Abstract
DNA polymerase γ (pol γ) is required to maintain the genetic integrity of the 16,569-bp human mitochondrial genome (mtDNA). Mutation of the nuclear gene for the catalytic subunit of pol γ (POLG) has been linked to a wide range of mitochondrial diseases involving mutation, deletion, and depletion of mtDNA. We describe a heterozygous dominant mutation (c.1352G→A/p.G451E) in POLG2, the gene encoding the p55 accessory subunit of pol γ, that causes progressive external ophthalmoplegia with multiple mtDNA deletions and cytochrome c oxidase (COX)–deficient muscle fibers. Biochemical characterization of purified, recombinant G451E-substituted p55 protein in vitro revealed incomplete stimulation of the catalytic subunit due to compromised subunit interaction. Although G451E p55 retains a wild-type ability to bind DNA, it fails to enhance the DNA-binding strength of the p140-p55 complex. In vivo, the disease most likely arises through haplotype insufficiency or heterodimerization of the mutated and wild-type proteins, which promote mtDNA deletions by stalling the DNA replication fork. The progressive accumulation of mtDNA deletions causes COX deficiency in muscle fibers and results in the clinical phenotype.
Human mtDNA, which encodes 13 essential components of the respiratory chain, is replicated continuously in dividing cells and postmitotic tissues. Failure to preserve the genetic integrity of the mitochondrial genome during replication results in depletion, deletion, or mutation of mtDNA, which ultimately impairs oxidative phosphorylation and causes cellular dysfunction and disease.1 The accuracy of mtDNA replication depends on the coordinated action of many nuclear-encoded proteins and on the correct balance of nucleotides within the mitochondrial matrix. mtDNA is replicated by DNA polymerase γ (pol γ), which is composed of a 140-kDa catalytic subunit (p140) and a 55-kDa accessory subunit (p55).2 The catalytic subunit possesses DNA polymerase, 3′–5′ exonuclease, and 5′ deoxyribose phosphate lyase activities, whereas the accessory subunit is a DNA-binding factor that confers high processivity on the protein complex by increasing its affinity to DNA.3 Mutations in the gene encoding the catalytic subunit (POLG [GenBank accession number NM_007215]) found on chromosome 15q25 cause progressive external ophthalmoplegia (PEO)—both dominant (PEOA1 [MIM 157640]) and recessive (PEOB [MIM 258450])—Alpers syndrome (MIM 203700), and ataxia with peripheral neuropathy (MIM 607459).4–11 POLG mutations that cause PEO are linked to the formation of multiple mtDNA deletions in skeletal muscle. Mutation of other nuclear genes involved in mtDNA replication and nucleotide metabolism—including the adenine nucleotide translocase (ANT1), the mtDNA helicase Twinkle (C10orf2), thymidine kinase 2 (TK2), deoxyguanosine kinase (DGUOK), and thymidine phosphorylase (TP)—also cause human metabolic disorders accompanied by depletion and mutation of mtDNA. However, the primary nuclear mutation remains undefined in many patients with secondary mtDNA defects.12 Given the essential role of the p55 accessory subunit for highly processive mtDNA synthesis and enhanced DNA binding by the pol γ holoenzyme,3 POLG2 on chromosome 17q is an obvious candidate gene for these patients.
Here, we describe the identification and characterization of a novel point mutation in the POLG2 gene associated with autosomal dominant PEO. Biochemical characterization of the mutant form of p55 reveals intermediate stimulation and restoration of salt tolerance, intermediate N-ethylmaleimide (NEM) protection assays, no enhancement of DNA binding by the pol γ complex, and no significant physical or functional interaction of the mutant p55 with the catalytic subunit under physiological conditions.
Subjects and Methods
Subjects
We identified a female patient, aged 60 years, who developed exercise intolerance and muscle pain by age 40, followed by progressive drooping of the eyelids (ptosis), PEO, and mild weakness of the facial and limb muscles. She had impaired glucose tolerance, evidence of a cardiac conduction defect (left bundle branch block and intermittent bigeminy), a fasting blood lactate of 1.8 mM (normal <2), and a serum creatine kinase of 664 U/liter (normal <170). Her mother had been similarly affected but is deceased, and her two sisters are unaffected. The patient has two asymptomatic offspring in their 30s who declined assessment and further investigation.
Mitochondrial Studies and mtDNA Molecular Genetics
Southern blot and long-range PCR of mtDNA was performed using the Expand Long Template PCR System (Roche) on DNA that was extracted from a muscle homogenate from the index case with the use of an established protocol amplifying an ∼9.9-kb fragment across the major arc.13 Cytochrome c oxidase (COX)–succinate dehydrogenase (SDH) histochemistry was performed on cryostat sections of the left quadriceps muscle biopsy. The activity of individual respiratory chain complexes was determined in relation to citrate synthase activity. Cryostat muscle sections were also cut and stained on membrane slides for laser microdissection (Leica ASLMD). Each cell was lysed, and both the total amount of mtDNA and the amount of nondeleted mtDNA were quantified in each fiber by real-time PCR, with use of the iQ Sybr Green on the BioRad ICycler (BioRad).14 Total mtDNA was determined in triplicate on a single cell from a target template spanning nucleotides 3459–3569 of the mtDNA MTND1 gene. Nondeleted mtDNA was determined in triplicate on a single cell from a target template spanning nucleotides 11145–11251 of the mtDNA MTND4 gene. The results were expressed as the percentage of deleted mtDNA by convention.15
Molecular Genetics
Total DNA was extracted by standard methods from either muscle or blood from 100 patients with sporadic or familial PEO with histochemical evidence of mitochondrial disease (i.e., COX-deficient muscle fibers and/or ragged-red fibers, in greater proportions than would be expected for age-matched healthy controls,16 and multiple deletions in skeletal muscle demonstrated with at least two of the following methods: Southern blot, long-range PCR, and real-time PCR).15 POLG2 was sequenced using published primer sequences incorporating forward and reverse M13 tags, a fluorescent chain-terminating sequencing kit (Beckman Coulter Quickstart), and a Beckman Coulter CEQ 8000 fluorescent DNA analyzer. Sequences were analyzed using Bioedit (v. 5.09) sequence alignment editor against reference sequences. One hundred forty-four healthy control subjects (288 control chromosomes) were screened for the 1352G→A mutation by denaturing high-performance liquid chromatography (DHPLC) of exon 8 PCR products generated from the sequencing primers (annealing temperature 57°C, Transgenomic 3500HT WAVE system that used a standard solvent gradient, melt temperature 60.6°C, and buffer B 59.8%).
