Abstract
Cathepsin D is a ubiquitously expressed lysosomal protease that is involved in proteolytic degradation, cell invasion, and apoptosis. In mice and sheep, cathepsin D deficiency is known to cause a fatal neurodegenerative disease. Here, we report a novel disorder in a child with early blindness and progressive psychomotor disability. Two missense mutations in the CTSD gene, F229I and W383C, were identified and were found to cause markedly reduced proteolytic activity and a diminished amount of cathepsin D in patient fibroblasts. Expression of cathepsin D mutants in cathepsin D−/− mouse fibroblasts revealed disturbed posttranslational processing and intracellular targeting for W383C and diminished maximal enzyme velocity for F229I. The structural effects of cathepsin D mutants were estimated by computer modeling, which suggested larger structural alterations for W383C than for F229I. Our studies broaden the group of human neurodegenerative disorders and add new insight into the cellular functions of human cathepsin D.
Cathespin D (CatD) belongs to the pepsin family of proteases and is one of the most studied aspartic proteases. It has been implicated in diverse biological processes. CatD promotes invasion and proliferation of cancer cells1 but is also involved in caspase-independent apoptosis.2,3 Increased concentrations of CatD are found during ischemic, inflammatory, and regenerative processes, such as coronary heart disease,4 inflammatory bowel disease,5 wound healing, and epidermal differentiation.6 CatD is a major component of lysosomes and functions as a highly active endopeptidase with an optimum pH between 3.0 and 5.0. CatD preferentially cleaves peptide bonds that are flanked by bulky hydrophobic amino acids and is strongly inhibited by pepstatin A (inhibition constant Ki=1–4 nM). Human CatD is synthesized and translocated into the endoplasmic reticulum (ER) as an inactive precursor proenzyme (53 kDa), processed into an enzymatically active, intermediate proenzyme (48 kDa), and finally converted in the lysosomal compartment into a noncovalently associated two-chain form with an N-terminal 15-kDa light chain and a C-terminal 33-kDa heavy chain.7,8
Targeted disruption of the Ctsd gene in mice leads to weight loss in the 3rd wk of life that is associated with progressive atrophy of the intestinal mucosa. Massive intestinal necrosis and profound destruction of lymphocytes in the spleen and thymus are found just before the mice die in a state of anorexia at age 4 wk.9 In addition, CatD-deficient mice develop seizures and progressive retinal atrophy, which leads to blindness. Lysosomal storage of an autofluorescent material, ceroid lipofuscin, is found in neurons.10 Inactivation of the Drosophila CatD homologue causes age-dependent neurodegeneration and also progressive neuronal accumulation of autofluorescent storage material.11 In sheep, a naturally occurring missense mutation in the active-site aspartate (D293N) of ovine Ctsd results in early neurodegeneration. Because of the autofluorescent neuronal storage material, the disease was subsequently named “congenital ovine neuronal ceroid lipofuscinosis” (CONCL [MIM *116840]).12 Recently, a second natural animal model of CatD insufficiency was discovered in American bulldogs.13 Affected dogs had 36% of the CatD-specific enzymatic activity found in control dogs, and they presented with a milder phenotype than CatD-deficient mice and sheep. Similar to these animal models, all currently known human neuronal ceroid lipofuscinosis (NCL) forms are characterized by developmental regression, visual loss, and epilepsy in addition to the name-giving accumulation of autofluorescent lysosomal storage material. At present, six different disease genes have been identified for human NCL: CLN1, CLN2, CLN3, CLN5, CLN6, and CLN8.14,15
We screened a group of 25 infants and children with a nonidentified genetic cause of an NCL-like disorder and found the first case of human CatD deficiency. We determined the molecular mechanisms and functional consequences of two missense mutations in the human CTSD gene, and, in doing so, have extended our understanding of the biological functions of human CatD.
Material and Methods
Material
Cell culture medium (Dulbecco's modified Eagle medium [DMEM]), zeocin, hygromycin, Lipofectamin 2000, and Taq DNA polymerase were purchased from Invitrogen. Oligonucleotides were synthesized by MWG-Biotech. For amplification of genomic DNA fragments of CTSD, the primer combination hCDgDNA6960F (5′-GTGTAAACCGAGCCCTGATGACTT-3′) and hCDgDNA7525R (5′-CAGCAGCAGGGAGGGGGCAGCACT-3′) and the primer combination hCDgDNA10788F (5′-GGGGAGCCCCAAGGCCACCACTA-3′) and hCDgDNA11224R (5′-CTCGGCGAAGCCCACCCTGTTGTT-3′) were used. For amplification of the CTSD cDNA fragments, the primer combination hCDcDNA526F (5′-CGTGAAGAATGGTACCTCGTTTGA-3′) and hCDcDNA1319R (5′-CAGTGTAGTAGCGGCCGATGAAGAC-3′) was used. [α-32P]dCTP, [35S]methionine, and molecular-weight protein markers were obtained from Amersham Biosciences. Restriction enzymes were supplied by New England BioLabs. Goat anti-human CatD antibody and rabbit anti-mouse CatD antibody were a generous gift from Professor Kurt von Figura (University of Göttingen). Rat anti-mouse lysosomal-associated membrane protein 1 (LAMP-1) antibody was acquired from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City). Biotin SP–conjugated donkey anti-goat antibody, peroxidase-conjugated donkey anti-goat antibody, and peroxidase-conjugated streptavidin were purchased from Jackson ImmunoResearch Lab. For western blotting, peroxidase staining was done with tetramethyl benzidine (TMB) substrate (Seramun). Alexa Fluor 488 donkey anti-goat antibody and Alexa Fluor 546 goat anti-mouse and goat anti-rat antibodies were obtained from Molecular Probes. The BigDye terminator kit was used for semiautomated sequencing, in accordance with the recommendations of the manufacturer (Perkin Elmer Applied Biosystems). RNA was isolated from patient fibroblasts with the RNeasy kit as described by the manufacturer (Qiagen). Total RNA was fractionated by agarose-formaldehyde gel electrophoresis, the gel was blotted onto a Hybond-XL membrane (Amersham Biosciences), and the northern blot was hybridized in accordance with standard procedures by use of the Nick Translation System (Invitrogen) and the QuickHyb hybridization solution (Stratagene). The hybridization of RNA was performed with two independent control RNAs that showed very similar CatD expression. For cDNA synthesis, the Superscript III First-Strand Synthesis System (Invitrogen) was used. The substrate MOCAc-GKPIIFFRLK(Dnp)-R-NH2 was obtained from Calbiochem. All other reagents were purchased from Sigma-Aldrich.
