Abstract
Genetic variations in promoter sequences that alter gene expression play a prominent role in increasing susceptibility to complex diseases. Also, expression levels of APP are essentially regulated by its core promoter and 5′ upstream regulatory region and correlate with amyloid β levels in Alzheimer disease (AD) brains. Here, we systematically sequenced the proximal promoter (−766/+204) and two functional distal regions (−2634/−2159 and −2096/−1563) of APP in two independent AD series with onset ages ⩽70 years (Belgian sample, n=180; Dutch sample, n=111) and identified eight novel sequence variants. Three mutations (−118C→A, −369C→G, and −534G→A) identified only in patients with AD showed, in vitro, a nearly twofold neuron-specific increase in APP transcriptional activity, similar to what is expected from triplication of APP in Down syndrome. These mutations either abolished (AP-2 and HES-1) or created (Oct1) transcription-factor binding sites involved in the development and differentiation of neuronal systems. Also, two of these clustered in the 200-bp region (−540/−340) of the APP promoter that showed the highest degree of species conservation. The present study provides evidence that APP-promoter mutations that significantly increase APP expression levels are associated with AD.
The currently most-favored hypothesis advocates a pivotal role for the amyloid precursor protein (APP) in the molecular etiology of Alzheimer disease (AD [MIM 104300]). Clinical mutations in autosomal dominant AD were shown to increase relative concentrations of the 42-aa amyloid β (Aβ) peptide,1–3 a proteolysis product of APP, which shows increased propensity to aggregate4 and deposit in amyloid plaques in AD brains.5 Also, different missense mutations in the APP gene (APP [MIM 104760]) cause autosomal dominant early-onset familial AD6 (Alzheimer Disease & Frontotemporal Dementia Mutation Database). Standard molecular diagnostic screening of APP is currently limited to exons 16 and 17—coding, in part, for the Aβ peptide—and their flanking splice sites. However, it has not yet been excluded that genetic variation influencing transcriptional activity of APP also contributes to disease risk. Aβ peptide production depends largely on the amount of APP substrate; therefore, it is conceivable that regulation of APP transcription might indeed play an important role in AD susceptibility. In fact, several studies have identified higher levels of APP mRNA in AD brains (for review, see the work of Theuns and Van Broeckhoven7), and increased expression of APP has been correlated with Aβ deposition in brain in instances such as severe head injury.8 Perhaps the most convincing evidence came from the observation that APP triplication in patients with Down syndrome (DS) leads to an overexpression of APP9 and deposition of Aβ peptide in neuritic amyloid plaques,10 which results in a 50-year-earlier onset of AD symptoms.
APP is expressed in a variety of tissues, with the highest expression levels in neuronal cells of the CNS, and can be induced by a variety of agents, such as growth hormones and cytokines, as well as stress conditions (for review, see the work of Theuns and Van Broeckhoven7). Up-regulation of APP transcriptional activity11,12 corroborates the mRNA-expression studies,13,14 which suggests a major role for the APP promoter activity in APP expression. The proximal promoter region of APP is devoid of a functional TATA box and shows a high GC content, and transcription initiation is regulated by a strong initiator element (INR) surrounding the major transcription start site (TSS) +1.15–18 Further, APP promoter activation is mainly governed by two GC-rich elements, the −93/−82 fragment (APBβ) and the −65/−41 fragment (APBα).19 Transcriptional activation of APP can also be mediated by heat-shock factor-1 (HSF-1) binding to the heat-shock element (HSE) at position −317 after induction by numerous stress factors.20 Another transcriptional activator was mapped to −350/−366, which harbors an AP-1 binding site and flanks the GC box.21 Of further interest is the fact that members of the NFκβ/Rel family can specifically recognize two identical sequences at −2250/−2241 and −1837/−1822 in the distal promoter region of APP, referred to as “APPκβ sites.”22 Both the expression patterns and the proximal promoter region of APP are highly conserved between mammalian species (⩾80%).23–26
Linkage and association studies support the hypothesis that genetic variability at the APP locus might contribute to increased risk for late-onset AD,27–33 in the absence of coding mutations.34 These genetic data further suggest that increased susceptibility might result from genetic mutation in the 5′ regulatory region of APP. However, early on, screenings of the APP promoter in sporadic and familial early- and late-onset AD did not reveal AD-specific mutations.34–37 More-recent studies detected a +37G/C polymorphism in APP exon 1 while sequencing the −573/+125 fragment of the APP promoter in 20 individuals.38 The +37 C allele was overrepresented in patients with late-onset AD who lacked apolipoprotein E (APOE) ɛ4 alleles (frequency 17.2%), compared with elderly control individuals (10%) (odds ratio [OR] 2.08; 95% CI 1.26–3.45; adjusted for age, sex, and education). Subsequent sequencing of the −308/+124 fragment in 173 patients with late-onset AD and 840 control individuals revealed one more rare variant in control individuals, −9G/C (0.7%), that was absent in patients with AD. However, neither variant, −9G/C or +37G/C, showed allelic differences in promoter activity when tested in U-87 glioma cells with use of a reporter gene assay.
Although these initial results were disappointing, the existence of AD-related mutations that alter transcriptional activity could not yet be excluded, since only parts of the APP promoter were analyzed. The APP locus is known to be complex with several other active sites in the 5′ regulatory region, apart from the core promoter. In fact, functional elements that control activity of the human APP promoter are located in three regions: −2257/−2234, −2250/−2241, and −1837/−1822 (for review, see the work of Theuns and Van Broeckhoven7). Therefore, we engaged in a systematic analysis, in two independent early-onset AD patient and control samples, of the proximal APP promoter region, as well as two more-distal regions (−2634/−2159 and −2096/−1563) that were shown to encompass key elements contributing to high levels of APP expression in different cell types (for review, see the work of Theuns and Van Broeckhoven7).
Material and Methods
Patient and Control Groups
Patients with AD (n=180) and control individuals (n=180) from the Dutch-speaking Flanders region of Belgium were derived from a prospective study of dementia,39,40 whereas Dutch patients (n=111) and control individuals (n=270) were ascertained in a population-based study of early-onset AD in the four northern provinces of The Netherlands and in metropolitan Rotterdam. The patients were sampled during two study periods. The original sample was collected between 1980 and 198741 and was extended between 1997 and 2000 in a genetically isolated part of the area described above and with the same sampling criteria.42 Main characteristics of these study samples are summarized in table 1. Dutch patients received a probable diagnosis of AD before age 65 years. Belgian patients with AD were included if onset age was ⩽70 years; 80 patients had an onset age ⩽65 years. Clinical diagnosis of probable AD was based on consensus of at least two neurologists in the Belgian study or of a neurologist and a member of the research team in the Dutch study, in accordance with the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer’s Disease and Related Disorders Association criteria.43 For all patients, detailed data on family history of dementia in first-, second-, and third-degree relatives were collected by interviewing a next-of-kin relative of the patient. The criterion we used for classification as familial AD was the presence of at least one first-degree relative with dementia. The criteria for autosomal dominant inheritance were (1) at least three patients with clinically diagnosed AD in ⩾2 generations and (2) detailed medical records available on the clinical diagnosis of AD in at least two affected relatives.