Proteins
The catalytic subunit of human pol γ was overexpressed in baculovirus-infected Sf9 cells and was purified to homogeneity, as described elsewhere.17,18 Several modifications were made to the plasmid bearing the recombinant accessory subunit. The coding sequence was transferred from pQESL-Hp55 to a pET-based expression vector,3 and the N-terminal His6 affinity tag was removed. The new N-terminal amino acid sequence was Met1-Asp26-Ala27-Gly28, which deletes the 25 N-terminal amino acids that compose the mitochondrial targeting sequence.3 The amino acids Ser-His6 were added to the C-terminus to incorporate a new affinity tag. Additionally, the QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used to change seven rare codons (Arg36 from AGG to CGT, Pro39 from CCC to CCG, Gly41 from GGA to GGT, Arg75 from AGA to CGT, Arg76 from AGG to CGT, Leu79 from CTA to CTG, and Gly81 from GGA to GGT) to optimize translation in Escherichia coli. The resulting plasmid (pET-p55CHIS) was confirmed by DNA sequencing. The G451E derivative of p55 was constructed by site-directed mutagenesis, with the use of pET-p55CHIS as the template and the mutagenic primers 5′-CTACTTTGGAGAATGAATTAATACATCTGA-3′ and 5′-TCAGATGTATTAATTCATTCTCCAAAGTAG-3′ (Oligos Etc.). DNA sequencing confirmed the G451E substitution and also revealed a synonymous CTG→TTG mutation at Leu codon 347 in the mutant construct, which is not expected to affect the recombinant protein. E. coli BL21(DE3) that had been transformed with a plasmid expressing either wild-type (WT) p55 or G451E p55 was grown at 37°C in 1.5 liters of Luria-Bertani media containing 100 μg/ml ampicillin to an OD595 (optical density) of 1.0, and then cultures were chilled to 30°C and induced for 13 h with 1 mM isopropyl thiogalactoside. Cells were harvested by centrifugation, were washed once with 10 mM HEPES (pH 7.1) and 50 mM NaCl, were frozen with liquid nitrogen, and were stored at −80°C. Both WT p55 and G451E p55 were purified to homogeneity from frozen cell pellets as described elsewhere,3 except that the Triton X-100 in the third wash buffer for Ni-NTA agarose chromatography was replaced with 0.01% NP-40. Whereas the N-terminal His6-tagged p55 protein eluted from a MonoS column at ∼0.54 M NaCl,3 both the WT and G451E C-terminal His6-tagged derivatives eluted from MonoS at 0.34 M NaCl. Codon optimization enhanced protein production 4- to 10-fold relative to the original construct, which permitted purification of up to 1.5 mg p55 (fraction IV) from each liter of culture media. For the current study, both forms of p55 (fraction IV) were transferred into a buffer containing 10 mM HEPES (pH 7.4), 0.15 M NaCl, 3 mM EDTA, and 0.005% surfactant P20 (polysorbate 20), by passage through a 5-ml HiTrap desalting column (Amersham Biosciences), before being frozen with liquid nitrogen and stored at −80°C.
Enzymatic Assays
DNA polymerase activity was determined as described elsewhere3 and used poly(rA)-oligo(dT)12–18 (Amersham Biosciences) as the substrate. The two-subunit form of pol γ was reconstituted as described elsewhere,3 and reactions contained the indicated quantities of NaCl and/or the inhibitor NEM. The processivity of pol γ was determined by monitoring the extension of a 5′-end–labeled oligonucleotide primer hybridized to M13mp18 DNA, as described elsewhere.19 Reaction products were resolved by denaturing polyacrylamide gel electrophoresis and alkaline agarose gel electrophoresis3 and were visualized with a Typhoon 9400 PhosphorImager (Molecular Dynamics) and NIH Image 1.63 software.
Analytical DNA-Cellulose Chromatography
A 1-ml column of single-stranded DNA cellulose (Sigma) was equilibrated at 4°C in 0.025 M Tris-Cl (pH 7.5), 10% glycerol, 1 mM 2-mercaptoethanol, and 0.1 mM EDTA (buffer A) also containing 0.025 M NaCl. Samples of purified recombinant p140, WT p55, and G451E p55 were adjusted by dilution to the ionic strength of this buffer and were applied to the column, which was washed with 9.5 ml of equilibration buffer and was developed with a 15-ml linear gradient of NaCl (0.025–0.6 M) in buffer A. Samples of each gradient fraction were analyzed by SDS-PAGE on 4%–20% gradient gels (Invitrogen), with the use of a Tris-glycine buffer system, and proteins were stained with Coomassie Brilliant Blue.