The Patient
A patient with two heterozygous mutations in CTSD was identified from a total of 25 pediatric patients showing symptoms of an unidentified, NCL-like neurodegenerative disease including motor and visual disturbances. This group of patients was screened for possible NCL diseases. In patient fibroblasts, activities for palmitoyl protein thioesterase 1 and tripeptidyl peptidase 1 were normal, whereas CatD activity was clearly reduced. Sequencing the coding regions of CTSD resulted in the identification of the missense mutations g.6517T→A and g.10267G→C, whereas no pathogenic base alterations were revealed in the CLN3, CLN5, CLN6, and CLN8 genes.
After a normal early psychomotor development, the patient first showed neurodegenerative symptoms—in particular, ataxia and visual disturbances—at early-school age. The ocular fundus was found to show retinitis pigmentosa, and cranial magnetic resonance imaging (MRI) scans revealed cerebral and cerebellar atrophy. In the course of disease, she developed progressive cognitive decline, loss of speech, retinal atrophy, and loss of motor functions. Now, at age 17 years, she is wheel-chair bound and severely mentally retarded. Until now, there has been no evidence of hematological disease, immunodeficiency, intestinal abnormalities, or dystrophia. In fact, because of immobilization, the patient has become overweight during the past few years.
Expression of Wild-Type CatD and Mutants in CatD−/− Mouse Fibroblasts and Flp-In 3T3 Cells
CTSD cDNA was received from the IMAGE consortium (RZPD clone ID IRAUp969A1048D6) and was cloned into the expression vectors pcDNA3.1/Zeo and pEF5/FRT/V5-D-TOPO (Invitrogen). Site-directed mutagenesis was performed using a QuickChange site-directed mutagenesis kit (Stratagene), in accordance with the supplier’s protocol. All final constructs were verified by semiautomated sequencing.
CatD−/− mouse fibroblasts and Flp-In 3T3 cells were seeded on 35-mm plates at a density of 2×105 cells/well in DMEM supplemented with fetal calf serum (FCS) at 37°C and 5% CO2. After 24 h of culturing, the cells were transfected with 2 μg of plasmid and 5 μl of Lipofectamin 2000 in 0.5 ml of Opti-MEM. The medium was replaced with DMEM supplemented with FCS after 6 h of incubation. Selection with zeocin or hygromycin was started 48 h after transfection, and stable clones were achieved after 14 d of selection. Cellular homogenates from individual clones were assayed for CatD activity by use of the substrate MOCAc-GKPIIFFRLK(Dnp)-R-NH2, as described elsewhere.16 Alternatively, cell lysates were analyzed by western blotting with anti-human CatD antibody. The western-blotting experiments (fig. 1B and 1C) were repeated twice and led to very similar results.
Figure 1.
