.
The HsRad51 protein selects DNA rich in G-residues from a pool of random oligonucleotides
| 1. | GCGTGTGTGGTGGTGTGC |
| 6. | GGCGTGTGTGGTGGTGTG |
| 8. | GGGGGATGTGCGTGCCCG |
| 9. | GGGTGTGTGGTGGTGTGC |
| 13. | GGGTGGTTGGTTACTGCC |
| 14. | GGGGACCAGCATTTGCCC |
| 15. | GGGGGATGTACGTGCCCG |
| 16. | GGGGGGGGTGGTTGTGCC |
| 17. | GGGGGAGTGGGATGTCCC |
| 18. | GGGGGAAAGCTGCGTGCC |
| 19. | GGGGGGACGTACTGTGCC |
| 20. | GGGAAGCATGTTGGACCC |
| 21. | GGGGAATTACGTGGCCCG |
| 22. | GGGAGGTGTTGGCTGCCG |
| 23. | GGGAAGGTTGCGTGTCCC |
| 24. | GGGGGTAGTGCAGTGCCC |
| 25. | GGGGGATGGTGTGTGCCC |
| 26. | GGGGGTAGTGGTGTGCCC |
| 27. | GGGGGTAGTGGTGTGCCC |
| 28. | GGGGAATTAAGTTGTCCC |
| 29. | GGGGAGTGTGGTTGGCCC |
| 30. | GGGGCAAGGTCGGCCCGT |
| 31. | GGGGAGTGCTGTGTGCCC |
| 32. | GGGGGGAGTAGTGGCCCC |
| 33. | GGGGTATAGGGCTGGACC |
| 34. | GGTATGTGGGGGGTGTAC |
| 35. | GGGCAGGGATGTCGTGCC |
| 36. | GGGGGGAGTGGTGTGCCC |
| 37. | CTGTGTGGTGGTGTGTCC |
| 38. | GGGGGGTAGTGCTGTCCC |
| 39. | GGGGGGCAACTGGTGCTG |
| 40. | GGGGAGAGCTGCTGTGCC |
| 41. | GGGGGGAGCTACGTGGCG |
Totals: %A: 7.2; %C: 16.7; %G: 52.4; %T: 24.
Shown are the unique sequences of the 18 nt region after five rounds of selection and amplification, along with the average base composition for all of the sequences that were selected.
HsRad51 protein binds approximately 3 nt per protein monomer (14); therefore, we examined the trinucleotide distribution in the sequences selected by HsRad51 protein. Table 2 shows that the significantly over-represented trinucleotide sequences are exclusively composed of G- and T-residues; the same bias is seen for ScRad51 and E.coli RecA proteins (2,3). Correspondingly, many trinucleotides containing A- and C-residues are significantly under-represented, as also observed for RecA and ScRad51 proteins.
Table 2.
Trinucleotide occurrences in the sequences selected by HsRad51 protein
| Trinucleotides | Frequency |
|---|---|
| GTG | 17.3 |
| GGG | 12.8 |
| TGT | 9.3 |
| GGT | 7.8 |
| TGG | 6.6 |
| TGC | 5.3 |
| GGA | 3.3 |
| GCC | 3.2 |
| CCC | 3.0 |
| CGT | 2.3 |
| CAT | 2.0 |
| CTG | 1.8 |
| AGT | 1.8 |
| GGC | 1.7 |
| GCG | 1.5 |
| GTA | 1.5 |
| GTT | 1.4 |
| GCT | 1.4 |
| TTG | 1.4 |
| GAG | 1.4 |
| ATG | 1.0 |
| GTC | 1.0 |
| GCA | 0.9 |
| TAC | 0.9 |
| TAG | 0.9 |
| AGG | 0.8 |
| AAG | 0.8 |
| GAA | 0.8 |
| TCC | 0.8 |
| GAT | 0.8 |
| AGC | 0.8 |
| TCC | 0.8 |
| ACG | 0.6 |
| GAC | 0.6 |
| CAG | 0.4 |
| ACC | 0.4 |
| TTA | 0.4 |
| ATT | 0.4 |
| ACT | 0.4 |
| TCG | 0.3 |
| CAA | 0.3 |
| ATT | 0.3 |
| TAT | 0.3 |
| AAC | 0.2 |
| CGG | 0.2 |
| AGA | 0.2 |
| AAA | 0.2 |
| ATA | 0.2 |
| TAA | 0.2 |
| CTA | 0.2 |
| CCA | 0.2 |
| TTT | 0.2 |
The frequency was determined by the number of times a certain trinucleotide occurred in all of the sequences, and divided by the total number of trinucleotides in the selected sequences. The expected random statistical frequency is 1.6% (1/64).