Immunoprecipitation Assay
Rabbit polyclonal antibodies (DPg) against recombinant pol γ17 were immobilized on Protein G Sepharose beads (Amersham Biosciences), and the beads were then equilibrated in phosphate buffered saline NP-40 (PBSN)–BSA buffer consisting of 0.05 M KPO4 (pH 7.5), 0.15 M NaCl, 0.1% NP-40, and 0.1 mg/ml BSA. Prepared DPg Protein G Sepharose beads (10 μl) were mixed as indicated with purified WT pol γ (3 μg), A467T mutant pol γ (3 μg), and/or the p55 accessory subunit (3 μg) in a 1.5-ml polypropylene microfuge tube and were brought to a final volume of 0.4 ml with PBSN-BSA. Tubes were rotated end over end for 45 min at 4°C, and beads were collected by microcentrifugation at 5,000 rpm for 2 min at 4°C. The supernatant was removed and beads were washed twice with PBSN-BSA and once with PBSN lacking BSA. Beads were resuspended in 25 μl 2× lithium dodecyl sulfate (LDS) loading buffer (4× LDS loading buffer from Invitrogen, made 2× with PBSN lacking BSA), and the samples were heated for 10 min at 70°C before analysis on 4%–12% NuPage Bis-Tris polyacrylamide gels (Invitrogen). After electrophoresis, the proteins were electrotransferred to an Immobilon-P PVDF (polyvinylidene fluoride) membrane (Millipore). The membrane was then washed in TNT (50 mM Tris-HCl [pH 7.5], 0.5 M NaCl, and 0.1% Triton X-100) for 15 min and was blocked with 5% dried milk in TN (50 mM Tris-HCl [pH 7.5] and 0.5 M NaCl) at room temperature. The blot was incubated with 0.2 μg/ml anti-Penta-His monoclonal antibody (Qiagen) in TN containing 0.1 mg/ml BSA for 2 h, was washed three times with TN for 10 min, and was incubated in a 1/3,000 dilution of goat anti-mouse alkaline phosphatase–conjugated secondary antibody (Bio-Rad) for 1 h. After three 10-min washes in TNT and three 10-min washes in TN, bands were visualized with Western Blue reagent (Promega).
Other Methods
Protein concentration was determined in relation to a BSA standard, as described by Bradford.20
Results
Mitochondrial Biochemistry and mtDNA Analysis
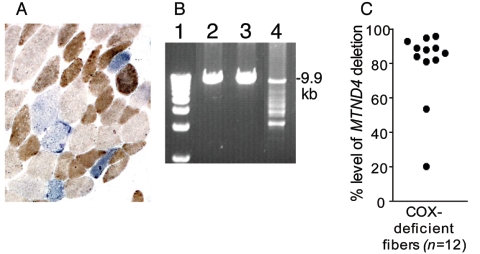
Skeletal muscle histochemistry on the index case revealed a mosaic COX defect with 6% COX-negative fibers (fig. 1A). Biochemical analysis of a skeletal muscle homogenate revealed normal respiratory chain complex activity. Southern blot analysis and long-range PCR of skeletal muscle mtDNA revealed multiple mtDNA deletions (fig. 1B). Real-time PCR of single muscle fibers detected high-percentage levels of deleted mtDNA in the majority of COX-defective muscle fibers, which is typical of a multiple mtDNA deletion disorder (fig. 1C).15
Figure 1.
Skeletal muscle histochemistry and mtDNA analysis. A, Dual COX-SDH histochemistry (see the “Subjects and Methods” section) showing a mosaic distribution of COX-deficient muscle fibers (blue) among fibers with normal COX activity (brown). B, Long-range PCR of skeletal muscle mtDNA (extracted from a tissue homogenate). Lane 1, 1-kb ladder; lane 2, young control muscle; lane 3, age-matched control muscle; lane 4, patient muscle showing multiple mtDNA deletions. C, Real-time PCR analysis of COX-negative muscle fibers. The majority of fibers contain high-percentage levels of mtDNA deletions that remove the ND4 region (see the “Subjects and Methods” section).
Identification of a Heterozygous Mutation in POLG2
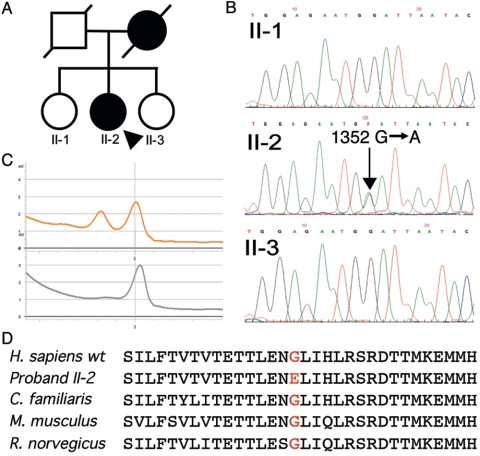
Sequence analysis revealed no mutations in ANT1, C10orf2, or POLG. Given the critical role of the p55 subunit in pol γ function in vitro, we sequenced the eight coding exons and adjacent intronic regions of POLG2 in this patient, as well as in 100 other patients with PEO and multiple mtDNA deletions in skeletal muscle and who were known to not have mutations in ANT1, C10orf2, or POLG. A single heterozygous transition (1352G→A) was identified in the patient described above, whereas her two unaffected sisters did not harbor the mutation (fig. 2A and 2B). The mutation was not detected in 288 control chromosomes from the same geographic region by DHPLC analysis (fig. 2C). The 1352G→A mutation is predicted to alter a highly conserved glycine to glutamate at codon 451 (G451E) (fig. 2D).