Expression of human mutant and wild-type CatD. A, Northern-blot analysis of CatD mRNA expression. Five micrograms of total RNA purified from control fibroblasts and patient fibroblasts was fractionated by agarose-formaldehyde gel electrophoresis, was blotted, and then was hybridized with a 32P-labeled CatD-specific probe. The amount of CatD-specific mRNA was similar for control and patient RNA, and no additional transcript was detectable in the patient. The quality of mRNA was controlled by equal expression levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). B, Immunodetection of CatD in control and patient fibroblasts. Cell homogenates (50 μg protein per lane) from control and patient human skin fibroblasts were separated by SDS-PAGE, were transferred to nitrocellulose membrane, and were incubated with goat anti-human CatD antibody, then with biotin SP–conjugated donkey anti-goat antibody, and finally with peroxidase-conjugated streptavidin. The blot was stained with TMB substrate to visualize the 53-kDa proenzyme precursor and the 33-kDa heavy chain of mature CatD. The 33-kDa band of patient cells has only 15% of the intensity of band for control cells. The level of β-actin expression is equal for both. C, Expression of human wild-type (wt) CatD and mutants in CatD−/− mouse fibroblasts. Lysates (30 μg protein per lane) from CatD−/− mouse fibroblasts stably transfected with either vector alone or constructs containing human wild-type CatD or mutant F229I and W383C CatD were subjected to SDS-PAGE, were transferred to nitrocellulose membrane, and were stained with TMB substrate after incubation with goat anti-human CatD antibody and peroxidase-conjugated donkey anti-goat antibody. No immunostaining was detectable for vector-transfected cells. All others showed immunoreactivity for the 53-kDa CatD precursor. Wild type–transfected cells had the strongest signal for the 33-kDa heavy chain of mature CatD, mutant F229I showed about 50% of the intensity of the wild type, and no 33-kDa protein was detectable for mutant W383C. The positions of the precursor proenzyme (prepro) and the heavy chain of mature CatD are indicated. D, Metabolic labeling of human wild-type (wt) and mutant CatD. Vector-transfected CatD−/− cells and CatD−/− cells transfected with human wild-type CatD or mutant F229I and W383C CatD were metabolically labeled with [35S]Met for 30 min and were chased for 30 min and 4 h in DMEM supplemented with 10% (v/v) FCS. The CatD was immunoprecipitated from cell lysate using goat anti-human CatD antibody in combination with pansorbin cells and was resolved on SDS-PAGE. Mutant F229I is incompletely processed to the 33-kDa heavy chain of the mature protease when compared with wild-type CatD, whereas mutant W383C is hardly processed at all. After addition of E64 and leupeptin (E64+Leu), the intensity of the 33-kDa heavy chain slightly decreased for all three constructs.
Kinetic Analysis of Wild-Type and Mutant CatD in Cell Lysates
For kinetic studies, 0.5 μg of cellular lysates were incubated with 1.3, 2, 3.3, 6.7, 10, 13.3, 23.3, 33.3, and 45 μM of substrate MOCAc-GKPIIFFRLK(Dnp)-R-NH2. Reaction velocities were measured at defined time intervals with a microplate reader (Synergy HT [BIO-TEK Instruments]). Linear phase velocities were plotted against the substrate concentrations; Km and Vmax were calculated by nonlinear regression using the equation V=Vmax×[S]/(Km+[S]), where V is linear phase velocity, Vmax is maximal enzyme velocity, [S] is the substrate concentration, and Km is the Michaelis-Menten constant. Data obtained from one of two independent experiments were measured in triplicate, and the means and SDs were plotted.
Metabolic Labeling and Immunoprecipitation of Wild-Type and Mutant CatD
CatD−/− mouse fibroblasts stably transfected with vector, wild-type CatD, and F229I or W383C mutant CatD were grown to 70% confluence and then were washed twice with methionine-free DMEM (Invitrogen); were incubated at 37°C for 1 h in the same medium, with or without proteinase inhibitor E64 (18 μM) and leupeptin (10 μM); and then were incubated again for 30 min in the same medium supplemented with 100 μCi/ml [35S]Met (Amersham Biosciences). The radioactive medium was removed. Cells were washed twice with Hank’s balanced salt solution and were chased in DMEM supplemented with FCS with or without proteinase inhibitor E64 (18 μM) and leupeptin (10 μM). At 30 min and 4 h, cells were washed three times with cold PBS and were solubilized in cold Tris-buffered saline with 0.1% TX-100 and a protease inhibitor cocktail. After precipitation of DNA with protaminsulfate (0.3 mg/ml), the supernatant was adapted to an immunoprecipitation buffer containing 150 mM NaCl, 10 mM NaH2PO4/K2HPO4 (pH 7.4), 0.5% Triton-X 100, 0.3% sodium deoxycholate, 0.2% SDS, 1% BSA, and proteinase inhibitor mix (Sigma-Aldrich). The supernatants were centrifuged at 13,000 g for 10 min and were incubated for 4 h with 1/10 volume of preimmune serum and Pansorbin cells (Calbiochem). The suspension was centrifuged at 20,000 g for 15 min, and the supernatant was incubated overnight with 1/20 volume of goat anti-human CatD antibody and with 1/2 volume of washed Pansorbin cells. The suspension was centrifuged again and washed three times with immunoprecipitation buffer. The proteins bound to the anti-CatD antibody were separated from Pansorbin cells by boiling in sample buffer containing 60 mM Tris (pH 6.8), 25% glycerol, 2% SDS, 14 mM 2-mercaptoethanol, and 0.1% bromphenol blue. The denatured proteins were subjected to SDS-PAGE by use of the Laemmli method. The gels were fixed in an aqueous solution of 10% acetic acid and 50% methanol, were soaked for 30 min in Amplify solution (Amersham Biosciences), were vacuum-dried at 60°C, and were analyzed by quantitative radioactive imaging. The pulse chase was performed four times; displayed is one representative experiment (fig. 1D).
Electron Microscopy
A skin biopsy specimen from the patient was taken from the axilla and was immediately fixed in 2.5% glutaraldehyde and postfixed in buffered 1% osmium tetroxide for 1 h. After two 5-min rinses in PBS, the nerve segments were dehydrated in graded concentrations of alcohol (50%, 70%, 80%, 96%, and 100%), were incubated twice for 20 min in propylene oxide, and were embedded in Araldite. Semithin sections (1 μm) were stained with toluidine blue. Thin sections, contrasted with uranylacetate and lead citrate, were examined with a Zeiss EM 10B electron microscope. Patient lymphocytes were purified from whole blood by use of Ficoll separation, were fixed in 2.5% glutaraldehyde, and then were processed as described for the skin biopsy. A total of 250 lymphocytes were examined for intracellular inclusions.
