Abstract
We describe four unrelated children who were referred to two tertiary referral medical genetics units between 1991 and 2005 and who are affected with juvenile polyposis of infancy. We show that these children are heterozygous for a germline deletion encompassing two contiguous genes, PTEN and BMPR1A. We hypothesize that juvenile polyposis of infancy is caused by the deletion of these two genes and that the severity of the disease reflects cooperation between these two tumor-suppressor genes.
Juvenile polyposis syndrome (JPS [MIM 174900]) is a condition, first defined in 1964 by McColl et al.,1 that predisposes to multiple juvenile hamartomatous polyps of the gastrointestinal tract. Although the presence of an isolated juvenile polyp is common in childhood, occurring in perhaps as many as 5% of all children, JPS occurs in 1 in 100,000 individuals.2 The diagnosis of JPS is often considered one of exclusion.3 Indeed, other heritable syndromes with hamartomatous polyps, such as Cowden syndrome (CS [MIM 158350]) and Bannayan-Riley-Ruvalcaba syndrome (BRRS [MIM 153480]) have to be ruled out first.2 Germline heterozygous loss-of-function mutations of the PTEN gene are identified in 85% and 65% of patients with CS and BRRS, respectively.4 Sachatello et al.5 have defined three types of juvenile polyposis: juvenile polyposis coli, generalized juvenile polyposis, and juvenile polyposis of infancy. Juvenile polyposis coli and generalized juvenile polyposis result from the variable expression of the same disease, since some cases of both forms have been reported to segregate according to a dominant mode in the same family.6 These conditions are more prevalent in the first 2 decades of life but not before age 6 years for the generalized form.7 Juvenile polyposis of infancy is characterized by its very early manifestation in the first 2 years of life and by a generalized polyposis with a severe course due to recurrent gastrointestinal bleeding, diarrhea, exudative enteropathy, and inanition. The expected lifespan of affected children is therefore limited.5 Only rare cases of juvenile polyposis of infancy have been reported in the literature. In contrast to colon-restricted or generalized juvenile polyposis, for which a family history of the disease is positive in 20%–50% of cases, individuals with juvenile polyposis of infancy do not present with a family history.5,8–12 About 50% of colon-restricted or generalized JPS cases are caused by heterozygous germline loss-of-function mutations, usually point or small size mutations, within the SMAD4 or BMPR1A gene.9–11 So far, no intragenic germline SMAD4 or BMPR1A mutations have been identified in juvenile polyposis of infancy.13 Because of the lack of family history, it was suggested that juvenile polyposis of infancy is an autosomal recessive condition.5,12 In the present study, we sought to determine whether JPS is a recessive condition or, instead, is due to de novo large gene deletions or rearrangements that result in such a severe phenotype that the shortened lifespan is incompatible with childbearing.
The four patients described herein were born from nonconsanguineous, healthy parents. They all presented in the first months of life with extensive gastrointestinal juvenile hamartomatous polyposis.
Patient 1 was a girl who presented with macrocephaly (+2 SD) at birth that was more severe (+3.5 SD) at age 3 years (table 1). She presented, in the 1st year of life, with two subcutaneous lipomas of the abdominal wall and two hemangiomas of the back. She had down-slanting palpebral fissures and a large forehead. No mental retardation was observed. The severity of gastrointestinal bleeding and diarrhea led to colectomy at age 10 mo. The child died at age 3 years because of recurrent bleeding and inanition. The extradigestive features of the disease and, especially, the macrocephaly led to the diagnosis of BRRS. A search for a germline mutation of the PTEN gene by denaturing gradient gel electrophoresis scanning of the coding sequence did not uncover any point or small size mutation. Because a de novo large deletion of the gene was suspected, a segregation analysis of genetic markers of the PTEN locus (indirect analysis) from parents to the child was performed and showed absence of paternal markers (data not shown), indicating a de novo deletion of the PTEN locus.
Table 1.