Figure 2.
A novel mutation in POLG2. A, Pedigree of the proband (II-2, arrow) and her unaffected siblings. B, Sequence traces showing a heterozygous base substitution in the proband, which is absent in her unaffected sisters. C, DHPLC traces for an exon 8 amplicon from the proband (upper) and control individual (lower). D, Amino acid conservation across species in exon 8 of POLG2, with codon 451 identified in red.
Reduced Stimulation of p140 by G451E p55
Given the central role of p55 in mtDNA replication, we tested whether the phenotypic changes caused by the G451E substitution would also result in measurable biochemical defects. For comparison of the biochemical properties of WT p55 and G451E p55, both forms of the protein lacking the mitochondrial targeting sequence were overproduced in E. coli and were purified to apparent homogeneity, as described in the “Subjects and Methods” section. The catalytic activities of isolated p140 on natural DNA substrates are inhibited by physiological concentrations of salt, and association of p140 with WT p55 both stimulates polymerase activity and significantly raises the ionic strength for optimal pol γ activity.3,17 The ability of G451E p55 to affect the polymerase activity of p140 on poly(rA)-oligo(dT) was assessed at a variety of salt concentrations (fig. 3). Whereas WT p55 stimulated p140 activity almost threefold and raised the salt optimum from ∼75 mM to ∼150 mM NaCl (compare squares and triangles in fig. 3), the G451E form of p55 enhanced polymerase activity only about twofold, with merely a 25-mM increase in the salt optimum (circles in fig. 3). Neither protein was able to stimulate p140 in the absence of supplemental salt. Because stimulation and restoration of salt tolerance by p55 occur through a mechanism of enhanced DNA binding by the p140-p55 complex,3 we hypothesized that the incomplete stimulation and limited restoration of salt tolerance exhibited by G451E p55 were caused by altered DNA binding by G451E p55 and/or an impaired interaction with the p140 catalytic subunit.
Figure 3.
Effects of WT p55 and G451E p55 on pol γ activity. DNA polymerase activity was measured on poly(rA)-oligo(dT), as described in the “Subjects and Methods” section. Reactions contained 12 ng (88 fmol) p140, either alone (squares) or with 9.6 ng (178 fmol) of WT p55 (triangles) or G451E p55 (circles), and the indicated amounts of NaCl. Values are the averages of two independent measurements.
DNA Binding by G451E p55
The WT accessory subunit binds double-stranded DNA and folded single-stranded DNA.3,21 Single-stranded DNA–cellulose chromatography was used to measure the intrinsic strength with which WT p55 and G451E p55 bind to DNA (fig. 4A). As expected, WT p55 dissociated from the resin at 0.22 M NaCl (dashed line in fig. 4A). In a separate test, G451E p55 also eluted at 0.22 M NaCl, demonstrating that the mutant protein retains the ability to bind DNA with the same strength as WT p55 (solid line in fig. 4A). Elution of isolated p140 at ∼0.20 M NaCl is also shown (dotted line in fig. 4A). Since the DNA-binding properties and the chromatographic behavior during purification of G451E p55 were indistinguishable from that of WT p55, the analysis was extended to search for potential defects in subunit interaction. When purified p140 and WT p55 were mixed briefly and were applied to the DNA-cellulose column (fig. 4B), more than half of the protein in the mixture bound the resin more tightly and eluted at ∼0.32 M NaCl (dashed line in fig. 4B) in fractions 19 and 20 (fig. 4C). The enhanced DNA-binding properties of this salt-stable pol γ complex has been documented elsewhere.3 In contrast, G451E p55 and p140 failed to form a more tightly binding complex (solid line in fig. 4B). Instead, each protein in the mixture chromatographed independently, with peak elution of both proteins at 0.22 M NaCl in fractions 17 and 18 (fig. 4D). This apparent inability of the G451E p55 and p140 subunits to assemble into a pol γ holoenzyme complex prompted additional experiments to evaluate the functional and physical interactions of the subunits.
Figure 4.
Analysis of pol γ subunits by analytical single-stranded DNA–cellulose chromatography. Purified recombinant p140, WT p55, and G451E p55 were resolved by single-stranded DNA–cellulose chromatography, as described in the “Subjects and Methods” section. Elution of proteins from the column was monitored by absorbance at 280 nm. A, WT p55 (dashed line), G451E p55 (solid line), or p140 (dotted line) were applied separately and eluted with an NaCl gradient. B, The p140 catalytic subunit (29 μg) was mixed with 31 μg WT p55 (dashed line) or 27 μg G451E p55 (solid line) and was eluted as before. Samples of the indicated fractions of the p140-WT p55 profile (C) and the p140-G451E p55 profile (D) were resolved by SDS-PAGE and were stained with Coomassie Brilliant Blue, as described in the “Subjects and Methods” section. Arrows indicate the individual migration positions of p140 and p55 (C and D).