Table 1.
Characteristics of Belgian and Dutch Early-Onset AD Study Groups
|
Belgian Sample |
Dutch Sample |
|||
| Characteristic | Subjects with AD (n=180) |
Control Subjects (n=180) |
Subjects with AD (n=111) |
Control Subjects (n=270) |
| Mean age, in years, at onset/inclusion (±SD) | 63.8 ± 5.9 | 59.2 ± 16.7 | 56.1 ± 5.5 | 57.5 ± 2.8 |
| Percentage female | 55 | 56 | 76 | 60 |
| Percentage of familial ADa | 41.5 | NA | 69 | NA |
Familial was defined as the presence of at least one affected first-degree relative. NA = not applicable.
On the basis of clinical examination, Belgian and Dutch control individuals had no neurological or psychiatric antecedents and were subjects without organic disease involving the CNS. Genomic DNA of patients was systematically screened for mutations in the coding exons of four dementia genes—PSEN1 (MIM 104311), PSEN2 (MIM 600759), MAPT (MIM 157140), and PRNP (MIM 176640)—and exons 16 and 17 of APP. We identified putative causal missense mutations in eight Belgian patients (4% of the Belgian sample; four mutations in PSEN1, three in PSEN2, and one in APP [N.B., unpublished data]), and in seven Dutch patients (6% of the Dutch sample; five mutations in PSEN1 and two in PSEN2).42,44 This study was approved by the medical ethics committee of the University of Antwerp.
Sequence Analysis of the APP 5′ Regulatory Region
The proximal promoter region of APP (−766/+204) was amplified by PCR with use of three overlapping primer sets (fig. 1): APP−766F (5′-cccccgccccgcaaaatc-3′) and APP−218R (5′-tgggcttcgtgaacagtgggagggagag-3′), APP−356F (5′-atgattcaagctcacggggacgag-3′) and APP−25R (5′-gctcagagccaggcgagtcagc-3′), and APP−99F (5′ggcggcgccgctaggggtctct-3′) and APP+204R (5′-ctccagcgcccgagccgtccag-3′). The distal promoter fragments (−2634/−2159 and −2096/−1563) were amplified using two additional primer sets: APP−2634F (5′-gacgcaatcagcagcataatca-3′) and APP−2159R (5′-ctgggaaggaggaggcaact-3′) and APP−2096F (5′-catgcttggtttaacgctctgc-3′) and APP−1563F (5′-gttcactttctgcaccacatttacc-3′). Oligonucleotide primers for PCR amplification of the APP promoter were based on GenBank accession number D87675.1. Numbering is relative to the major TSS +1 at nt 9001 in D87675.1. About 20 ng genomic DNA was amplified in a total reaction volume of 25 μl containing 10 pmol of each primer, 0.2 mM dNTPs (dATP, dCTP, and dTTP) (Amersham), 0.5 mM 7-deaza-dGTP (Amersham), 0.5 U Platinum or Titanium Taq DNA polymerase (Invitrogen), and 1× Platinum/Titanium Taq reaction buffer. Further reaction conditions were thoroughly optimized for each primer set (available on request). PCR products were screened for mutations by direct sequencing with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on the ABI3730 automated DNA sequencer (Applied Biosystems) with use of the PCR primers. Sequences were analyzed using the NovoSNP software.45 Each of the variations was confirmed by RFLP or pyrosequencing (table 2) in the respective carriers. To screen 450 age- and sex-matched healthy control individuals, we used deaza sequencing to detect the variations located between −766 and −218 and pyrosequencing assays for the remaining variations (table 2). Because of the high GC content of the APP promoter, it was necessary to perform a nested PCR with the pyrosequencing primers on the respective promoter PCR products.
Figure 1.
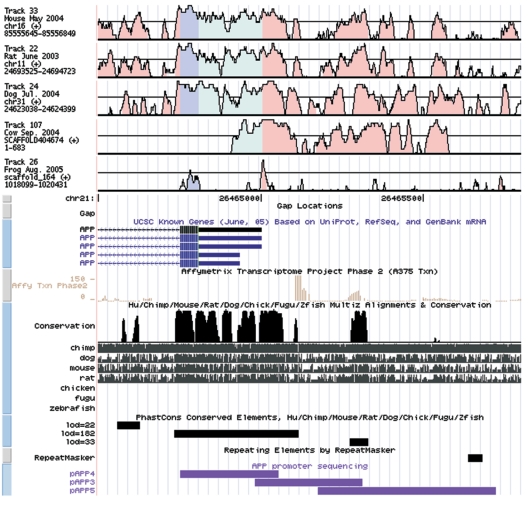
Conservation plot of the APP 5′ upstream region. Plots were generated using the VISTA software, and tracks were presented on the UCSC Genome Bioinformatics Browser.53 Conserved regions are defined as regions with a conservation score ⩾50% that are ⩾20 bp. Horizontal black lines crossing the conservation plots mark the 70% conservation boundary. Regions of high conservation are colored as exons (dark blue), UTRs (light blue), or noncoding sequences (pink).
Table 2.
Mutation Detection Assays[Note]
|
Primer(5′→3′) |
|||||
| PCR |
Pyrosequencing/Sequencing |
||||
| Variation(s) | Forward | Reverse | Forward | Reverse | Pyrosequencing |
| −2335C/T | gacgcaatcagcagcataatca | ctgggaaggaggaggcaact | bio-gatctcggctcacttcaagc | aaattagccgggcgtcgt | gtagtcccagctac |
| −1901G/T | catgcttggtttaacgctctgc | gttcactttctgcaccacatttacc | attctcctgcctcagcctct | bio-gtgaaacccccatctctactaaaaat | gctgggattacaggca |
| −1750G/A | catgcttggtttaacgctctgc | gttcactttctgcaccacatttacc | bio-cctgacctcaggtgatctgc | gcaaacgtgagaccctttgt | attattaagaattttaaggc |
| −534G/A | gaaattccaggttgctcgtg | bio-ggcgtttctggaagagaatg | gggggttaaaaaatgag | ||
| −479C/T | ctgtctcaacaagcaaagaaaatcct | bio-gtggggcaggcgtttctg | ttaagcttcactcgtt | ||
| −371G/A and −369C/G | cccccgccccgcaaaatc | tgggcttcgtgaacagtgggagggagag | cccccgccccgcaaaatc | tgggcttcgtgaacagtgggagggagag | |
| −118C/A | atgattcaagctcacggggacgag | gctcagagccaggcgagtcagc | bio-agggcgctgcacctg | ctcggcacccgagaga | gaactgcgcccgct |
| +37G/C | ggcggcgccgctaggggtctct | ctccagcgcccgagccgtccag | bio-gggctccgtcagtttcct | ccgcgtccttgctctg | gggcccccgcgca |
Note.— Variants −371G/A and −369C/G were detected by sequencing and were confirmed by StuI RFLP. For all other variations, we designed a pyrosequencing assay.