Clinical and Genetic Characteristics of Patients with Juvenile Polyposis Who Carry 10q Deletions[Note]
| Characteristic | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient of Tsuchiya et al.14 |
Patient of Jacoby et al.15 |
Patient of Arch et al.16 and Marsh et al.17 |
Patient 1 of Zigman et al.18 |
Patient 2 of Zigman et al.18 |
| Pregnancy | Normal | Normal | Normal | Normal | ? | Normal | Normal | ? | ? |
| Juvenile polyposis: | |||||||||
| Discovered by | Rectal bleeding | Rectal bleeding | Rectal bleeding | Rectal bleeding | Rectal bleeding | Rectal bleeding | Rectal bleeding | ? | ? |
| Age at onset | 1 mo | 2.5 mo | 3 mo | 18 mo | 2 years | 1 year | 18 mo | ? | ? |
| Localization | Stomach→rectum | Stomach→rectum | Stomach→rectum | Duodenum→pancolonic | Duodenum→rectum | Colona | Duodenum, colon | ? | ? |
| Histological nature | Hamartomas | Hamartomas | Hamartomas | Hamartomas | Hamartomas | Hamartomas | Hamartomas | Hamartomas | Hamartomas |
| Colectomy | Yes | Yes | Yes | No (at age 18 mo) | ? | ? | ? | + | − |
| Macrocephaly | +3.5 SD at age 3 years | +6 SD at age 3 years | +2.5 SD at birth; +2 SD at age 13 years | +4 SD | + | − | +7 SD | + | ? |
| Lipoma | + | − | − | ? | ? | ? | + | ? | ? |
| Cutaneous abnormalities | Hemangiomas | − | Hemangiomas, speckled penis | ? | + | + | + | + | ? |
| Mental retardation | − | − | − | − | + | + | + | + | + |
| Facial dysmorphism | + | + | − | + | + | + | + | ? | ? |
| Cytogenetics | 46,XX.ish del(10)(q23.2;q23.3) | 46,XX.ish del(10)(q23.2;q23.3) | 46,XY.ish del(10)(q23.2;q23.3) | 46,XX,t(2;10)(q31;p15) | del(10)(q23.2;q23.3) | del(10)(q22.3;q24.1) | del(10)(q23.2;q24.1) | der(9)t(9;10)(p24.1;q24.1),der(10)del(10)(q23.2;q24.1)t(9;10)(p24.1;q23.2) | del(10)(q23.1;q24.2) |
| Deletion of PTEN | + | + | + | + | + | + | + | + | + |
| Deletion of BMPR1A | + | + | + | + | + | Probable | Probable | Probable | − |
Note.— None of these patients had a family history of juvenile polyposis. + = positive finding; − = negative finding; ? = unknown.
Exploration of the upper digestive tract was not mentioned in the case report.
Patient 2 is a girl who presented with macrocephaly (+2 SD) at birth that was more severe (+6 SD) at age 3 years (table 1). She presented with hypertelorism, flat nasal bridge, large forehead, low-set ears, and small mouth and chin. She did not present with cutaneous abnormalities. No mental retardation was observed. The severity of bleeding and diarrhea led to colectomy at age 17 mo. The child was 4 years old at the time of the last observation. She presented with adenomatous polyps and low-grade dysplasia foci in the small bowel. A search for point and small size mutations in the coding sequences of the PTEN and SMAD4 genes had negative results. Because of the similarity of this patient's condition with the disease observed in patient 1, an indirect analysis of the PTEN locus was also performed, and a lack of maternal marker contribution was detected (data not shown), thus indicating a complete hemizygous deletion of the PTEN locus.
Patient 3 is a boy who presented with macrocephaly (+2.5 SD) at birth that was less severe at age 13 years (table 1). He presented with a speckled penis and hemangiomas of the abdominal wall. He did not present with facial dysmorphy. The severity of bleeding and diarrhea led to colectomy at age 8 years. He was 14 years old at the time of the last observation. Foci with severe dysplasia and an adenocarcinoma located in a polyp were detected in duodenum. A search for point or small size mutations in the coding sequences of the PTEN and SMAD4 genes had negative results. Because parental DNAs were not available, segregation analysis of PTEN locus markers was not performed.