G451E p55 Exhibits Reduced Protection of p140 against Inactivation by NEM
NEM inhibits certain enzymes, including human pol γ, through covalent modification of solvent-accessible cysteine residues. Because cysteine has not been identified as a catalytic residue in any DNA polymerase, we presume that NEM inhibits pol γ by interfering with the formation of a ternary complex among the enzyme, the primer template, and the incoming dinucleotide triphosphate substrate. Association of p55 with p140 protects the catalytic subunit from inactivation by NEM,3,22 and this protective effect has been used as a chemical probe to assess association of the pol γ subunits.19 Inhibition of pol γ at different concentrations of NEM was reestablished in the standard polymerase assay, in which 1.0 mM NEM inhibited the polymerase activity of isolated p140 by ∼95% (fig. 5). Brief preincubation of p140 with a twofold molar excess of WT p55 protected >80% of polymerase activity at all concentrations of NEM that were tested. A reduced level of protection was observed on preincubation with the mutant form of p55, and protection by G451E p55 was only 65%–80% as effective as protection by WT p55 at NEM concentrations >0.1 mM. This intermediate protection of p140 against attack by NEM further supports the notion that the G451E substitution in p55 compromises association with the p140 subunit.
Figure 5.
G451E p55 only partially protects p140 against inactivation by NEM. DNA polymerase activity was measured as described in the “Subjects and Methods” section, except that 2-mercaptoethanol was excluded. Reactions contained 75 mM NaCl, 8.0 ng (58 fmol) p140—either alone (squares) or in the presence of 6.3 ng (116 fmol) WT p55 (triangles) or G451E p55 (circles)—and the indicated amounts of NEM. Values are the average of at least two independent measurements.
Impaired Subunit Interaction Revealed by Failure to Immunoprecipitate G451E Subunit
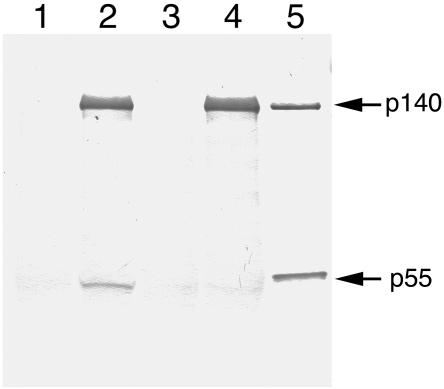
Physical association of the subunits was evaluated further by immunoprecipitation assay. Protein G Sepharose beads were loaded with polyclonal antibodies raised against purified recombinant p140, were washed thoroughly, and were used to test the ability of immobilized p140 to capture the WT and mutant forms of p55. Coimmunoprecipitation of WT p55 with p140 was straightforward, and capture of p55 was clearly dependent on the presence of p140 (fig. 6, lanes 1 and 2). However, p140 was unable to capture an appreciable quantity of G451E p55 (fig. 6, lanes 3 and 4). In separate control experiments, increasing available G451E p55 to 5 μg and increasing the amount of beads by 250% did not permit detection of G451E p55 in the immunoprecipitates (data not shown). Failure to coimmunoprecipitate G451E p55 further suggests a weakened interaction between the G451E form of p55 and the p140 subunit.
Figure 6.
G451E p55 fails to coimmunoprecipitate with p140. The p140-dependent coimmunoprecipitation of WT p55 or G451E p55 with immobilized anti-p140 rabbit antibodies was assayed as described in the “Subjects and Methods” section. Samples contained WT p55 alone (lane 1), p140 catalytic subunit and WT p55 (lane 2), G451E mutant p55 (lane 3), p140 and G451E mutant p55 (lane 4), or 0.2 μg each of p140 and p55 loaded directly as positive controls for the western blot (lane 5). The positions of p140 and p55 standards are indicated with arrows.
G451E p55 Fails to Enhance the Processivity of p140
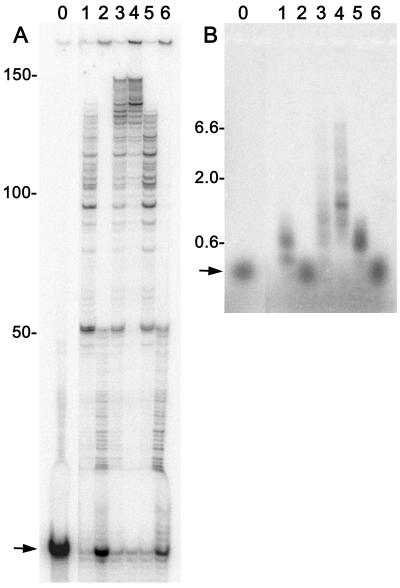
On binding productively to primer-template DNA, the pol γ catalytic subunit exhibits moderate processivity by synthesizing 50–75 nt before dissociating from the DNA.17 Association of the WT accessory subunit promotes tighter binding to DNA (fig. 4), and reduced dissociation from the primer template significantly increases the processivity of the pol γ complex to several thousand nucleotides.3 Because processivity is a sensitive functional assessment of subunit interaction during DNA synthesis, the effects of G451E p55 on the processivity of p140 were measured in gel-based primer extension assays (fig. 7). Under reaction conditions that limit DNA-binding events, the isolated p140 subunit extended the primer by 75–100 nt at 0 mM NaCl (fig. 7A, lane 1), and distinct pausing due to template secondary structure at ∼50 and ∼85 nt was apparent, as observed elsewhere.17–19 Primer extension activity of p140 was inhibited at 150 mM NaCl (fig. 7A, lane 2). Inclusion of WT p55 permitted primers to be extended by ∼150 nt at low salt concentrations (fig. 7A, lane 3). The addition of 150 mM NaCl greatly reduced polymerase stalling near the primer, and roughly half the products became too large to enter the polyacrylamide gel (fig. 7A, lane 4). In contrast, G451E p55 was unable to stimulate primer extension by p140 at either salt concentration (compare fig. 7A lanes 1 and 2 with lanes 5 and 6). Whereas physiological salt conditions are optimal for activity of the p140-WT p55 complex,3 150 mM NaCl may be high enough to mask stimulation of p140 by G451E p55. However, when the experiment was repeated at 75 mM NaCl, no stimulatory effect was observed for G451E p55 (data not shown). Similarly, inclusion of a DNA trap in the reactions to absolutely restrict pol γ to a single DNA-binding event did not change our interpretation. Reaction products were also separated on alkaline agarose gels to more fully resolve the high–molecular-weight reaction products (fig. 7B). Again, isolated p140 was more active at low salt concentrations, and the shorter products were not well resolved by this method (fig. 7B, lanes 1 and 2). When in complex with WT p55, p140 synthesized products as long as 7 kb or the full length of the M13 template (fig. 7B, lanes 3 and 4). The absence of high–molecular-weight primer extension products again demonstrates the failure of G451E p55 to enhance the processivity of p140 (fig. 7B, lanes 5 and 6).