Immunofluorescent Microscopy
Flp-In 3T3 cells and CatD−/− mouse fibroblasts stably expressing wild-type or mutant CatD were grown on coverslips to 80% confluence. The cells were washed twice with PBS and were fixed with 3% paraformaldehyde in PBS for 20 min at 37°C. Cells were washed again four times with PBS and then were permeabilized and blocked with 5 mg/ml saponin in PBS for 1 h at room temperature. Subsequently, the permeabilized cells were incubated with primary antibodies, goat anti-human CatD antibody, and rabbit anti-mouse CatD or rat anti-mouse LAMP-1 antibody for 2 h at a dilution of 1:200 in PBS containing 1 mg/ml saponin. After three washes with PBS, cells were incubated with the appropriate secondary antibodies conjugated with Alexa Fluor 488 or 546 for 1 h in the dark. Flp-In 3T3 cells were costained with anti-human CatD antibody and anti-mouse CatD antibody, and CatD−/− mouse fibroblasts were costained with anti-human CatD antibody and anti-mouse LAMP-1 antibody. Finally, coverslips were mounted on object glasses with Fluoromount medium (Polysciences). The fluorescence was viewed using a Leica NTSC laser confocal microscope (Leica) in sequential scan mode. At least 50 cells per coverslip were examined under the microscope, and representative cells were photographed. The transfection and staining experiments with the Flp-In 3T3 cells and those with the CatD−/− mouse fibroblasts gave similar results; the expression level was only slightly higher in the CatD−/− mouse fibroblasts.
Sequence Alignment and Molecular Modeling
Amino acid sequences from members of the pepsin family of peptidases were retrieved from the MEROPS database and were aligned using the program CLUSTALW with the protein weight matrix GONNET. The crystal structure of CatD (Protein Data Bank ID 1LYA)8 was used as a template for modeling of the described variants. Model building was done with Swiss-PdbViewer,17 version 3.7. Energy minimizations and initial molecular dynamics simulations were performed using the AMBER 8 package.18 Molecular dynamics were calculated at 300 K with explicit solvent for up to 1.9 ns until the root mean SDs of coordinates remained constant. Images were created using the program PyMOL,19 version 0.98.
Results
Identification of a Patient Compound Heterozygous for Two Missense Mutations in the CTSD Gene
In a group of 25 infants and childs with an unidentified, NCL-like neurodegenerative phenotype, we identified a child with two mutations (g.6517T→A and g.10267G→C) in exons 5 and 9 of the CTSD gene (fig. 2). The first mutation results in the amino acid substitution F229I of the CatD precursor proenzyme (which corresponds to F165I of the mature CatD); the second mutation causes the amino acid replacement W383C (which corresponds to W319C of the mature CatD). Mutational analyses of the parents showed that mutation F229I was derived from the mother, and mutation W383C from the father. No further genetic alterations were detected in the CTSD coding sequences of the affected child and her parents. DNA analyses of 110 controls revealed neither of the two mutations.
Figure 2.
Sequencing data of the two CTSD mutations g.6517T→A and g.10267G→C. By use of genomic DNA templates and either the primer combination hCDgDNA6960F and hCDgDNA7525R or hCDgDNA10788F and hCDgDNA11224R, PCR fragments of the human CTSD gene were amplified and sequenced with both primers independently. The g.6517T→A allele was detected in the mother, the g.10267G→C allele in the father. The patient is compound heterozygous for both alleles. Amplification of CTSD cDNA fragments of the patient (with primers hCDcDNA526F and hCDcDNA1319R) disclosed the presence of the c.685T→A and c.1149G→C alleles in heterozygous form, confirming the results obtained from the sequencing of genomic DNA. The mutations c.685T→A and c.1149G→C result in missense mutations F229I and W383, respectively, of the precursor proenzyme of human CatD (pre) and correspond to amino acid exchanges F165I and W319C, respectively, of the mature human CatD (mat).
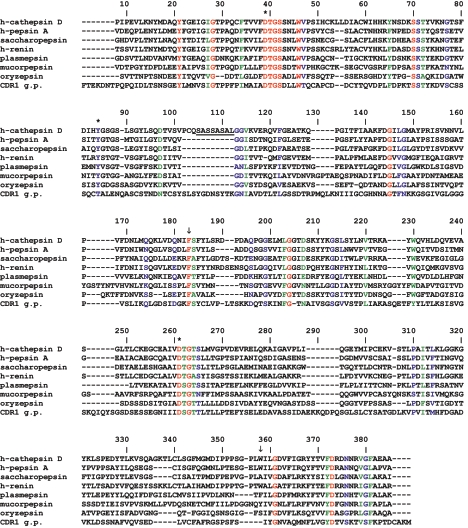
Alignment of mature human CatD with other active peptidases from humans (pepsin A and renin), fungi (saccharopepsin, mucorpepsin, and oryzepsin), sporozoa (plasmepsin), and plants (CDR1 g.p.) demonstrated highly conserved regions within the pepsin family of peptidases (fig. 3). The residue F229 (F183 in fig. 3) belongs to a group of 15 aa (of the total 389 aa positions displayed in fig. 3) that are strictly conserved among the members of this protein family. In contrast, residue W383 (W358 in fig. 3) is conserved among all 12 human pepsin peptidases and nearly all other mammalian members of this family but is not conserved within pepsin peptidases from more distantly related species.