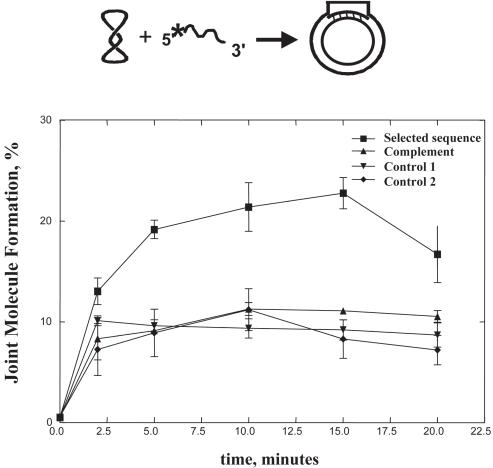
HsRad51 protein can promote the invasion of supercoiled DNA by homologous ssDNA to form a joint molecule (14). We therefore investigated the preference for joint molecule formation catalyzed by HsRad51 protein with respect to sequence content. Previously, it was demonstrated that both RecA and ScRad51 proteins show an enhanced rate and extent of joint molecule formation with selected DNA (2,3). HsRad51 protein was tested in the same fashion using oligonucleotides that contained a selected sequence (SKBT16); its complement (SKBT17), which pairs in the same place on the plasmid; or two other sequences (SKBT19, SKBT20), which are homologous to the supercoiled DNA in a region outside of the selected sequence. Figure 1 shows that HsRad51 protein pairs DNA containing selected sequence 1 (SKBT16) to a homologous supercoiled plasmid [pBT54CN1; (2)] with about a 2-fold greater yield (20%) than DNA containing the other control DNA sequences (7–10%). Therefore, the selected DNA is also a preferred pairing DNA, which is the same result observed for both RecA and ScRad51 proteins (2,3).
Figure 1.
The HsRad51 protein promotes joint molecule formation more efficiently with DNA containing a selected sequence. Joint molecules were formed with a 6-fold molar (molecule) excess of oligonucleotide relative to the supercoiled plasmid, pBT54CN1. The percentage of joint molecule formation was determined relative to the limiting amount of plasmid DNA. The graph shows joint molecule formation promoted by HsRad51 protein with pBT54CN1 and the following oligonucleotides (2): SKBT16 (squares; contains selected sequence 1), SKBT17 (triangles; contains the complement of SKBT16), ‘Control 1’ is SKBT19 (inverted triangle; pairs on the opposite side of the plasmid) and ‘Control 2’ is SKBT20 (diamond; pairs adjacent to SKBT16). Error bars represent the standard deviation.