Figure 7.
G451E p55 does not enhance processive DNA synthesis of pol γ. Primer-extension reactions were performed as described in the “Subjects and Methods” section. Reactions contained p140 catalytic subunit (lanes 1–6), WT p55 (lanes 3 and 4), G451E p55 (lanes 5 and 6), and singly primed M13 DNA. Activity was measured at 0 mM NaCl (odd-numbered lanes) or 150 mM NaCl (even-numbered lanes). Lane 0 had no enzyme. Reaction products were resolved by denaturing polyacrylamide gel electrophoresis (A) and alkaline agarose gel electrophoresis (B). The arrows in panels A and B mark the unextended 35-mer primer. Markers indicate the number of nucleotides synthesized past the primer.
The dominant inheritance pattern suggests that G451E p55 may interfere with the natural function of WT p55 in vivo. To investigate this possibility, we undertook primer-extension assays to search for a dominant negative biochemical effect in vitro. Consistent with recent work indicating that the pol γ holoenzyme may function as a heterotrimer,23,24 reactions were made to contain two p55 subunits for every p140 subunit, and the processivity of DNA synthesis was monitored as the ratio of WT p55 to G451E p55 was varied. Over a wide range of salt concentrations, the presence of G451E p55 did not diminish the stimulatory effects of WT p55 beyond that expected from simple dilution (data not shown). In separate experiments, increasing amounts of G451E p55 were titrated into standard reactions to test for inhibition of processive DNA synthesis catalyzed by p140-WT p55. An eightfold stoichiometric excess of G451E p55 had no effect on p140-WT p55 activity, suggesting that exchange of p55 subunits within the heterotrimer was not occurring in the 30-min time frame of the experiment. Although the WT component of p55 dimers within the mitochondria of an individual heterozygous for POLG2 cannot easily be determined, the failure of our mixing experiments to reveal a dominant negative effect in vitro suggests that the phenotype of the affected heterozygote may be due to simple haploinsufficiency, as discussed below.
Discussion
This work describes the first example, to our knowledge, of a pathogenic mutation in POLG2. Clinically, the patient had typical late-onset PEO, with no specific clinical features that were different from the other 100 patients who were screened for POLG2 mutations and had a WT sequence. One goal of this study was to identify biochemical defects in the G451E p55 that might help to explain the phenotypic changes caused by the G451E substitution in vivo. Biochemical characterization of the purified, recombinant G451E p55 protein in vitro revealed incomplete stimulation of the catalytic subunit and limited restoration of salt-tolerant polymerase activity, as compared with the effects of WT p55. Although G451E p55 retains a WT ability to bind DNA, it fails to enhance the DNA-binding strength of the p140-p55 complex. A weakened subunit interaction between G451E p55 and p140 was further demonstrated by intermediate protection of p140 from inactivation with NEM, by the failure of G451E p55 to coimmunoprecipitate with p140, and by the inability of G451E p55 to enhance the processivity of p140. Elsewhere, we characterized a mutant form of the pol γ catalytic subunit associated with Alpers syndrome, juvenile spinocerebellar ataxia-epilepsy syndrome, and recessive forms of PEO, and this pathogenic A467T substitution both caused a severe catalytic defect and interfered with association of the subunits.19 Since the G451E substitution in p55 disrupts only association of the subunits, we propose that any mutation that impairs assembly of the pol γ holoenzyme is sufficient to stall the mtDNA replication fork, thereby inducing mtDNA deletions and the attendant disease phenotypes. Additionally, the G451E mutation in POLG2 and pathogenic mutations in POLG offer a unique opportunity to define the p140-p55 subunit interface.
The proband in the current study is heterozygous for the G451E substitution in p55, prompting the simple assumption that haplotype insufficiency of POLG2 causes disease by reducing the availability of functional pol γ holoenzyme. Such a partial loss of holoenzyme function is consistent with the delayed onset of symptoms (at age 40 years) in the proband. In an extreme example of haplotype insufficiency that affects the bioavailability of pol γ, we recently documented a similar gene-dose effect for the pol γ holoenzyme in which homozygous patients with two A467T POLG alleles usually present with late-onset Alpers syndrome or early ataxia in their teens, whereas monoallelic expression of an A467T POLG allele was accompanied by very early onset of Alpers syndrome.25 In contrast, evidence that the mother was similarly affected suggests autosomal dominant inheritance of PEO due to the G451E mutation in POLG2. In this situation, the presence of the mutant G451E p55 should be able to interfere with the normal functions of WT pol γ holoenzyme. A dominant negative effect was not revealed by mixing experiments in vitro, which supports the notion that symptoms in affected heterozygotes are caused by haplotype insufficiency.