MatInspector46 was applied to investigate the effect of the variations on putative transcription-factor binding sites (TFBSs), with use of a core similarity cutoff value of 0.75 and an optimized matrix similarity threshold. Conserved sequences were detected using the VISTA tools.
Luciferase Reporter Gene Constructs
Genomic fragments of the APP proximal promoter were obtained by PCR amplification of DNA of patients or asymptomatic mutation carriers with use of the APP-766F and APP+204R primers, as described above, and were cloned into the pCR2.1-TOPO vector (Invitrogen). The integrity of all inserts was confirmed by sequence analysis with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), with use of vector-specific primers. Mutant clones were selected and consequently recloned into the promoterless pGL3 basic vector (Promega) upstream of the firefly luciferase gene, by use of KpnI and XhoI.
Transient Transfection in Cultured Mammalian Cells
Human SH-SY5Y neuroblastoma cells were propagated in a minimal essential medium with Earle’s salt, 10% fetal bovine serum, 2 mM l-glutamine, 200 IU/ml penicillin, 200 g/ml streptomycin, and 0.1 mM nonessential amino acids (Invitrogen). Human HEK293 embryonic kidney cells were propagated in OptiMem (Invitrogen), with 10% fetal bovine serum, 200 IU/ml penicillin, and 200 g/ml streptomycin. For transient transfection, SH-SY5Y and HEK293 cells were seeded in 24-well tissue-culture dishes, at 7.5×105 and 6×105 cells per well, respectively, and were allowed to recover for 24 h. Cells were cotransfected with 32 ng (HEK293) or 80 ng (SH-SY5Y) of pRL-TK plasmid that contained the herpes simplex virus thymidine kinase promoter upstream of the Renilla luciferase gene (Promega) and 800 ng of either one of the APP promoter constructs or one of the control plasmids, with use of 2.4 μl Lipofectamine 2000 (Invitrogen). Empty pGL3-basic vector was used as a negative control, and pGL3-promoter plasmid containing the SV40 early promoter upstream of the firefly luciferase gene (Promega) was used as a positive control.
Luciferase Activity
Transfected cells were cultured for 24–36 h, were washed with 1 ml PBS (Invitrogen), and were lysed with Passive lysis buffer (Promega). Firefly luciferase activities (LAF) and Renilla luciferase activities (LAR) were measured sequentially by use of a Dual-Luciferase reporter assay system (Promega) and a Veritas Microplate Luminometer with Dual Reagent Injectors Luminometer (Promega). To correct for transfection efficiency and DNA uptake, the relative luciferase activity (RLA) was calculated as RLA=LAF/LAR.
Electrophoretic Mobility–Shift Assays (EMSAs)
Nuclear factors were extracted from SH-SY5Y cells with use of the NucBuster Protein Extraction Kit (Novagen). DIG-labeled single-stranded oligonucleotides (31 bp) spanning each variant of APP (−534G→A, −369C→G, and −118C→A) were designed and HPLC purified. Blunt-ended double-stranded probes were obtained by annealing the specific oligonucleotides with their respective reverse complements and were checked on a nondenaturing 15% polyacrylamide gel in 0.25× Tris-borate-EDTA (TBE). For the binding reactions, a 200-fmol DIG-labeled double-stranded probe was added to a total reaction volume of 20 μl containing 10 μg SH-SY5Y nuclear extract, 1× binding buffer (12% glycerol, 20 mM HEPES or TRIS, 50 mM KCl, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF), and 1 μg poly (dI-dC) (Roche Applied Science). For competition assays, unlabeled double-stranded probes were added to the reaction mixture prior to addition of the labeled probe. Binding reactions were incubated at room temperature for 20 min. Protein-DNA complexes were analyzed by electrophoresis on nondenaturing 6% polyacrylamide gels in 0.25× TBE and were visualized by chemiluminescent detection with the DIG gel-shift kit (Roche Applied Science).
Real-Time PCR mRNA Quantification
mRNA was isolated from cultured lymphoblast cells of mutation carriers and control individuals by use of the mRNA Chemagic isolation system (Chemagen), and first-strand cDNA was synthesized from 300 ng of mRNA by use of the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). APP expression levels were quantified using a Taqman-MGB real-time PCR assay on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Primers and probes were designed with the PrimerExpress software (Applied Biosystems); sequences are available on request. Human APP mRNA quantities were normalized for the three housekeeping genes ubiquitine C (hUBC), β2-microglobulin (hB2M), and tyrosine 3-mono-oxygenase/tryptophan 5-mono-oxygenase activation protein, ζ polypeptide (hYWHAZ), as recommended by Vandesompele et al.47 Then, 1/20 fold-diluted cDNA was amplified in a 20-μl reaction containing 1× qPCR Mastermix Plus (no UNG [Eurogentec]), 300 nM primers, and 250 nM probe, by use of the universal amplification protocol (Applied Biosystems).
Statistical Analysis
The Mann-Whitney U test, a nonparametric analogue to the unpaired t test, was used to compare the RLA produced by the wild-type (WT) and mutant transfectants, as well as real-time PCR quantifications of APP expression levels.
Results
Variation in the 5′ Regulatory Region of APP
We sequenced the −766/+204 region of the proximal APP promoter (fig. 1) and exon 1 and two more-distal promoter regions −2634/−2159 and −2096/−1563, in two independently ascertained Dutch-speaking groups with early-onset AD composed of 180 Belgian and 111 Dutch patients (table 1). In total, we identified nine heterozygous sequence variants and confirmed their presence in the respective carriers by a second method; that is, PCR-RFLP analysis or pyrosequencing (tables 3–5).46,48 Analysis of 450 age- and sex-matched healthy control individuals (180 Belgian; 270 Dutch) demonstrated that three (33%) of the nine variants were known polymorphisms: −2335C/T (rs364091), −1901G/T (rs1235879), and +37G/C (rs459543) (table 3); the last is the same one reported by Athan and colleagues.38 Also in agreement with that study, the −9G/C variant was not observed in the patients with AD. Six (67%) of the nine variations were present in patients only (table 4). One Dutch patient, d807, whose age at onset was 50 years, was a compound heterozygote for two promoter mutations, −1750G→A and −118C→A. One other mutation, −369C→G, was identified in two familial patients, d811 (Dutch) and d1081 (Belgian), with very similar onset ages: 63 and 61 years, respectively. Haplotype analysis in these patients identified shared alleles at five neighboring microsatellite markers in the 210-kb APP region, suggestive of a potential common ancestor, although the shared haplotype had a frequency of 1% in control individuals. Another familial Dutch patient, d786, carried the −534G→A mutation. In two Belgian patients, d4605 and d5165, with ages at onset of 55 and 62 years, respectively, the −371G→A and −479C→T mutations were identified, respectively. Neither of the patients with familial AD fulfilled our criteria for autosomal dominant AD.