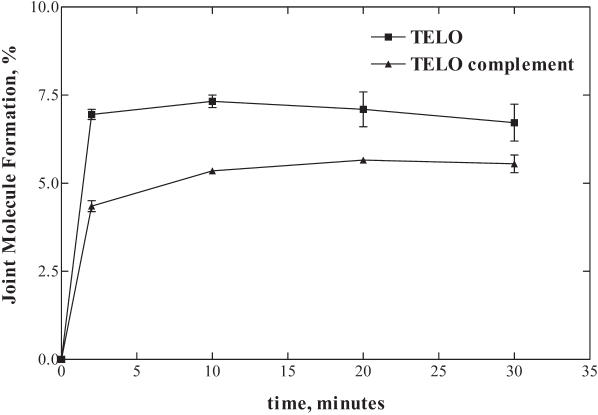
We also tested joint molecule formation by HsRad51 protein with an oligonucleotide containing three tandemly repeated TTAGGG human telomeric sequences (Figure 2). Telomeres are required for stable maintenance of eukaryotic chromosomes and both telomeric and sub-telomeric repeat sequences constitute hot-spots for genetic recombination when telomerase function is absent (15,16). Figure 2 shows that human Rad51 pairs DNA containing the three tandem telomeric repeat sequences (TELO) to a homologous supercoiled plasmid with a 1.6-fold greater extent (7%) than the complement of this sequence (TELO complement; 4.3%). The yield of DNA pairing product was reproducibly and significant lower for these sequences than for the preferred pairing sequences used in Figure 1. We cannot explain the low yield, but it is common to observe locus- (i.e. sequence-) dependent variation in DNA pairing efficiencies in vitro. It is for this reason that a comparison of a paring sequence to its complement is more meaningful. Thus, although the GT-rich strand of the telomeric sequence used here shows a low absolute pairing yield, it is an almost 2-fold better pairing sequence than its complement, just as was observed for selected sequence #1.
Figure 2.
The HsRad51 protein promotes joint molecule formation more efficiently with DNA containing three tandem telomeric repeat sequences (TTAGGG) than with the complement of this sequence. Joint molecules were formed with a 6-fold molar (molecule) excess of oligonucleotide relative to the homologous supercoiled plasmid. The percentage of joint molecule formation was determined relative to the limiting amount of plasmid DNA. The graph shows joint molecule formation promoted by HsRad51 protein with supercoiled plasmid DNA and homologous oligonucleotides containing the telomeric repeat sequence TTAGGG three times in tandem (TELO) as well as the complement of this sequence (TELO complement). Error bars represent the standard deviation.
DISCUSSION
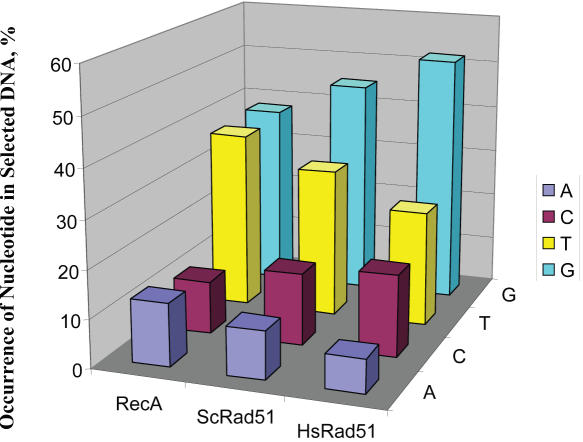
Previously, it was noted that the set of GT-rich sequences selected by both a bacterial and a eukaryotic (yeast) DNA strand exchange protein were similar to genetically unstable DNA sequences within mammalian genomes (2,3). Here we establish that the human DNA strand exchange protein displays a similar bias, with regard both to binding and pairing activity. Figure 3 shows the mononucleotide distribution for the sequences that were selected by DNA strand exchange proteins from bacteria, yeast and humans. Most striking is the over-representation of G- and T-residues (although, as noted above, when only mononucleotides are considered, the occurrence of T-residues is at statistically expected value for HsRad51). Interestingly, the table shows two unexplained trends when the bias displayed by proteins from the increasingly complex organisms is compared: the frequencies of the G- and C-residues increase at the expense of T- and A-residues, respectively, in going from RecA to HsRad51 proteins. Additionally, Table 3 shows the four most over-represented trinucleotides in the sequences selected by each of these proteins. All three proteins display a similar bias for GT-richness in this set of trinucleotides. Due to the universality of this preference and the fact that these sequences constitute recombination hot-spots in bacterial and eucaryal genomes, the possibility is presented that DNA strand exchange proteins may play an important role in contributing to the genetic instability at these loci.
Figure 3.
DNA strand exchange proteins select for sequences that are primarily G-rich, and under-represented for A- and C-residues. Shown are the nucleotide frequencies for each of the DNA strand exchange proteins examined: E.coli RecA, S.cerevisiae Rad51 (2,3), and H.sapiens Rad51 protein.