Patient 4, whose case was recently reported in brief, is a girl who presented with macrocephaly in the 1st year of life (+4 SD) (table 1).19 She exhibited frontal bossing, depressed nasal bridge, and high arched palate in addition to broad thumbs and toes. Notably, this patient was found to have atrial and ventricular septal defects, with atresia of the portal vein. No mental retardation was observed. She presented with >50 juvenile polyps throughout the entire colon and duodenum by age 18 mo, which was her age at the last follow-up. A search for point and small size mutations in PTEN, BMPR1A, and SMAD4 was performed, with negative results. However, because it was known that 10% of patients found to be “negative” for PTEN mutations have large deletions of PTEN and that these patients have an excess of hamartomatous polyps, deletion analysis by quantitative PCR of each exon of PTEN was performed and revealed a whole-gene deletion (data not shown).20
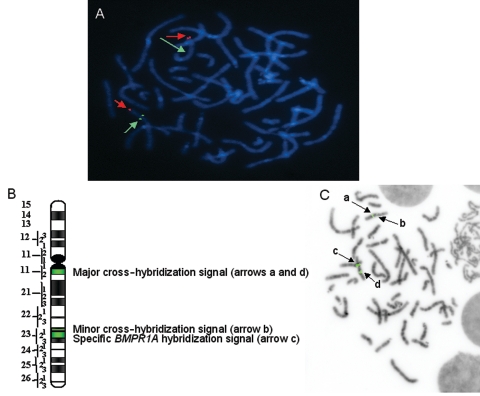
In patients 1, 2, and 3, the germline deletion of one copy of the PTEN locus was confirmed by FISH with BAC RP11-60C5, encompassing the PTEN gene (figs. 1A and 2C). In patient 3, however, the deletion was detected in only 17% of blood-circulating lymphocytes, indicating that a de novo deletion occurred in the postzygote; thus, patient 3 is mosaic for this deletion. We were not able to determine the percentage of cells carrying the PTEN deletion at the digestive-tract level, because cells from oral mucosa and the upper digestive tract were not available. Nevertheless, because of the extent and severity of the polyposis, we hypothesize that most of the cells of the gastrointestinal tract carry this deletion.
Figure 1.
Deletion of the PTEN and BMPR1A loci evidenced by FISH in patient 2. A, PTEN FISH. The hybridization of BAC RP11-60C5 (green) and control probe (YAC 889C10, 10qtel) (red) shows, in one chromosome copy, the lack of the BAC RP11-60C5 signal, indicating the heterozygous deletion of the PTEN locus. B, Scheme of BAC RP11-411A19 hybridization on chromosome arm 10q. BAC RP11-411A19 contains the BMPR1A gene. The specific signal encompassing the BRMR1A locus is located in 10q23. In addition to the specific signal, a major cross-hybridization signal was detected in 10q11.2. When the specific hybridization signal is missing, a second, minor cross-hybridization signal appears. It is due to a 25-kb region located 13 Mb centromeric to BMPR1A, which presents 96% homology with a region of BAC RP11-411A19. C, BMPR1A FISH. The major cross-hybridization signal of BAC RP11-411A19, centromeric to the specific signal, was used as control (arrows a and d). The specific signal (arrow c) appears largely decreased in one chromosome (arrow b), suggesting the complete or partial deletion of the BMPR1A gene. As mentioned above, the remaining signal is due to a second cross-hybridization (arrow b).
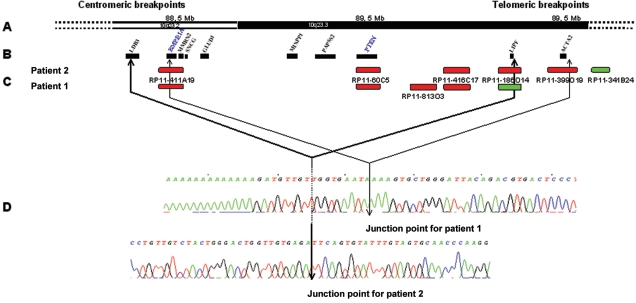
Figure 2.
Deletion breakpoints of patients 1 and 2. A, Scheme of the physical map of the 10q23 region. B, Genes located in the regions of deletion. C, Main BACs used for FISH. Deleted BACs are red; undeleted BACs are green. D, Junction points defined in patients 1 and 2. For patient 1, the proximal breakpoint is located 2.5 kb upstream of the BMPR1A coding sequence (chromosome position 88623207), and the telomeric breakpoint is located 5 kb downstream of the ACTA2 gene (chromosome position 90706778). For patient 2, the proximal breakpoint is located in intron 1 of LDB3 (chromosome position 88421964), and the telomeric breakpoint is located 19 kb downstream of the LIPF gene (chromosome position 90446243).