Figure 3.
Alignment of the mature human CatD with selected active peptidases of the pepsin family. The amino acid sequence of human CatD (h-cathepsin D [aa 66–410]) was aligned with the following pepsin peptidases: human pepsin A and human renin (h-pepsin A [aa 63–388] and h-renin [aa 73–406]), saccharopepsin (aa 78–405) from the yeast Saccharomyces cerevisiae, mucorpepsin (aa 76–424) from the fungus Rhizomucor miehei, oryzepsin (aa 72–390) from the fungus Aspergillus oryzae, plasmepsin-1 (aa 126–449) from the sporozoan Plasmodium falciparum, and CDR1 g.p. (aa 70–437) from the plant Arabidopsis thaliana. The sequences were retrieved from the MEROPS database and were ordered by degree of homology. Identical amino acid residues are red, highly homologous amino acids are green, and low-homology amino acids are blue. A characteristic feature of the pepsin family is the three amino acids D39, Y84, and D261 (asterisks [*]). In the h-cathepsin D sequence, the 8-aa residues that separate the N-terminal light chain (97 aa) from the C-terminal heavy chain (244 aa) are underlined. During the maturation of human CatD, these 8 aa are cleaved off. The positions of the two human missense mutations found in the patient are indicated by arrows. F183 (F229 in the full-length h-cathepsin D sequence) is strictly conserved, W358 (W383 in the full-length h-cathepsin D sequence) is merely conserved among human peptidases of the pepsin family (shown only for h-pepsin A and h-renin).
Electron Microscopic Illustration of Storage Material in Schwann Cells
Ultrastructural examination of the skin biopsy material from the patient revealed pathological inclusions in nonmyelinated Schwann cells. Within the cytoplasm of these Schwann cells, two main types of pathological inclusions were found—namely, granular osmiophilic-like deposits and myelin-like lamellar structures (fig. 4). In comparison with the granular deposits found in patients with infantile NCL (CLN1 defect), these granular inclusions appear more heterogeneously and are less abundant within cells. The indicated myelin-like lamellar structures are less specific for NCL diseases because these inclusions are also found in other storage diseases, such as the mucopolysaccharidoses. Inclusions were not detected in patient endothelial cells, fibroblasts, and sweat glands. In addition, no storage material was identified in peripheral lymphocytes.
Figure 4.
Electron microscopy of storage material in Schwann cells from skin biopsy specimen from the patient. A, At a magnification of 20,000×, the image displays two nonmyelinated Schwann cells (left) and one myelinated Schwann cell. The two nonmyelinated Schwann cells contain various osmiophilic inclusions (arrow and arrowhead). B, Displayed details at 50,000× magnification show two types of intracellular inclusions: granular osmiophilic deposits (arrowhead) and myelin-like lamellar structures (arrow).
Reduced Stability, Delayed Processing, and Decreased Enzymatic Activity Due to Missense Mutations F229I and W383C in Human CTSD
Northern-blot analysis of patient RNA revealed levels of CatD-specific transcription similar to those in a control sample (fig. 1A). In contrast, western-blot analyses of fibroblast cell lysates from the patient showed a markedly decreased intensity of the 33-kDa band of mature human CatD (fig. 1B). The enzymatic activity measured with a fluorogenic substrate in the patient fibroblasts was decreased to 6.7% of the mean activity of 24 controls (data not shown).
To investigate the functional consequences of mutant CatD, we cloned the cDNAs of wild-type, mutant F229I, and mutant W383C CatD in expression vectors and stably expressed all three in CatD−/− mouse fibroblasts. Western-blot analyses of cell lysates from transfected cells disclosed a reduction (F229I) or absence (W383C) of mature CatD (fig. 1C). The reduced intensities of mature CatD indicate differences in processing of mutant CatD. Therefore, we metabolically labeled cells with radioactive methionine for 30 min and performed pulse-chase experiments. After the medium was exchanged with a nonradioactive one, wild-type CatD as well as mutants F229I and W383C were harvested after chase times of 30 min and 4 h, respectively. Wild-type and mutant CatD were immunoprecipitated from homogenates of transfected cells with anti-human CatD antibodies. The intensity of the 53-kDa precursor proenzyme CatD signal decreased with the length of chase time (fig. 1D). After 4 h of chase, all wild-type precursor CatD was processed to the 33-kDa heavy chain of mature CatD, ∼96% of mutant F229I precursor, but only ∼7% of mutant W383C precursor. The addition of inhibitors of thiol and serine proteases (E64 and leupeptin) resulted in increased intensity of the 53-kDa precursor and slightly decreased intensity of the 33-kDa form. The data thus indicate that the maturation of mutant F229I is slightly delayed, whereas the processing of mutant W383C is severely disturbed. Furthermore, they suggest that the stability of the mature 33-kDa CatD protein is not dependent on thiol or serine proteases.