Table 3.
Common Sequence Variants in the APP 5′ Regulatory Region[Note]
Table 4.
Sequence Variants in the APP 5′ Regulatory Region of Patients with Early-Onset AD
| Variationa and Patient ID |
Nationality | Age at Onset (years) |
Ageb (years) |
Family History |
APOE Genotypec |
TFBS Alterationsd (Core/Matrix Similarity) |
| −1750G→Ae: | No major changes | |||||
| d807 | Dutch | 50 | *64 | − | 34 | |
| −534G→A: | +OCT1 (.89/.92) | |||||
| d768 | Dutch | 52 | *70 | + | 34 | |
| −479C→T: | −GAGA (.750/.789) and +OCT1 (.771/.843) | |||||
| d5165 | Belgian | 62 | *69 | − | 33 | |
| −371G→A: | −AP2 (.976/.916) and −STAF (.904/.799) | |||||
| d4605 | Belgian | 55 | 64 | − | 33 | |
| −369C→G: | −AP-2 (1/.92) | |||||
| d811 | Dutch | 63 | *Unknown | + | 34 | |
| d1081 | Belgian | 61 | 75 | + | 44 | |
| −118C→Ae: | −AP-2 (1/.91) and −HES-1 (.83/.92) | |||||
| d807 | Dutch | 50 | *64 | − | 34 |
Table 5.
Sequence Variants in the APP 5′ Regulatory Region of Control Individuals[Note]
To investigate to what extent genetic variability in the APP promoter was patient related, we sequenced the −766/+204 proximal promoter region in 48 control individuals. We identified only two additional rare polymorphisms, −343A/C and −375G/C, that were each present in one control individual (table 5). Also, since four of six mutations identified in patients were located in the −766/−218 region, we sequenced this 548-bp region in all 450 control individuals but did not detect any additional variants.
Transcriptional Activity of APP-Promoter Mutations
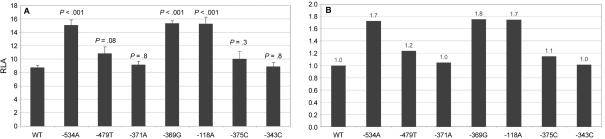
We cloned the −766/+204 APP promoter fragment that contained WT or mutant −534A, −479T, −371A, −369G, or −118A alleles into the pGL3 basic vector upstream of a firefly luciferase reporter gene. As controls, we used the −375C and −343C alleles, which were identified in control individuals. Reporter gene assay of transiently transfected human embryonic kidney (HEK293) cells showed no significant differences in expression level between (1) −534A, −479T, −371A, −369G, and −118A and (2) WT promoter sequences. Also, real-time PCR APP mRNA quantification in lymphoblasts from the mutation carriers (n=2) did not reveal significant differences in expression levels, compared with control individuals (data not shown). However, in human neuroblastoma (SH-SY5Y) cells, a significant (P<.001), nearly twofold increase in APP transcriptional activity was observed for three of the five mutant alleles identified in patients with early-onset AD: −534A, −369G, and −118A (fig. 2). The other two mutant alleles, −479T and −371A, did not significantly increase transcriptional activity (P=.08 and P=.8, respectively). Neither of the two variant alleles found in control individuals significantly altered transcriptional activity of the APP promoter in either of the studied cell types (fig. 2).
Figure 2.
Transcriptional activity of APP promoter variants. A, Bars represent firefly/Renilla luciferase ratios for the different constructs (RLA). Values are mean (±SEM) of at least eight independent measurements. The significance of differences in expression was calculated using the Mann-Whitney U test. P values are presented above the bars. B, Relative increase of APP promoter activity compared with WT.
Allele-Specific Transcription-Factor Binding
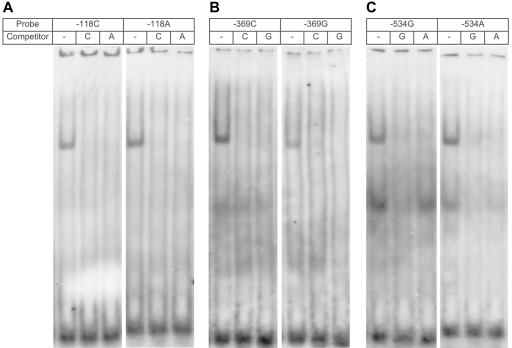
Using EMSA, we examined whether the three mutant alleles affecting APP expression (−118C→A, −369C→G, and −534G→A) interfered with the specific recognition of the APP promoter by nuclear factors extracted from SH-SY5Y cells. We used 31-bp double-stranded oligomers that contained one of six possible promoter alleles. We observed that SH-SY5Y nuclear extracts contained nuclear proteins binding specifically to all three regions of the APP promoter, which resulted in the formation of one or two major complexes (fig. 3). Competition with 50-fold excess of the respective unlabeled oligomers resulted in complete inhibition of complex formation. For −118C→A, one major complex bound to both alleles; however, it had a higher binding affinity for the mutant A allele. The major complex binding to the −369C→G probe showed a decrease in binding affinity or even a slight shift in complex mobility for the mutant G allele. Two major complexes were formed on the oligomers of the −534G→A variant; however, there were clear differences in binding affinity for the faster migrating complex, which preferentially bound to the WT G allele. Also, competition with a 50-fold excess of the cold −534A oligomer did not completely inhibit binding of the smaller complex to the −534G allele, which supports the hypothesis of higher binding affinity of this complex for the G allele.
Figure 3.
Allele-specific binding of transcription factors. EMSA analysis of allele-specific effect of −118C→A (A), −369C→G (B), and −534G→A (C) on the interaction of nuclear protein complexes extracted from SH-SY5Y cells. DIG-labeled double-stranded probes (200 fmol) were incubated with 10 μg nuclear extract from SH-SY5Y cells. In competition experiments, 50-fold excess of unlabeled probe was added before the addition of the labeled probes.