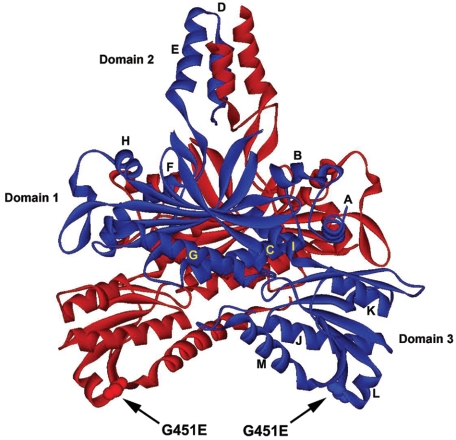
A more complex analysis involving physical and structural studies of pol γ also merits consideration. The crystal structure of the mouse pol γ accessory subunit indicates that the isolated accessory subunit exists as a homodimer23 (fig. 8). The structure shows that each monomer has three distinct domains, where domain 1 consists of a seven-stranded β-sheet and all strands except one are antiparallel. One face of the twisted β-sheet is covered with helices G and C, whereas the other solvent-accessible β-sheet forms a pocket lined with helices F and H. Domain 2 encompasses secondary structural elements that interact with their symmetric counterparts in the other monomer.23 Within this domain, three strands from each monomer form a six-stranded antiparallel β-sheet across the dimer interface. Helices D and E of each monomer form a four-helix bundle across the homodimer interface, and the helical axes are roughly parallel to the twofold axis relating both monomers. Domain 3 contains a five-stranded mixed β-sheet located between helices J and M on one side and a β-hairpin (formed by two other strands) and helix K on the other side. On the basis of the crystal structure of the mouse p55, Gly451 in the human protein is located on the inner face of domain 3 and is not involved in homodimerization of the accessory subunit (fig. 8). Accordingly, the G451E substitution in human p55 is unlikely to prevent heterodimerization of G451E p55 and WT p55. However, deletion of 30 residues near the C-terminus of p55 (deletion of amino acids 456–485) disturbs the inner face of domain 3 and prevents interaction with p140 in electrophoretic mobility shift assays,26 which suggests that this portion of p55 constitutes part of the interface with the catalytic subunit. Also, recent biochemical evidence demonstrates that the human p55 homodimer associates with the catalytic subunit to form a 1:2 heterotrimer (p140:p55:p55).24 The dissociation constant for p55 homodimers is very low (<0.1 nM).24 This predicts that G451E and WT p55 monomers did not exchange during our in vitro mixing experiments, which prevented us from measuring any dominant negative effect in vitro. Although factors affecting assembly of the pol γ holoenzyme after importation of the subunits into mitochondria are not well studied, in vivo heterodimerization of G451E and WT p55 remains a distinct possibility. By extension, only one fourth of the pol γ heterotrimers in individuals heterozygous for POLG2 would be WT, which may compromise mtDNA replication even more effectively than simple haplotype insufficiency.
Figure 8.
Homologous position of the G451E side chain on the ribbon structure of the homodimeric mouse accessory subunit. The ribbon drawing was generated with Swiss PDB viewer from Protein Data Bank file 1G5H.23 Each monomer is colored either blue or red. Amino acids in domains 1 and 2 constitute the dimer interfaces, which are far removed from the homologous position of the G451E substitution in human PEO (arrows). Helices A–M are explained in the “Discussion” section.
In summary, we have described the first pathogenic mutation in POLG2 that causes a disorder of mtDNA maintenance. The G451E mutation disrupts interaction between the accessory subunit and the catalytic subunit, ultimately causing the multiple deletions in the mtDNA of this patient. We therefore believe that POLG2 should be included when searching for nuclear genes with mutations that cause disorders of mtDNA maintenance, especially when POLG, C10orf2, ANT1, TK, DGUOK, and TP are determined to have WT sequences in affected patients.
Acknowledgments
We thank Dr. Rachelle Bienstock for generating the p55 structure figure from the Protein Data Bank file. We also thank Drs. L. Worth and M. DellaVecchia, for critical review of this manuscript, and Emma Blakely and Langping He, for the diagnostic mtDNA analysis. P.F.C. is a Wellcome Trust Senior Fellow in Clinical Science. This research was supported by The Wellcome Trust, the Association Française contre les Myopathies, the United Mitochondrial Diseases Foundation, the European Union Framework Programmes EUMitocombat and Mitocircle (to P.F.C.), and the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health.