Since the BMPR1A gene is located on chromosome 10q23, close to the PTEN gene,9 and since the main feature of the disease associated with BMPR1A mutations is digestive polyposis, we searched in patients 1, 2, and 3 for an associated deletion of the BMPR1A gene by FISH with BAC RP11-411A19, encompassing its locus (figs. 1B, 1C, and 2C). BAC RP11-411A19 gave two signals: one at the expected BMPR1A locus on 10q23 and another on 10q11.2. The intensity of one 10q23 signal was clearly decreased in the three patients, in agreement with at least a partial deletion of the BMPR1A locus. In patient 4, the deletion of the BMPR1A gene was demonstrated using exon-by-exon quantitative PCR.19,20 In all four patients, high-resolution karyotype was normal on the 10q22-q23 region. Interestingly, however, patient 4 showed cytogenetic abnormality 46,XX,t(2;10)(q31;p15), to which cardiac abnormalities could be related.
In patient 4, the length of the deletion encompassing both PTEN and BMPR1A was estimated to be at least 1.2 Mb. The estimation was done using serial microsatellite typings and multiple serial semiquantitative PCR19 (X. P. Zhou and C. Eng, unpublished data). In patients 1 and 2, we precisely localized the breakpoints of the deletions and thus their extent by combining array-based comparative genomic hybridization (array CGH) at 1-Mb resolution, FISH, and molecular genetics21 (fig. 2). Array CGH showed that the two deletions were ∼2 Mb long and that they were different but overlapping (fig. 2A–2C). The centromeric region in both cases appeared to be located in BAC RP11-411A19. Subsequently, in patient 1, we narrowed the region of the telomeric breakpoint by FISH with BACs RP11-168O10, RP11-149I23, and RP11-341B24, which is conserved (fig. 2C). For patient 2, we used BACs RP11-813O3, RP11-416C17, and RP11-186O14, which is conserved (fig. 2C). The centromeric breakpoints were located in regions of ∼40 and 200 kb in patients 1 and 2, respectively; the telomeric breakpoints were located in regions of 120 and 220 kb. To further narrow the breakpoint regions, we used the multiplex PCR/liquid chromatography method developed by Dehainault et al.22 In this method, multiple small genomic regions of 200–300 bp are coamplified using unlabeled primers. Then, PCR products are separated by ion-pair reverse-phase high-performance liquid chromatography. They are quantified by fluorescent detection using a postcolumn intercalation dye. The relative peak intensities for each target directly reflect the copy number of each studied region. By choosing, step by step, a set of 100 pairs of primers for studying the four breakpoint regions, we were able to narrow the breakpoint regions to ∼5 kb (primer sequences available on request). Also, long-range PCR was performed for each patient by use of the forward primer of the first centromeric undeleted PCR fragment of the centromeric breakpoint region and the reverse primer of the first telomeric undeleted PCR fragment of the telomeric breakpoint region. PCR fragments of 1.4 and 1.7 kb length were obtained for patients 1 and 2, respectively (data not shown). Both fragments were directly sequenced on both DNA strands, which allowed the deletion breakpoints to be defined at the nucleotide level (fig. 2D). According to the Ensembl database (version 35), the deletion should span 2,083,571 and 2,024,279 nt for patients 1 and 2, respectively (fig. 2). The analysis of the sequences surrounding the deletion breakpoints did not give any clues with respect to the chromosomal mechanisms involved in the rearrangement. The faint hybridization signal of BAC RP11-411A19, encompassing the BMPR1A gene on the deleted chromosome, resulted from a minor cross-hybridization. Indeed, a region of 25 kb located 13 Mb centromeric to BMPR1A showed 96% homology with a region of BAC RP11-411A19 (fig. 1B and 1C).