The kinetic properties of CatD were determined for patient and control fibroblasts as well as for CatD−/− mouse fibroblasts transfected with wild-type CatD, F229I and W383C mutant CatD, and the vector. Increasing concentrations of the fluorogenic substrate MOCAc-GKPIIFFRLK(Dnp)-R-NH2 were incubated with equal amounts of cell lysates derived from the fibroblasts. The measured reaction velocity was plotted in relation to the concentration of substrate; Km and Vmax were determined by nonlinear curve fitting (fig. 5). The Km value for patient fibroblast lysates was comparable to that of control lysates; however, Vmax for patient lysates was only 7.7% of that measured for control lysates (fig. 5A). Lysates derived from CatD−/− mouse fibroblasts transfected with wild-type CatD or mutant F229I had comparable Km values as well, but Vmax for mutant F229I was only 26% of that for wild-type CatD (fig. 5B). For mutant W383C, only negligible substrate turnover could be detected, and the estimated Vmax was 0.3% of that for wild-type CatD. No substrate cleavage was detected for vector-transfected CatD−/− mouse fibroblasts (data not shown).
Figure 5.
Kinetic analysis of human CatD in patient fibroblasts and transfected CatD−/− mouse fibroblasts. The kinetic properties of human CatD were measured for patient and control lysates (A) and for lysates derived from CatD−/− mouse fibroblasts transfected with wild-type CatD (wt) and mutant F229I (B). Increasing concentrations of the fluorogenic substrate MOCAc-GKPIIFFRLK(Dnp)-R-NH2 were incubated for 20 min with 0.5 μg of cell lysates each. The increase in fluorescence was plotted in relation to the concentration of substrate, and Km and Vmax were determined by nonlinear curve fitting. The calculated Km values were similar to the reported value (Km=3.7 μM) determined with purified CatD. Vmax for patient lysates was only 7.7% of that measured for control lysates. Lysates derived from CatD−/− mouse fibroblasts transfected with mutant F229I had a Vmax of 26% when compared with those transfected with wild-type CatD.
Intracellular Localization of Human CatD Mutants
Since both missense mutations were associated with a significant decrease in enzymatic activity, we compared intracellular trafficking of these human CatD mutants with endogenous mouse CatD. We transfected 3T3 fibroblasts with the cDNA of wild-type CatD, of mutant F229I, and of mutant W383C. Single stable integration of the expression cassettes was achieved using Flp-In technology (Invitrogen). Human and mouse CatD were immunolabeled with specific antibodies and were double-stained with different secondary antibodies (fig. 6). We observed colocalization of anti-human CatD immunostaining with the anti-mouse CatD immunoreactivity in the wild-type CatD and the F229I mutant. However, the W383C mutant showed a distinct staining pattern. Only a small amount of mutant protein colocalized with mouse CatD, whereas most of the protein was detected in nonlysosomal compartments, in particular within the ER (fig. 6).
Figure 6.
Immunohistochemical localization of CatD mutants expressed in Flp-In 3T3 cells and CatD−/− mouse fibroblasts. The cells were cultivated on glass coverslips. Washed and fixed cells were costained with goat anti-human CatD antibody and secondary Alexa Fluor 488 conjugates (green) and with either rabbit anti-mouse CatD antibody or with mouse anti–protein disulfide isomerase (PDI) antibody and secondary Alexa Fluor 546 conjugates (red). Vector, No signal for human CatD (hCD); only mouse CatD (mCD) is detectable with typical pattern (red). WT, In 3T3 cells transfected with human wild-type CatD, human and mouse CatD colocalize. F229I, In 3T3 cells transfected with mutant F229I CatD, human and mouse CatD colocalize. W383C, In 3T3 cells transfected with mutant W383C, the staining pattern for human CatD (green) clearly differs from the signal for mouse CatD (red). However, minor signals show colocalization of mutant W383C with mouse CatD (arrow). W383C, In CatD−/− cells transfected with mutant W383C, the staining pattern for human CatD (green) mostly colocalizes with the ER marker (red; anti-PDI antibody). The length of the bars corresponds to 8 μm.
Corresponding experiments were performed with CatD−/− fibroblasts transfected with wild-type or mutant CatD. In this case, double-labeling was done with anti-CatD antibody and anti–LAMP-2 antibody. These studies confirmed the results obtained with transfected 3T3 fibroblasts, showing lysosomal targeting of mutant F229I but almost complete missorting of mutant W383C (fig. 6; data shown only for mutant W383C).
Discussion
We report, for the first time, that human CatD deficiency is responsible for an autosomal recessive neurodegenerative disease that can manifest in early childhood. We elucidated the underlying pathogenetic mechanisms leading to functional loss of CatD. We studied protein stability, posttranslational processing, enzymatic activity, and intracellular trafficking of mutant CatD in heterologous expression systems.
CatD belongs to the pepsin family of peptidases that presently comprises 442 known members from various species. The strict conservation of F229 (F183 in fig. 3) underlines the crucial role of this residue in protease function. Although residue W383 (W358 in fig. 3) is only conserved among human and mammalian pepsin peptidases, it also must have a central role in CatD function, because mutant W383C leads to disturbed posttranslational processing, intracellular mistargeting, and complete loss of proteolytic activity.