Table 3.
DNA strand-exchange proteins select for DNA sequences that are primarily G-rich, and under-represented for A and C residues
| Trinucleotide frequency (%) | ||||
|---|---|---|---|---|
| RecAaa | GTG (7.6) | TGG (7.6) | GTT (6.3) | GGT (6.0) |
| ScRad51b | GTG (10.4) | TGG (7.2) | TGT (7.4) | GGT (7.0) |
| HsRad51c | GTG (17.3) | GGG (12.8) | TGT (9.3) | GGT (7.8) |
Shown are the four most frequently occurring trinucleotides found in the selected DNA for the three DNA strand exchange proteins examined: E.coli RecA, S.cerevisiae Rad51 and H.sapiens Rad51 proteins. The bias for trinucleotides composed exclusively of G and T residues is evident.
aTracy and Kowalczykowski (2); bTracy, et al. (3); and cthis work.
To further examine the implications of our results, the HsRad51-selected ssDNA sequences were compared to sequences located in the NCBI (National Center for Biotechnology Information) database using a BLAST search. Many of the sequences identified in the search are found at genetically unstable loci in Bacteria and Eucarya. One class of these closely related sequences are the minisatellite DNA sequences, which frequently show germ-line instability, and involve the transfer of blocks of repeats from one allele to another. Human minisatellite instability occurs specifically during meiosis and this instability also appears to be a by-product of meiotic recombination (5). Jeffreys et al. (5) proposed that a dsDNA break is introduced at the beginning of a minisatellite repeat array, and the resulting gap is then bridged by strand invasion from the other allele to provide a template for gap repair, leading to conversion. They identified meiotic crossover events that they propose were created by resolution and isomerization of the recombination junctions formed by this strand invasion event. These data supported other evidence that minisatellite DNA sequences serve as recombination ‘warm-spots’.
Another identified sequence-related class are the Alu repetitive elements, which contain a common 26 bp consensus sequence (5′-CCTGTAATCCCAGCACTTTGGGAGGC-3′), whose complement contains a region (5′-GTGCTGGGATT-3′) that is GT-rich and bears some similarity to the GT-rich E.coli recombinational hot-spot, Chi (5′-GCTGGTGG-3′) (17). Alu elements are associated with genetic rearrangements and, thus, were suggested to be recombinational hot-spots. Recombination in human cells mediated by repetitive elements is observed, with some of these events being due to Alu-Alu mediated ectopic recombination (18,19).
A third class of sequences with similarity to the selected DNA is the telomeric sequences, which are required for stable maintenance and segregation of eukaryotic chromosomes. Telomeric repeat sequences are also hot-spots for genetic recombination and are generally G(T)-rich (15,16). Alternative pathways for telomere maintenance exist in the absence of telomerase, which depend on telomere–telomere recombination (20,21). Work by Griffith et al. has shown that mammalian telomeres, composed of TTAGGG repeats, form a large duplex loop structure that is referred to as a T-loop. This structure is presumably formed by the invasion of the telomeric 3′-ssDNA overhang into the duplex telomeric repeat array, and its formation can be catalyzed in vitro by the telomeric repeat-binding factor TRF2 protein (4,22). It is unclear at this point whether or not other proteins are involved in coordination with TRF2 protein for the formation of T-loops, but the fact that the human recombination protein, HsRad51, shows a preference for D-loop formation with sequences similar to those found at telomere ends, could suggest a possible role in T-loop formation.
Additionally, in both yeast and mammals, short stretches of telomere-like poly(GT) sequences increase the rate of recombination (23). Finally, many of the selected sequences are similar to the DNA triplets that are associated with triplet expansion diseases in humans. Recently it was demonstrated that the expansion of the triplet repeat CTG/CAG sequence which can lead to hereditary neurological diseases is caused, at least in part, by genetic recombination between the repeat sequences (6). While evidence exists that the instability of these loci also can result from DNA polymerase slippage after formation of unusual DNA structures, such as hairpins, it is conceivable that these sequences serve as targets for DNA strand exchange proteins, which could contribute to this increase in recombination. The effect of the many additional proteins involved in eukaryotic homologous DNA recombination, specifically other members of the Rad52 epistasis group, on HsRad51 function at G(T)-rich sequences, must be addressed in future studies.