Web Resources
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for POLG2 [accession number NM_007215])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PEOA1, PEOB, Alpers syndrome, and ataxia with peripheral neuropathy)
References
- 1.DiMauro S, Schon EA (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348:2656–2668 10.1056/NEJMra022567 [DOI] [PubMed] [Google Scholar]
- 2.Kaguni LS (2004) DNA polymerase γ, the mitochondrial replicase. Annu Rev Biochem 73:293–320 10.1146/annurev.biochem.72.121801.161455 [DOI] [PubMed] [Google Scholar]
- 3.Lim SE, Longley MJ, Copeland WC (1999) The mitochondrial p55 accessory subunit of human DNA polymerase γ enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem 274:38197–38203 10.1074/jbc.274.53.38197 [DOI] [PubMed] [Google Scholar]
- 4.Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C (2001) Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet 28:211–212 10.1038/90034 [DOI] [PubMed] [Google Scholar]
- 5.Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi G, Zeviani M (2002) Mutations of mitochondrial DNA polymerase γ are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann Neurol 52:211–219 10.1002/ana.10278 [DOI] [PubMed] [Google Scholar]
- 6.Van Goethem G, Martin JJ, Dermaut B, Lofgren A, Wibail A, Ververken D, Tack P, Dehaene I, Van Zandijcke M, Moonen M, Ceuterick C, De Jonghe P, Van Broeckhoven C (2003) Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul Disord 13:133–142 10.1016/S0960-8966(02)00216-X [DOI] [PubMed] [Google Scholar]
- 7.Naviaux RK, Nguyen KV (2004) POLG Mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann Neurol 55:706–712 10.1002/ana.20079 [DOI] [PubMed] [Google Scholar]
- 8.Van Goethem G, Luoma P, Rantamaki M, Al Memar A, Kaakkola S, Hackman P, Krahe R, Lofgren A, Martin JJ, De Jonghe P, Suomalainen A, Udd B, Van Broeckhoven C (2004) POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology 63:1251–1257 [DOI] [PubMed] [Google Scholar]
- 9.Ferrari G, Lamantea E, Donati A, Filosto M, Briem E, Carrara F, Parini R, Simonati A, Santer R, Zeviani M (2005) Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-γA. Brain 128:723–731 10.1093/brain/awh410 [DOI] [PubMed] [Google Scholar]
- 10.Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC (2005) Consequences of mutations in human DNA polymerase γ. Gene 354:125–131 10.1016/j.gene.2005.03.029 [DOI] [PubMed] [Google Scholar]
- 11.Nguyen KV, Ostergaard E, Ravn SH, Balslev T, Danielsen ER, Vardag A, McKiernan PJ, Gray G, Naviaux RK (2005) POLG mutations in Alpers syndrome. Neurology 65:1493–1495 [DOI] [PubMed] [Google Scholar]
- 12.Hudson G, Deschauer M, Taylor RW, Hanna MG, Fialho D, Schaefer AM, He L-P, Blakely E, Turnbull DM, Chinnery PF. POLG1, C10ORF2 and ANT1 mutations are uncommon in sporadic PEO with multiple mtDNA deletions. Neurology (in press) [DOI] [PubMed] [Google Scholar]
- 13.Blakely EL, He L, Taylor RW, Chinnery PF, Lightowlers RN, Schaefer AM, Turnbull DM (2004) Mitochondrial DNA deletion in “identical” twin brothers. J Med Genet 41:e19 10.1136/jmg.2003.011296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durham SE, Bonilla E, Samuels DC, DiMauro S, Chinnery PF (2005) Mitochondrial DNA copy number threshold in mtDNA depletion myopathy. Neurology 65:453–455 [DOI] [PubMed] [Google Scholar]
- 15.He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, Taylor RW, Turnbull DM (2002) Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res 30:e68 10.1093/nar/gnf067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brierley EJ, Johnson MA, Lightowlers RN, James OF, Turnbull DM (1998) Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann Neurol 43:217–223 10.1002/ana.410430212 [DOI] [PubMed] [Google Scholar]
- 17.Longley MJ, Ropp PA, Lim SE, Copeland WC (1998) Characterization of the native and recombinant catalytic subunit of human DNA polymerase γ: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry 37:10529–10539 10.1021/bi980772w [DOI] [PubMed] [Google Scholar]
- 18.Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC (2004) Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat Struct Mol Biol 11:770–776 10.1038/nsmb805 [DOI] [PubMed] [Google Scholar]
- 19.Chan SSL, Longley MJ, Copeland WC (2005) The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J Biol Chem 280:31341–31346 10.1074/jbc.M506762200 [DOI] [PubMed] [Google Scholar]
- 20.Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 21.Carrodeguas JA, Pinz KG, Bogenhagen DF (2002) DNA binding properties of human pol γ B. J Biol Chem 277:50008–50014 10.1074/jbc.M207030200 [DOI] [PubMed] [Google Scholar]
- 22.Longley MJ, Copeland WC (2002) Purification, separation and identification of the catalytic and accessory subunits of the human mitochondrial DNA polymerase. In: Copeland WC (ed) Mitochondrial DNA: methods and protocols. Vol 197 in: Methods in molecular biology. Humana Press, Totowa, NJ, pp 245–258 [Google Scholar]
- 23.Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C (2001) Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase γ, Pol γ B, functions as a homodimer. Mol Cell 7:43–54 10.1016/S1097-2765(01)00153-8 [DOI] [PubMed] [Google Scholar]
- 24.Yakubovshaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF (2006) Functional human mitochondrial DNA polymerase γ forms a heterotrimer. J Biol Chem 281:374–382 10.1074/jbc.M509730200 [DOI] [PubMed] [Google Scholar]
- 25.Chan SSL, Longley MJ, Naviaux RK, Copeland WC (2005) Mono-allelic POLG expression resulting from nonsense-mediated decay and alternative splicing in a patient with Alpers syndrome. DNA Repair 4:1381–1389 10.1016/j.dnarep.2005.08.010 [DOI] [PubMed] [Google Scholar]
- 26.Carrodeguas JA, Bogenhagen DF (2000) Protein sequences conserved in prokaryotic aminoacyl-tRNA synthetases are important for the activity of the processivity factor of human mitochondrial DNA polymerase. Nucleic Acids Res 28:1237–1244 10.1093/nar/28.5.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]