Thus, in patient 1, from centromere to telomere, the following genes are deleted: BMPR1A, MMRN2, SNCG, GLUD1, MINPP1, PAPSS2, PTEN, LIPF, and ACTA2. In patient 2, the deletion also involves LDB3 in the centromeric region but does not include ACTA2 (fig. 2). The main features of the putative proteins corresponding to these genes are described in appendix A (online only). Moreover, the following transcribed sequences are deleted in patient 1: C10orf116, Q5VTM1, FAM35A, FAM22A, PAPSS2, ATAD1, CFLP1, C10orf59, ANKRD22, Q9P2H4, and Q8N9Z5. The same transcribed sequences, except for ANKRD22, Q9P2H4, and Q8N9Z5, are also deleted in patient 2. Although we cannot exclude the role of other genes, we hypothesize that the deletion of both BMPR1A and PTEN play a major role in the pathogenesis of the disease in the four children described here. Also, the role of MINPP1, located between PTEN and BMPR1A, deserves to be discussed. MINPP1 encodes a multiple inositol polyphosphate phosphatase that can catalyze similar reactions as PTEN in vitro, but little is known of its function in humans in vivo.23 However, the chicken homologue has been shown to play a role in the maturation and proliferation of chondrocytes,24 and, presumably, a role in cell growth and differentiation might be attributable to the function of human MINPP1. Paradoxically, the rat Minpp1 knockout model only results in rectal prolapse, without evidence of gastrointestinal polyps or other phenotypes.25 Nonetheless, we should not underestimate what the additive or synergistic role of MINPP1 may be when considered in the context of conjoint deletion of BMPR1A and PTEN.
The functions of the PTEN protein include a lipoprotein phosphatase activity that downregulates the PI3K/AKT pathway.26 The PI3K/AKT pathway plays an important part in the survival, growth, and proliferation of cells. The BMPR1A gene encodes a type 1 receptor of the binding proteins of the BMP pathway. This pathway downregulates cell proliferation, especially the proliferation of cells of the gastrointestinal tract.27,28 Thus, the deletion of both BMPR1A and PTEN should lead to increased proliferation of gastrointestinal cells, through modification of the regulation of two different cellular pathways. Moreover, we hypothesize that the joint deletion of these two genes has a greater than additive effect on the severity of the disease, leading to the phenotype of juvenile polyposis of infancy. Indeed, molecular interactions have been documented between the TGF-β/BMP and the PI3K/AKT pathways. In vitro exposure of the MCF-7 breast cancer cell line to BMP2, one of the BMPR1A ligands, induces an increase in PTEN protein levels by inhibiting its proteasome degradation.27 Furthermore, it has been shown that the BMP pathway downregulates the β-catenin protein level in intestinal stem cells through positive regulation of PTEN.28
Many contiguous-gene syndromes have been described.29 However, to our knowledge, only one contiguous-gene syndrome is associated with a severe expression of one of the diseases whose gene is deleted, thus suggesting a cooperative effect between two deleted genes. The deletion of the TSC2 and PKD1 genes leads to a severe infantile polycystic kidney disease.30 It should be noted that TSC2 is the gene involved in tuberosis sclerosis, a disease defined by the presence of hamartomas. Interestingly, a search for loss of heterozygosity at the PTEN locus in a few hamartomatous polyps of both patient 1 and patient 2 showed the retention of the remaining wild-type allele (data not shown). Although inactivation of the second allele by mutation or epigenetic event cannot be excluded, this finding favors the hypothesis of a haploinsufficient effect of PTEN and BMPR1A in the development of hamartomas, as reported elsewhere.31
The lack of loss of heterozygosity at the hamartoma level, however, does not preclude a high risk of cancer, because the loss of the second wild-type allele was presumably associated with the transformation reported in the breast tumor of a woman with CS.32 Furthermore, it is possible that the inactivation of the second allele is associated with transformation as well, as shown consistently for certain types of sporadic solid tumors, including colorectal cancers.33,34 Relatedly, the risk of colon cancer in individuals with JPS is well known, although not precisely estimated.2 This raises the question as to whether cancer risk is increased with the deletion of both BMPR1A and PTEN, compared with the cancer risk associated with JPS. The detection of grade 3 dysplasia foci in patient 2 at age 3 years and the presence of an adenocarcinoma in patient 3 at age 14 years suggest that the risk of cancer is major.