Discussion
The identification of missense mutations in APP led to the amyloid cascade hypothesis that supports a key role for increased levels of the more aggregatable Aβ42 amyloid peptide in the molecular pathogenesis of AD.1,2 The fact that triplication of APP in patients with DS also leads to AD pathology suggested that mutations in the 5′ regulatory region increasing APP transcription might exert a similar effect by increasing the levels of APP substrate and, by doing so, increase the risk of AD. Although initial studies failed to identify mutations that affected APP transcription in patients with AD, we identified six putative mutations in patients with early-onset AD that were absent from control individuals, by an extensive analysis of all sequences of the APP 5′ regulatory region that were shown elsewhere to contain functional elements. Further, luciferase reporter-gene analyses of five proximal APP promoter mutations revealed that three mutations (−534G→A, −369C→G, and −118C→A) increased APP transcriptional activity significantly by nearly twofold in cultured neuroblastoma cells, compared with two rare variants (−375G/C and −343A/C) that we observed in the same promoter segment in control individuals only. This increased promoter activity correlated with allele-specific binding of nuclear factors. The neuron-specific doubling of expression is comparable to the increased level of APP expected from the genomic APP triplication in patients with DS. Therefore, one could predict that promoter mutations also cause a critical elevation of APP in vivo in the patients with AD. We could not expand on this hypothesis, because we did not have autopsied brain material available for any of the mutation carriers, since they either were deceased without autopsy (n=4) or are still alive (n=2) (tables 3–5). In line with the absence of an elevation of APP promoter activity in nonneuronal cells, increased mRNA levels were not observed in lymphoblasts either. The reliability of our findings is, moreover, supported by the fact that all mutation carriers received a follow-up diagnosis of probable AD. Further, we observed 100% (n=55) correlation in the Belgian prospective study and 88% (n=17) correlation in the Dutch study, with pathological diagnoses for patients with probable AD (authors' unpublished data). Also, quantification by ELISA of the abundance of amyloid β, tau, and phospho-tau in the cerebrospinal fluid of two Belgian APP mutation carriers showed a decrease of Aβ42 and a slight increase of tau and phospho-tau that is typical for AD.
The observation that two proximal promoter mutations (−479C→T and −371G→A) did not significantly affect APP expression might have two explanations. These mutations are unrelated to AD and were missed from our control group because they have a population frequency <0.11% (<1 of 900 control chromosomes). Alternatively, the effect of these mutations could not be observed in vitro because of absence of other necessary regulatory elements outside the cloned promoter fragment. None of the six proximal promoter variations affected previously proven active TFBSs; however, MatInspector analysis revealed that all promoter mutations significantly altered one or more TFBS (table 4), with the exception of distal −1750G→A, which was therefore not analyzed in vitro. The −369G and −118A alleles abolish a predicted activator-protein-2 (AP-2) binding site. AP-2 is a cell type–specific transcription factor critical for neural gene expression in mammals.49 Several genes involved in CNS transmitter systems of fundamental importance for human behavior have multiple AP-2 binding sites in their 5′ regulatory regions.50 Abolition of a repressing AP-2 site in the APP promoter might significantly increase its expression. The −118A allele also abolishes a predicted HES-1 binding site. HES-1 is a mammalian basic helix-loop-helix (HLH) transcriptional repressor and a downstream target of the Notch signaling pathway. Notch signaling and HLH factors have been demonstrated to regulate numerous stages of nervous system development, including proliferation of stem and progenitor cells, differentiation of individual populations of neurons, and maintenance of mature phenotype and synaptic connectivity of neuronal circuits.51 Neuronal injury induces suppression of HES-1 expression that may lead to stimulation of neurite growth and regeneration. The −534A allele creates a binding site for the octamer binding factor 1 (Oct1), a member of the POU domain family of transcription factors. The POU proteins play essential roles in the development of highly specialized tissues, such as complex neuronal systems. More specifically, the POU domain was shown to be essential for neurite outgrowth. EMSA demonstrated that the three promoter mutations that increased APP expression in vitro showed allele-specific binding affinities. It will, however, be necessary to determine whether these particular TFBSs are active elements that, if altered by the mutation, increase APP expression. Therefore, it is of interest to mention that Lahiri et al. reported a correlation between altered TFBS affinities for two known 5′ upstream polymorphisms that alter APP expression.52 One of these variants (−1023T/C), although different from those we described (−2335C/T, −1901G/T, and +37G/C), was located in same the 87-kb linkage disequilibrium (LD) block and was suggested to be associated with late-onset AD.
Together, our sequencing efforts resulted in a 1.2% (7 of 582) (table 4) allelic frequency of APP proximal promoter variation in patients with early-onset AD, significantly higher (P=.037) than the allelic frequency of APP coding mutations—one Belgian patient with early-onset AD carried the London APP Val717Ile mutation—of 0.17% (1 of 582) in the same AD groups. Also of interest is the fact that four mutations were located in a 200-bp sequence (−540 to −340) of the proximal promoter that showed the highest degree of interspecies conservation (sequence similarity >95%) of the overall 5′ regulatory region (fig. 1). Moreover, the conservation of this 200-bp fragment was comparable to the degree of conservation observed for exon 1. Therefore, replication studies might focus their attention on this part of the 5′ regulatory region of APP in an attempt to identify AD-associated mutations. However, finding new mutations should necessarily be accompanied by reporter-gene and EMSA analyses, since the two mutations we identified in control individuals were also located in this promoter segment and did not affect APP transcriptional activity. Besides the rare mutations, we also identified three known polymorphisms, −2335C/T (rs364091), −1901G/T (rs1235879), and +37G/C (rs459543) (table 5); the last is the same as that reported by Athan et al.38 The allele frequency of 4% in control individuals was also similar to that reported for white individuals.38 All three were in complete LD and were located within the same 86-kb LD block spanning the APP 5′ upstream region, from 11 kb upstream of the initiator (rs1235889) to intron 2 (rs2830053) (International HapMap Project). Whereas the study of Athan et al.38 showed a weak association of +37G/C with late-onset AD, we did not find significant association in either the Dutch group with early-onset AD (OR 0.7; 95% CI 0.2–2.0; P=.5; adjusted for age and sex) or when stratified for APOE genotype (OR 1.6; 95% CI 0.2–14.3; P=.7). The absence of genetic association with the APP promoter in our Dutch sample with early-onset AD is not surprising, given the low number of patients carrying mutations in the APP promoter (n=6) (table 4) and since five of six patients presented with a different mutation.
It has now widely been accepted that genetic causes of susceptibility to complex diseases reflect a different spectrum of sequence variants than mutations that dominate monogenic54–56 disorders. This spectrum includes mutations that alter gene expression; in particular, promoter mutations have been shown to result in inherited diseases, including neurodegenerative brain diseases. In Parkinson disease (PD [MIM 168600]), two mutations were identified in the 5′ regulatory region of NR4A2 (MIM 601828) that cosegregated with familial PD57 and markedly reduced NR4A2 mRNA levels. Also, multiple association studies showed that variations in 5′ regulatory regions of SNCA (MIM 163890)58,59 and PARK2 (MIM 602544)60 increased PD susceptibility, with some variations increasing disease risk by modulating gene transcription. In AD, we provided evidence elsewhere that promoter mutations might explain the increased risk for early-onset AD associated with PSEN1 by decreasing expression levels of PSEN1 in neurons.61,62 Low levels of PSEN1 can lead to impairment of memory and to synaptic plasticity, followed by age-dependent neurodegeneration.63
In conclusion, our study of three functionally active sequences in the APP 5′ regulatory region with use of highly sensitive sequencing methods confirmed that the APP promoter region indeed has a very low genetic variability, with complete absence of common variants. Most importantly, our study revealed the presence of six AD-associated mutations, five of which created or abolished TFBSs in the APP promoter in six patients with early-onset AD. Three of these mutations increased APP expression in vitro in neuroblastoma cells by nearly twofold, because of differential TF binding, comparable to that observed in the case of a genomic triplication of APP, as in patients with DS. Our data are also in line with the recent report of APP locus duplication in families with autosomal dominant early-onset AD.64
The frequency of promoter mutations in our population was seven times higher than that of coding mutations in exons 16 and 17 of APP. Together, our data support a role for rare promoter mutations in increasing risk of early-onset AD. It will, however, be mandatory to analyze the APP promoter and particularly its 200-bp conserved proximal promoter fragment, in which most mutations cluster, in additional independently ascertained samples of both early- and late-onset AD.