The results obtained by kinetic analyses of the human CatD mutants were consistent with and complementary to our findings from western blotting and pulse-chase experiments. Almost none of the 53-kDa precursor proenzyme of mutant W383C was processed to the mature 33-kDa peptidase, and hardly any enzymatic activity was detectable (fig. 1C and 1D). In contrast, mutant F229I was correctly processed, although with some delay, to an active peptidase. However, for mutant F229I, the maximal enzyme velocity, Vmax, was only 26% the amount for wild-type CatD (fig. 5B). Since Vmax=kcat×[E]total (where kcat is the catalytic rate and [E]total is the total enzyme concentration), a smaller catalytic rate as well as a diminished concentration of active enzyme may contribute to a decreased Vmax. The substantially lower relative Vmax (7.7% of control) for the patient fibroblasts can be explained by the more profoundly reduced concentration of active enzyme (figs. 1B and 5A). The second patient allele, W383C, had no significant residual enzymatic activity.
These findings were extended by experiments to investigate the subcellular localization of human wild-type and mutant CatD. Heterologous expression of human wild-type CatD, F229I mutant, and W383C mutant demonstrated colocalization with mouse CatD for the wild-type and F229I mutant CatD but major mistargeting for the W383C mutant. Together with the results obtained from western blotting, pulse-chase experiments, and kinetic studies, these data indicate a partial loss of function for mutant F229I but an almost complete loss of function for mutant W383C.
To further explain our experimental results, we predicted the conformational effects of F229I and W383C mutations on wild-type CatD by computer modeling based on the published x-ray structure of CatD8 (fig. 7). Preliminary molecular dynamics modeling suggested larger structural alterations for the W383C mutant than for the F229I mutant when compared with the wild-type CatD structure (fig. 7B). The first 11 N-terminal aa of mature CatD were shown to play a key role in pH-dependent enzymatic activation of CatD.20 In the active form of CatD, these amino acids build the first strand of the interdomain β-sheet. For neutral pH values, a translocation of this strand inserting into the active-site cleft is reported. Since F229 is located in the third strand of the interdomain β-sheet, the reduced enzymatic activity of mutant F229I might be explained by conformational destabilization of the interdomain β-sheet (fig. 7A). In addition to spatial changes, the mutation W383C may induce an alternative disulfide bond formation because it is in close proximity to residue C290, which forms a disulfide bond with C286.
Figure 7.
Structural effects of CTSD mutations. A, Cartoon structure of CatD showing the positions of mutations within the enzyme domains. The active site is painted as a blue transparent surface. N and C labels indicate the N and C termini. The interdomain β-sheet of CatD and its flanking regions are in pink. This interdomain β-sheet plays a crucial role in the regulation of the enzymatic activity of CatD. Its N-terminal strand (numbered 1) was shown to relocate at neutral pH and to insert into the active site cleft, effectively blocking substrate access and keeping CatD in an inactive state.20 The residue F165 (F [red]) is located within strand 3 of the interdomain β-sheet. The N-terminal region of strand 4 is flanked by a short α-helix (aa G322 to Y329 [light green]) and by W319 (W [green]). B, Least-squares fit for the three averaged structures of wild-type CatD (blue), F165I (green), and W319C (red). N and C labels indicate the N and C termini. There are no major structural differences between the backbone coordinates of wild-type CatD and the F165I mutant (average root mean SD 1.31 Å). The comparison of main-chain atom coordinates of wild-type CatD and those of the W319C mutant revealed higher differences (average root mean SD 2.02 Å), with a maximum deviation of ∼10.1 Å for G298 (red arrow). Note that amino acid residues are assigned on the basis of the published crystal structure,8 and these residues correspond to residues of the full-length CatD precursor proenzyme sequence as follows: F165I = F229I, G298 = G362, W319C = W383C, G322 = G386, and Y329 = Y393.
Several animal models indicate that CatD deficiency leads to fatal neurodegeneration. CatD-deficient mice and sheep exhibit cerebral atrophy and show neuronal accumulation of autofluorescent storage material—mainly granular osmiophilic deposits.10,12 Our findings are in agreement with these observations. The CatD-deficient patient described here has similar granular deposits in Schwann cells derived from skin biopsy material and shows cerebral and cerebellar atrophy on cranial MRI scans. However, whereas CatD-deficient mice and sheep die very early in the course of disease, this patient presents a slower progress of disease, as does the recently described bulldogs homozygous for the missense mutation M199I.13 This might be because of significant residual enzymatic activities measured for human mutant F229I, as well as canine mutant M199I, versus a complete loss of CatD function in mice and sheep. Therefore, one may speculate about the more severe clinical phenotypes of human CatD deficiency that would be caused by a complete loss of CatD function. The level of residual proteolytic activity in distinct CatD mutants is likely to determine the clinical phenotype of this neurodegenerative disease.
In summary, mutations in the human CTSD gene are associated with a novel autosomal recessive disorder, CatD-deficient NCL. CatD has comparable functions in humans and other mammals and seems to play a key role in the homeostasis of neuronal structures. However, the variation in phenotypic presentation of CatD deficiency between mice, sheep, dogs, and humans points to additional species-specific differences in CatD function and thus disease mechanism. The study of other cases of this novel human neurodegenerative disease will further elucidate the pathology in CatD-related neurodegeneration and will promote attempts to design therapeutic strategies for this devastating group of disorders.