Acknowledgments
The authors are very grateful to Dr Patrick Sung for providing us with the purified HsRad51 protein. The authors would like to thank the following members of the Kowalczykowski lab for providing comments on this manuscript: Piero Bianco, Carole Bornarth, Deana Arnold, Julie Mirshad, Alex Mazin, Susan Shetterly and special thanks to Frederic Chédin and Jim New. The work described in this manuscript was supported by NIH grant AI-18987 and HFSP-RG63 to S.C.K. and by NIH training grant GM07377 to E.M.S. Funding to pay the Open Access publication charges for this article was provided by GM-62653.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kowalczykowski S.C., Dixon D.A., Eggleston A.K., Lauder S.D., Rehrauer W.M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tracy R.B., Kowalczykowski S.C. In vitro selection of preferred DNA pairing sequences by the Escherichia coli RecA protein. Genes Dev. 1996;10:1890–1903. doi: 10.1101/gad.10.15.1890. [DOI] [PubMed] [Google Scholar]
- 3.Tracy R.B., Baumohl J.K., Kowalczykowski S.C. The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev. 1997;11:3423–3431. doi: 10.1101/gad.11.24.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 5.Jeffreys A.J., Murray J., Neumann R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell. 1998;2:267–273. doi: 10.1016/s1097-2765(00)80138-0. [DOI] [PubMed] [Google Scholar]
- 6.Wells R.D., Jakupciak P. Genetic instabilities in (CTG/CAG) repeats occur by recombination. J. Biol. Chem. 1999;274:23468–23479. doi: 10.1074/jbc.274.33.23468. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa T., Yu X., Shinohara A., Egelman E.H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 8.Benson F.E., Stasiak A., West S.C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petes T.D., Malone R.E., Symington L.S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Broach J.R., Jones E., Pringle J., editors. Vol. I. NY: Cold Spring Harbor Laboratory; 1991. pp. 407–521. [Google Scholar]
- 10.Game J.C. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 11.Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim D.S., Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M., Matsushiro A., Yoshimura Y., Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumann P., Benson F.E., West S.C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 15.Pryde F.E., Louis E.J. Saccharomyces cerevisiae telomeres. A review. Biochemistry. 1997;62:1232–1241. [PubMed] [Google Scholar]
- 16.Stavenhagen J.B., Zakian V.A. Yeast telomeres exert a position effect on recombination between internal tracts of yeast telomeric DNA. Genes Dev. 1998;12:3044–3058. doi: 10.1101/gad.12.19.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudiger N.S., Gregersen N., Kielland-Brandt M.C. One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res. 1995;23:256–260. doi: 10.1093/nar/23.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffs A.R., Benjes S.M., Smith T.L., Sowerby S.J., Morris C.M. The BCR gene recombines preferentially with Alu elements in complex BCR-ABL translocations of chronic myeloid leukaemia. Hum. Mol. Genet. 1998;7:767–776. doi: 10.1093/hmg/7.5.767. [DOI] [PubMed] [Google Scholar]
- 19.Richardson C., Moynahan M.E., Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McEachern M.J., Blackburn E.H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 21.Rogan E.M., Bryan T.M., Hukku B., Maclean K., Chang A.C., Moy E.L., Englezou A., Warneford S.G., Dalla-Pozza L., Reddel R.R. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol. Cell Biol. 1995;15:4745–4753. doi: 10.1128/mcb.15.9.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greider C.W. Telomeres do D-loop-T-loop. Cell. 1999;97:419–422. doi: 10.1016/s0092-8674(00)80750-3. [DOI] [PubMed] [Google Scholar]
- 23.Majewski J., Ott J. GT repeats are associated with recombination on human chromosome 22. Genome Res. 2000;10:1108–1114. doi: 10.1101/gr.10.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]