Acknowledgments
We are grateful to the study participants for their cooperation. We also acknowledge the Genetic Service Facility. This work was supported by the Fund for Scientific Research–Flanders (FWO-F), the Medical Research Foundation Antwerp, Neurosearch Antwerp, the Interuniversity Attraction Poles program P5/19 of the Belgian Federal Science Policy Office, EU contract LSHM-CT-2003-503330 (APOPIS), and the Netherlands Organization for Scientific Research. J.T. and S.E. are postdoctoral fellows of the FWO-F. N.B. and V.B. are Ph.D. fellows of the FWO-F.
Web Resources
The accession number and URLs for data presented herein are as follows:
- Alzheimer Disease & Frontotemporal Dementia Mutation Database, http://www.molgen.ua.ac.be/ADMutations/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for APP [accession number D87675.1])
- Genetic Service Facility, http://www.vibgeneticservicefacility.be/
- International HapMap Project, http://www.hapmap.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AD, APP, PSEN1, PSEN2, MAPT, PRNP, PD, NR4A2, SNCA, and PARK2)
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for gene position and sequence information)
- VISTA, http://www-gsd.lbl.gov/vista/index.shtml (for detection of conserved sequences
References
- 1.Suzuki N, Cheung TT, Cai X-D, Odaka A, Otvos L Jr, Eckman C, Golde TE, Younkin SG (1994) An increased percentage of long amyloid β protein secreted by familial amyloid β protein precursor (βAPP717) mutants. Science 264:1336–1340 [DOI] [PubMed] [Google Scholar]
- 2.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S (1996) Secreted amyloid β-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med 2:864–870 10.1038/nm0896-864 [DOI] [PubMed] [Google Scholar]
- 3.Citron M, Westaway D, Xia W, Carlson G, Diehl T, Levesque G, Johnson-Wood K, Lee M, Seubert P, Davis A, Kholodenko D, Motter R, Sherrington R, Perry B, Yao H, Strome R, Lieberburg I, Rommens J, Kim S, Schenk D, Fraser P, St George H, Selkoe DJ (1997) Mutant presenilins of Alzheimer’s disease increase production of 42-residue amyloid beta-protein in both transfected cells and transgenic mice. Nat Med 3:67–72 10.1038/nm0197-67 [DOI] [PubMed] [Google Scholar]
- 4.Burdick D, Soreghan B, Kwon M, Kosmoski J, Knauer M, Henschen A, Yates J, Cotman C, Glabe C (1992) Assembly and aggregation properties of synthetic Alzheimer’s A4/beta amyloid peptide analogs. J Biol Chem 267:546–554 [PubMed] [Google Scholar]
- 5.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y (1994) Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43). Neuron 13:45–53 10.1016/0896-6273(94)90458-8 [DOI] [PubMed] [Google Scholar]
- 6.Goate A, Chartier-Harlin M-C, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349:704–706 10.1038/349704a0 [DOI] [PubMed] [Google Scholar]
- 7.Theuns J, Van Broeckhoven C (2000) Transcriptional regulation of Alzheimer’s disease genes: implications for susceptibility. Hum Mol Genet 9:2383–2394 10.1093/hmg/9.16.2383 [DOI] [PubMed] [Google Scholar]
- 8.Gentleman SM, Nash MJ, Sweeting CJ, Graham DI, Roberts GW (1993) Beta-amyloid precursor protein (beta APP) as a marker for axonal injury after head injury. Neurosci Lett 160:139–144 10.1016/0304-3940(93)90398-5 [DOI] [PubMed] [Google Scholar]
- 9.Rumble B, Retallack R, Hilbich C, Simms G, Multhaup G, Martins R, Hockey A, Montgomery P, Beyreuther K, Masters CL (1989) Amyloid A4 protein and its precursor in Down’s syndrome and Alzheimer’s disease. N Engl J Med 320:1446–1452 [DOI] [PubMed] [Google Scholar]
- 10.Wisniewski KE, Dalton AJ, Crapper-McLachlan DR, Wen GY, Wisniewski HM (1985) Alzheimer’s disease in Down’s syndrome: clinicopathologic studies. Neurology 35:957–961 [DOI] [PubMed] [Google Scholar]
- 11.Siman R, Card JP, Nelson RB, Davis LG (1989) Expression of beta-amyloid precursor protein in reactive astrocytes following neuronal damage. Neuron 3:275–285 10.1016/0896-6273(89)90252-3 [DOI] [PubMed] [Google Scholar]
- 12.Sola C, Garcia-Ladona FJ, Mengod G, Probst A, Frey P, Palacios JM (1993) Increased levels of the Kunitz protease inhibitor-containing beta APP mRNAs in rat brain following neurotoxic damage. Brain Res Mol Brain Res 17:41–52 10.1016/0169-328X(93)90071-V [DOI] [PubMed] [Google Scholar]
- 13.Wirak DO, Bayney R, Kundel CA, Lee A, Scangos GA, Trapp BD, Unterbeck AJ (1991) Regulatory region of human amyloid precursor protein (APP) gene promotes neuron-specific gene expression in the CNS of transgenic mice. EMBO J 10:289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahiri DK, Nall C (1995) Promoter activity of the gene encoding the beta-amyloid precursor protein is up-regulated by growth factors, phorbol ester, retinoic acid and interleukin-1. Brain Res Mol Brain Res 32:233–240 10.1016/0169-328X(95)00078-7 [DOI] [PubMed] [Google Scholar]
- 15.Salbaum JM, Weideman A, Lemaire H-G, Masters CL, Beyreuther K (1988) The promoter of Alzheimer’s disease amyloid A4 precursor gene. EMBO J 7:2807–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Fauci G, Lahiri DK, Salton SRJ, Robakis NK (1989) Characterization of the 5′-end region and the first two exons of the β-protein precursor gene. Biochem Biophys Res Commun 159:297–304 10.1016/0006-291X(89)92437-6 [DOI] [PubMed] [Google Scholar]
- 17.Yoshikai S, Sasaki H, Doh-ura K, Furuya H, Sakaki Y (1990) Genomic organization of the human amyloid beta-protein precursor gene. Gene 87:257–263 10.1016/0378-1119(90)90310-N [DOI] [PubMed] [Google Scholar]
- 18.Quitschke WW, Matthews JP, Kraus RJ, Vostrov AA (1996) The initiator element and proximal upstream sequences affect transcriptional activity and start site selection in the amyloid β- protein precursor promoter. J Biol Chem 271:22231–22239 10.1074/jbc.271.36.22231 [DOI] [PubMed] [Google Scholar]