Acknowledgments
We thank K. von Figura (University of Göttingen) and T. Dierks (University of Bielefeld) for the cathepsin D antibodies. We are grateful to Dirk Isbrandt (Centre of Molecular Neurobiology Hamburg), Jozsef Dudas (Department of Gastroenterology and Endocrinology, University of Göttingen), and A. Hasilik (University of Marburg) for their support. We thank T. Wilke, E. Krämer, and S. Hagen for their technical assistance. We are thankful to R. Schuh and H. Jäckle (Max-Planck-Institute for Biophysical Chemistry) for the use of the confocal microscope.
Web Resources
The URLs for data presented herein are as follows:
- MEROPS—the Peptidase Database, http://merops.sanger.ac.uk/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CONCL) [PubMed]
- Protein Data Bank, http://www.rcsb.org/pdb/
- PyMOL, http://pymol.sourceforge.net/
References
- 1.Nomura T, Katunuma N (2005) Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J Med Invest 52:1–9 10.2152/jmi.52.1 [DOI] [PubMed] [Google Scholar]
- 2.Broker LE, Kruyt FA, Giaccone G (2005) Cell death independent of caspases: a review. Clin Cancer Res 11:3155–3162 10.1158/1078-0432.CCR-04-2223 [DOI] [PubMed] [Google Scholar]
- 3.Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prebois C, Rochefort H, Vignon F (2005) Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett (http://www.sciencedirect.com/science/journal/03043835) (electronically published July 18, 2005; accessed February 24, 2006) [DOI] [PubMed] [Google Scholar]
- 4.Vivanco F, Martin-Ventura JL, Duran MC, Barderas MG, Blanco-Colio L, Darde VM, Mas S, Meilhac O, Michel JB, Tunon J, Egido J (2005) Quest for novel cardiovascular biomarkers by proteomic analysis. J Proteome Res 4:1181–1191 10.1021/pr0500197 [DOI] [PubMed] [Google Scholar]
- 5.Hausmann M, Obermeier F, Schreiter K, Spottl T, Falk W, Scholmerich J, Herfarth H, Saftig P, Rogler G (2004) Cathepsin D is up-regulated in inflammatory bowel disease macrophages. Clin Exp Immunol 136:157–167 10.1111/j.1365-2249.2004.02420.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egberts F, Heinrich M, Jensen JM, Winoto-Morbach S, Pfeiffer S, Wickel M, Schunck M, Steude J, Saftig P, Proksch E, Schutze S (2004) Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J Cell Sci 117:2295–2307 10.1242/jcs.01075 [DOI] [PubMed] [Google Scholar]
- 7.Horst M, Hasilik A (1991) Expression and maturation of human cathepsin D in baby-hamster kidney cells. Biochem J 273:355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldwin ET, Bhat TN, Gulnik S, Hosur MV, Sowder RC 2nd, Cachau RE, Collins J, Silva AM, Erickson JW (1993) Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design. Proc Natl Acad Sci USA 90:6796–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saftig P, Hetman M, Schmahl W, Weber K, Heine L, Mossmann H, Koster A, Hess B, Evers M, von Figura K, Peters C (1995) Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J 14:3599–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, Schulz-Schaeffer W, Watanabe T, Waguri S, Kametaka S, Shibata M, Yamamoto K, Kominami E, Peters C, von Figura K, Uchiyama Y (2000) Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci 20:6898–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myllykangas L, Tyynela J, Page-McCaw A, Rubin GM, Haltia MJ, Feany MB (2005) Cathepsin D-deficient Drosophila recapitulate the key features of neuronal ceroid lipofuscinoses. Neurobiol Dis 19:194–199 10.1016/j.nbd.2004.12.019 [DOI] [PubMed] [Google Scholar]
- 12.Tyynela J, Sohar I, Sleat DE, Gin RM, Donnelly RJ, Baumann M, Haltia M, Lobel P (2000) A mutation in the ovine cathepsin D gene causes a congenital lysosomal storage disease with profound neurodegeneration. EMBO J 19:2786–2792 10.1093/emboj/19.12.2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Awano T, Katz ML, O’Brien DP, Taylor JF, Evans J, Khan S, Sohar I, Lobel P, Johnson GS (2005) A mutation in the cathepsin D gene (CTSD) in American bulldogs with neuronal ceroid lipofuscinosis. Mol Genet Metab (http://www.sciencedirect.com/science/journal/10967192) (electronically published December 28, 2005; accessed February 24, 2006) [DOI] [PubMed] [Google Scholar]
- 14.Hofmann SL, Peltonen L (2001) The neuronal ceroid lipofuscinoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 3877–3894 [Google Scholar]
- 15.Mole SE (2004) The genetic spectrum of human neuronal ceroid-lipofuscinoses. Brain Pathol 14:70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K (1999) Characterization of new fluorogenic substrates for the rapid and sensitive assay of cathepsin E and cathepsin D. J Biochem (Tokyo) 125:1137–1143 (erratum 126:260) [DOI] [PubMed] [Google Scholar]
- 17.Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- 18.Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA [Google Scholar]
- 20.Lee AY, Gulnik SV, Erickson JW (1998) Conformational switching in an aspartic proteinase. Nat Struct Biol 5:866–871 10.1038/2306 [DOI] [PubMed] [Google Scholar]