The disease observed in the four children described here corresponds to juvenile polyposis of infancy as described by Sachatello et al.5 So far, no SMAD4 or BMPR1A point or small size mutations have been identified.9–11,13 On the basis of our data, therefore, we propose that cases of juvenile polyposis of infancy are due to de novo germline deletions of a chromosomal region encompassing the PTEN and BMPR1A genes. Indeed, five cases in children carrying a deletion of the 10q23 region, identified by karyotype, have been reported elsewhere14–18 (table 1). In the case reported by Tsuchiya et al.,14 the deletion of both genes was confirmed using FISH with YAC 771B4 and YAC 738B12, which was shown to encompass the two genes. These data were obtained after publication of the case.14 For Zigman et al.’s case 2, deletion of the BMPR1A locus may be excluded.18 In the three remaining cases, the cytogenetic characteristics of the deletions make a BMPR1A deletion probable. Two children, one a carrier and the other probably a carrier of BMPR1A and PTEN deletions, were clearly described as affected with juvenile polyposis of infancy. Both diagnoses were made in the first 2 years of life on the basis of rectal bleeding. Very early onset of disease and rectal bleeding are hallmarks of juvenile polyposis of infancy as defined by Sachatello et al.5 (table 1). Moreover, similar to the four cases reported here and to the four cases presenting with a cytogenetic deletion compatible with a deletion of the PTEN and BMPR1A genes, the seven cases of juvenile polyposis of infancy reported by Sachatello et al.12 are not associated with a family history. This argues strongly for a de novo deletion in each case. It should be noted that Grotsky et al.35 described a large family with 26 members thought to be affected with juvenile polyposis. Among them, two children died before age 4 years due to diarrhea, which suggests the diagnosis of juvenile polyposis of infancy. However, data were obtained by retrospective interview, and the diagnosis of juvenile polyposis of infancy was uncertain.
The clinical features associated with BRRS (macrocephaly, facial dysmorphy, speckled penis, and early-onset lipomas) were not all present in each patient, although macrocephaly was present in our four patients (table 1). This is not unexpected, since PTEN gene inactivation may lead to either BRRS or CS. Most of the clinical features of CS are absent in childhood and occur only later in life. Furthermore, three of our four patients are female and thus cannot manifest speckled penis. The description of families affected with both CS and BRRS suggests that modifying factors influence the expression of the PTEN gene defect.17 It is thus not surprising that the clinical presentation of juvenile polyposis of infancy may alternatively suggest the diagnosis of BRRS or JPS. Of the seven patients described by Sachatello et al.,12 only two presented with macrocephaly.
In summary, we have shown that the four unrelated cases of isolated juvenile polyposis of infancy reported in the present study are due to de novo germline deletions of 10q encompassing PTEN and BMPR1A. On the basis of this observation, we suggest that this contiguous gene deletion is the etiologic basis for the subset of juvenile polyposis termed “juvenile polyposis of infancy.” To confirm our hypothesis, it would be interesting to study at the molecular level the seven cases reported by Sachatello et al.,12 as well as other similar cases.
Acknowledgments
We gratefully thank the patients and their families for their participation in our study. We are grateful to C. Dehainault, M. C. Weill-Perrier, X. P. Zhou, and S. Chevalier, for excellent technical support, and S. Gad, A. Lauge, S. Olschwang, and K. Sweet, for participation in the early stages of the study. C.E. thanks K. Zbuk for helpful discussions. D.S. was supported by the Marie Curie host fellowship and the Programme Hospitalier de Recherche Clinique AOM 02-122. C.E. is a recipient of the Doris Duke Distinguished Clinical Scientist Award and is partially supported by the American Cancer Society (RSG02-151-01CCE).
Appendix A
Table A1.