- 19.Pollwein P, Masters CL, Beyreuther K (1992) The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res 20:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dewji NN, Do C (1996) Heat shock factor-1 mediates the transcriptional activation of Alzheimer’s β-amyloid precursor protein gene in response to stress. Brain Res Mol Brain Res 35:325–328 10.1016/0169-328X(95)00214-D [DOI] [PubMed] [Google Scholar]
- 21.Querfurth HW, Jiang J, Xia W, Selkoe DJ (1999) Enhancer function and novel DNA binding protein activity in the near upstream βAPP gene promoter. Gene 232:125–141 10.1016/S0378-1119(99)00091-8 [DOI] [PubMed] [Google Scholar]
- 22.Grilli M, Ribola M, Alberici A, Valerio A, Memo M, Spano P (1995) Identification and characterization of a κ B/Rel binding site in the regulatory region of the amyloid precursor protein gene. J Biol Chem 270:26774–26777 10.1074/jbc.270.45.26774 [DOI] [PubMed] [Google Scholar]
- 23.Yamada T, Sasaki H, Dohura K, Goto I, Sakaki Y (1989) Structure and expression of the alternatively-spliced forms of mRNA for the mouse homolog of Alzheimer’s disease amyloid beta protein precursor. Biochem Biophys Res Commun 158:906–912 10.1016/0006-291X(89)92808-8 [DOI] [PubMed] [Google Scholar]
- 24.Izumi R, Yamada T, Yoshikai S, Sasaki H, Hattori M, Sakaki Y (1992) Positive and negative regulatory elements for the expression of the Alzheimer’s disease amyloid precursor-encoding gene in mouse. Gene 112:189–195 10.1016/0378-1119(92)90375-Y [DOI] [PubMed] [Google Scholar]
- 25.Chernak JM (1993) Structural features of the 5′ upstream regulatory region of the gene encoding rat amyloid precursor protein. Gene 133:255–260 10.1016/0378-1119(93)90648-M [DOI] [PubMed] [Google Scholar]
- 26.Song W, Lahiri DK (1998) Functional identification of the promoter of the gene encoding the Rhesus monkey β-amyloid precursor protein. Gene 217:165–176 10.1016/S0378-1119(98)00340-0 [DOI] [PubMed] [Google Scholar]
- 27.Pericak-Vance MA, Bebout JL, Gaskell PC Jr, Yamaoka LH, Hung WY, Alberts MJ, Walker AP, Bartlett RJ, Haynes CA, Welsh KA, Earl NL, Heyman A, Clark CM, Roses AD (1991) Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet 48:1034–1050 [PMC free article] [PubMed] [Google Scholar]
- 28.Kehoe P, Wavrant-De VF, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ (1999) A full genome scan for late onset Alzheimer’s disease. Hum Mol Genet 8:237–245 10.1093/hmg/8.2.237 [DOI] [PubMed] [Google Scholar]
- 29.Wavrant-De Vrieze F, Crook R, Holmans P, Kehoe P, Owen MJ, Williams J, Roehl K, Laliiri DK, Shears S, Booth J, Wu W, Goate A, Chartier-Harlin MC, Hardy J, Perez-Tur J (1999) Genetic variability at the amyloid-β precursor protein locus may contribute to the risk of late-onset Alzheimer’s disease. Neurosci Lett 269:67–70 10.1016/S0304-3940(99)00417-6 [DOI] [PubMed] [Google Scholar]
- 30.Olson JM, Goddard KAB, Dudek DM (2001) The amyloid precursor protein locus and very-late-onset Alzheimer disease. Am J Hum Genet 69:895–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson JM, Goddard KAB, Dudek DM (2002) A second locus for very-late-onset Alzheimer disease: a genome scan reveals linkage to 20p and epistasis between 20p and the amyloid precursor protein region. Am J Hum Genet 71:154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, et al (2002) Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet 114:235–244 10.1002/ajmg.10183 [DOI] [PubMed] [Google Scholar]
- 33.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE (2003) Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet 12:23–32 10.1093/hmg/ddg007 [DOI] [PubMed] [Google Scholar]
- 34.Liddell MB, Bayer AJ, Owen MJ (1995) No evidence that common allelic variation in the amyloid precursor protein (APP) gene confers susceptibility to Alzheimer’s disease. Hum Mol Genet 4:853–858 [DOI] [PubMed] [Google Scholar]
- 35.Fidani L, Rooke K, Chartier-Harlin MC, Hughes D, Tanzi R, Mullan M, Roques P, Rossor M, Hardy J, Goate A (1992) Screening for mutations in the open reading frame and promoter of the β-amyloid precursor protein gene in familial Alzheimer’s disease: identification of a further family with APP717 Val→Ile. Hum Mol Genet 1:165–168 [DOI] [PubMed] [Google Scholar]
- 36.Rooke K, Goate A, Fidani L, Mullan M, Roques P, Rossor M, Hardy J, Chartier-Harlin MC (1992) Screening of the promoter and the β-amyloid sequence of the APP gene for polymorphisms in families with late onset Alzheimer’s disease. Neurodegeneration 1:237–240 [Google Scholar]
- 37.Rogaev EI, Lukiw WJ, Vaula G, Haines JL, Rogaeva EA, Tsuda T, Alexandrova N, Liang Y, Mortilla M, Amaducci L (1993) Analysis of the c-FOS gene on chromosome 14 and the promoter of the amyloid precursor protein gene in familial Alzheimer’s disease. Neurology 43:2275–2279 [DOI] [PubMed] [Google Scholar]
- 38.Athan ES, Lee JH, Arriaga A, Mayeux RP, Tycko B (2002) Polymorphisms in the promoter of the human APP gene: functional evaluation and allele frequencies in Alzheimer disease. Arch Neurol 59:1793–1799 10.1001/archneur.59.11.1793 [DOI] [PubMed] [Google Scholar]
- 39.Engelborghs S, Dermaut B, Goeman J, Saerens J, Marien P, Pickut BA, Van den Broeck M, Serneels S, Cruts M, Van Broeckhoven C, De Deyn PP (2003) Prospective Belgian study of neurodegenerative and vascular dementia: APOE genotype effects. J Neurol Neurosurg Psychiatry 74:1148–1151 10.1136/jnnp.74.8.1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engelborghs S, Dermaut B, Marien P, Symons A, Vloeberghs E, Maertens K, Somers N, Goeman J, Rademakers R, Van den Broeck M, Pickut B, Cruts M, Van Broeckhoven C, De Deyn PP (2006) Dose dependent effect of APOE ε4 on behavioral symptoms in frontal lobe dementia. Neurobiol Aging 27:285–292 10.1016/j.neurobiolaging.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 41.Hofman A, Schulte W, Tanja TA, van Duijn CM, Haaxma R, Lameris AJ, Otten VM, Saan RJ (1989) History of dementia and Parkinson’s disease in 1st-degree relatives of patients with Alzheimer’s disease. Neurology 39:1589–1592 [DOI] [PubMed] [Google Scholar]