Genes Located in the Regions of Deletions of Patients 1 and 2 and the Main Putative Protein Characteristics
| Gene | Main Putative Protein Characteristics |
| LDB3 (LIM domain binding 3) | Functions as a linker structure to maintain cytoskeletal structure during contraction in striated muscle. Probably involved in familial dilated cardiomyopathy36 and in myopathy myofibrillary.37 |
| MMR2 (multimerin 2) | Member of EMILIN family of proteins of the extracellular matrix.38 Expressed during development, especially in the cardiovascular system and in mesenchymal cells. |
| SNCG (synuclein gamma)a | Chaperone protein involved in the heat-shock protein-based multiprotein chaperone complex. Stimulates hormone-responsive mammary tumorigenesis.39 Highly expressed in breast and ovarian cancer progression. |
| GLUD1 (glutamate dehydrogenase) | Mitochondrial enzyme. Catalyzes the oxidative deamination of 1-glutamate to 2-oxoglutarate.40 Involved in hyperinsulinism-hyperammonemia syndrome, resulting from gain-of-function mutations within GLUD1.41 |
| MINPP1 (multiple inositol polyphosphate phosphatase) | Inositol polyphosphate phosphatase.23,25 Has a role in maturation and proliferation of chondrocytes. Its function in intracellular signalling is unknown.24 Potential role in the tumorigenesis of thyroid.42 |
| PAPSS2 (3′-phosphoadenosine 5′-phosphosulfate synthetase 2) | Involved in protein sulfatation, especially of extracellular matrix. Involved in recessively inherited spondyloepimetaphyseal dysplasia.43 |
| LIPF, or HGL (human gastric lipase) | Lipase secreted in stomach |
| ACTA2 (actin alpha 2 smooth-muscle aorta) | Member of actin family. Expressed in vascular smooth-muscle cells and fibroblast cells.44 Potential role in smooth-muscle cell differentiation45 and contractile activity of myofibroblasts.46 |
Previously identified as a breast cancer–specific gene (BCSG1).
Web Resources
The URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/index.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for JPS, CS, and BRRS)
References
- 1.McColl I, Busxey HJ, Veale AM, Morson BC (1964) Juvenile polyposis coli. Proc R Soc Med 57:896–897 [PubMed] [Google Scholar]
- 2.Merg A, Howe JR (2004) Genetic conditions associated with intestinal juvenile polyps. Am J Med Genet C Semin Med Genet 129:44–55 10.1002/ajmg.c.30020 [DOI] [PubMed] [Google Scholar]
- 3.Eng C (2003) PTEN: one gene, many syndromes. Hum Mutat 22:183–198 10.1002/humu.10257 [DOI] [PubMed] [Google Scholar]
- 4.Pilarski R, Eng C (2004) Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet 41:323–326 10.1136/jmg.2004.018036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachatello CR, Griffen WO Jr (1975) Hereditary polypoid diseases of the gastrointestinal tract: a working classification. Am J Surg 129:198–203 10.1016/0002-9610(75)90298-6 [DOI] [PubMed] [Google Scholar]
- 6.Stemper TJ, Kent TH, Summers RW (1975) Juvenile polyposis and gastrointestinal carcinoma: a study of a kindred. Ann Intern Med 83:639–646 [DOI] [PubMed] [Google Scholar]
- 7.Coburn MC, Pricolo VE, DeLuca FG, Bland KI (1995) Malignant potential in intestinal juvenile polyposis syndromes. Ann Surg Oncol 2:386–391 10.1007/BF02306370 [DOI] [PubMed] [Google Scholar]
- 8.Bronner MP (2003) Gastrointestinal polyposis syndromes. Am J Med Genet A 122:335–341 10.1002/ajmg.a.20476 [DOI] [PubMed] [Google Scholar]
- 9.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B (2001) Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet 28:184–187 10.1038/88919 [DOI] [PubMed] [Google Scholar]
- 10.Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA (1998) Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 280:1086–1088 10.1126/science.280.5366.1086 [DOI] [PubMed] [Google Scholar]