- 42.Dermaut B, Croes EA, Rademakers R, Van den Broeck M, Cruts M, Hofman A, van Duijn CM, Van Broeckhoven C (2003) PRNP Val129 homozygosity increases risk for early-onset Alzheimer’s disease. Ann Neurol 53:409–412 10.1002/ana.10507 [DOI] [PubMed] [Google Scholar]
- 43.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34:939–944 [DOI] [PubMed] [Google Scholar]
- 44.Cruts M, van Duijn CM, Backhovens H, Van den Broeck M, Wehnert A, Serneels S, Sherrington R, Hutton M, Hardy J, St George-Hyslop PH, Hofman A, Van Broeckhoven C (1998) Estimation of the genetic contribution of presenilin-1 and -2 mutations in a population-based study of presenile Alzheimer disease. Hum Mol Genet 7:43–51 10.1093/hmg/7.1.43 [DOI] [PubMed] [Google Scholar]
- 45.Weckx S, Del Favero J, Rademakers R, Claes L, Cruts M, De Jonghe P, Van Broeckhoven C, De Rijk P (2005) novoSNP, a novel computational tool for sequence variation discovery. Genome Res 15:436–442 10.1101/gr.2754005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034.01–0034.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brouwers N, Sleegers K, Engelborghs S, Bogaerts V, van Duijn CM, De Deyn PP, Van Broeckhoven C, Dermaut B (2006) The UBQLN1 polymorphism, UBQ-8i, at 9q22 is not associated with Alzheimer’s disease with onset before 70 years. Neurosci Lett 392:72–74 10.1016/j.neulet.2005.08.064 [DOI] [PubMed] [Google Scholar]
- 49.Mitchell PJ, Timmons PM, Hebert JM, Rigby PW, Tjian R (1991) Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev 5:105–119 [DOI] [PubMed] [Google Scholar]
- 50.Damberg M, Ekblom J, Oreland L (2000) Chronic pharmacological treatment with certain antidepressants alters the expression and DNA-binding activity of transcription factor AP-2. Life Sci 68:669–678 10.1016/S0024-3205(00)00969-3 [DOI] [PubMed] [Google Scholar]
- 51.Kabos P, Kabosova A, Neuman T (2002) Neuronal injury affects expression of helix-loop-helix transcription factors. Neuroreport 13:2385–2388 10.1097/00001756-200212200-00002 [DOI] [PubMed] [Google Scholar]
- 52.Lahiri DK, Ge YW, Maloney B, Wavrant-De Vrieze F, Hardy J (2005) Characterization of two APP gene promoter polymorphisms that appear to influence risk of late-onset Alzheimer’s disease. Neurobiol Aging 26:1329–1341 10.1016/j.neurobiolaging.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 53.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucl Acids Res 32:W273–W279 10.1093/nar/gkh053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackay TF (2001) Quantitative trait loci in Drosophila. Nat Rev Genet 2:11–20 10.1038/35047544 [DOI] [PubMed] [Google Scholar]
- 55.Peltonen L, McKusick VA (2001) Genomics and medicine: dissecting human disease in the postgenomic era. Science 291:1224–1229 10.1126/science.291.5507.1224 [DOI] [PubMed] [Google Scholar]
- 56.Toma DP, White KP, Hirsch J, Greenspan RJ (2002) Identification of genes involved in Drosophila melanogaster geotaxis, a complex behavioral trait. Nat Genet 31:349–353 [DOI] [PubMed] [Google Scholar]
- 57.Le WD, Xu P, Jankovic J, Jiang H, Appel SH, Smith RG, Vassilatis DK (2003) Mutations in NR4A2 associated with familial Parkinson disease. Nat Genet 33:85–89 10.1038/ng1066 [DOI] [PubMed] [Google Scholar]
- 58.Kruger R, Vieira-Saecker AM, Kuhn W, Berg D, Muller T, Kuhnl N, Fuchs GA, Storch A, Hungs M, Woitalla D, Przuntek H, Epplen JT, Schols L, Riess O (1999) Increased susceptibility to sporadic Parkinson’s disease by a certain combined alpha-synuclein/apolipoprotein E genotype. Ann Neurol 45:611–617 [DOI] [PubMed] [Google Scholar]
- 59.Pals P, Lincoln S, Manning J, Heckman M, Skipper L, Hulihan M, Van den Broeck M, De Pooter T, Cras P, Crook J, Van Broeckhoven C, Farrer MJ (2004) α-Synuclein promoter confers susceptibility to Parkinson’s disease. Ann Neurol 56:591–595 10.1002/ana.20268 [DOI] [PubMed] [Google Scholar]
- 60.West AB, Maraganore D, Crook J, Lesnick T, Lockhart PJ, Wilkes KM, Kapatos G, Hardy JA, Farrer MJ (2002) Functional association of the parkin gene promoter with idiopathic Parkinson’s disease. Hum Mol Genet 11:2787–2792 10.1093/hmg/11.22.2787 [DOI] [PubMed] [Google Scholar]
- 61.Theuns J, Del-Favero J, Dermaut B, van Duijn CM, Backhovens H, Van den Broeck M, Serneels S, Corsmit E, Van Broeckhoven C, Cruts M (2000) Genetic variability in the regulatory region of presenilin 1 associated with risk for Alzheimer’s disease and variable expression. Hum Mol Genet 9:325–331 10.1093/hmg/9.3.325 [DOI] [PubMed] [Google Scholar]
- 62.Theuns J, Remacle J, Killick R, Corsmit E, Vennekens K, Huylebroeck D, Cruts M, Van Broeckhoven C (2003) Alzheimer-associated C allele of the promoter polymorphism −22C→T causes a critical neuron-specific decrease of presenilin 1 expression. Hum Mol Genet 12:869–877 10.1093/hmg/ddg098 [DOI] [PubMed] [Google Scholar]
- 63.Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ III, Kandel ER, Duff K, Kirkwood A, Shen J (2004) Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron 42:23–36 10.1016/S0896-6273(04)00182-5 [DOI] [PubMed] [Google Scholar]
- 64.Rovelet-Lecrux A, Hannequin D, Raux G, Meur NL, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D (2006) APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 38:24–26 [DOI] [PubMed] [Google Scholar]