- 11.Howe JR, Sayed MG, Ahmed AF, Ringold J, Larsen-Haidle J, Merg A, Mitros FA, Vaccaro CA, Petersen GM, Giardiello FM, Tinley ST, Aaltonen LA, Lynch HT (2004) The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet 41:484–491 10.1136/jmg.2004.018598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachatello CR, Hahn IS, Carrington CB (1974) Juvenile gastrointestinal polyposis in a female infant: report of a case and review of the literature of a recently recognized syndrome. Surgery 75:107–114 [PubMed] [Google Scholar]
- 13.Friedl W, Uhlhaas S, Schulmann K, Stolte M, Loff S, Back W, Mangold E, Stern M, Knaebel HP, Sutter C, Weber RG, Pistorius S, Burger B, Propping P (2002) Juvenile polyposis: massive gastric polyposis is more common in MADH4 mutation carriers than in BMPR1A mutation carriers. Hum Genet 111:108–111 10.1007/s00439-002-0748-9 [DOI] [PubMed] [Google Scholar]
- 14.Tsuchiya KD, Wiesner G, Cassidy SB, Limwongse C, Boyle JT, Schwartz S (1998) Deletion 10q23.2-q23.33 in a patient with gastrointestinal juvenile polyposis and other features of a Cowden-like syndrome. Genes Chromosomes Cancer 21:113–118 [DOI] [PubMed] [Google Scholar]
- 15.Jacoby RF, Schlack S, Sekhon G, Laxova R (1997) Del(10)(q22.3q24.1) associated with juvenile polyposis. Am J Med Genet 70:361–364 [DOI] [PubMed] [Google Scholar]
- 16.Arch EM, Goodman BK, Van Wesep RA, Liaw D, Clarke K, Parsons R, McKusick VA, Geraghty MT (1997) Deletion of PTEN in a patient with Bannayan-Riley-Ruvalcaba syndrome suggests allelism with Cowden disease. Am J Med Genet 71:489–493 [DOI] [PubMed] [Google Scholar]
- 17.Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, et al (1999) PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8:1461–1472 10.1093/hmg/8.8.1461 [DOI] [PubMed] [Google Scholar]
- 18.Zigman AF, Lavine JE, Jones MC, Boland CR, Carethers JM (1997) Localization of the Bannayan-Riley-Ruvalcaba syndrome gene to chromosome 10q23. Gastroenterology 113:1433–1437 10.1053/gast.1997.v113.pm9352843 [DOI] [PubMed] [Google Scholar]
- 19.Sweet K, Willis J, Zhou XP, Gallione C, Sawada T, Alhopuro P, Khoo SK, Patocs A, Martin C, Bridgeman S, Heinz J, Pilarski R, Lehtonen R, Prior TW, Frebourg T, Teh BT, Marchuk DA, Aaltonen LA, Eng C (2005) Molecular classification of patients with unexplained hamartomatous and hyperplastic polyposis. JAMA 294:2465–2473 10.1001/jama.294.19.2465 [DOI] [PubMed] [Google Scholar]
- 20.Zhou XP, Waite KA, Pilarski R, Hampel H, Fernandez MJ, Bos C, Dasouki M, Feldman GL, Greenberg LA, Ivanovich J, Matloff E, Patterson A, Pierpont ME, Russo D, Nassif NT, Eng C (2003) Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am J Hum Genet 73:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vermeesch JR, Melotte C, Froyen G, Van Vooren S, Dutta B, Maas N, Vermeulen S, Menten B, Speleman F, De Moor B, Van Hummelen P, Marynen P, Fryns JP, Devriendt K (2005) Molecular karyotyping: array CGH quality criteria for constitutional genetic diagnosis. J Histochem Cytochem 53:413–422 10.1369/jhc.4A6436.2005 [DOI] [PubMed] [Google Scholar]
- 22.Dehainault C, Lauge A, Caux-Moncoutier V, Pages-Berhouet S, Doz F, Desjardins L, Couturier J, Gauthier-Villars M, Stoppa-Lyonnet D, Houdayer C (2004) Multiplex PCR/liquid chromatography assay for detection of gene rearrangements: application to RB1 gene. Nucleic Acids Res 32:e139 10.1093/nar/gnh137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi H, Tiller GE, Dasouki MJ, Romano PR, Wang J, O’Keefe RJ, Puzas JE, Rosier RN, Reynolds PR (1999) Multiple inositol polyphosphate phosphatase: evolution as a distinct group within the histidine phosphatase family and chromosomal localization of the human and mouse genes to chromosomes 10q23 and 19. Genomics 56:324–336 10.1006/geno.1998.5736 [DOI] [PubMed] [Google Scholar]
- 24.Romano PR, Wang J, O’Keefe RJ, Puzas JE, Rosier RN, Reynolds PR (1998) HiPER1, a phosphatase of the endoplasmic reticulum with a role in chondrocyte maturation. J Cell Sci 111:803–813 [DOI] [PubMed] [Google Scholar]
- 25.Chi H, Yang X, Kingsley PD, O’Keefe RJ, Puzas JE, Rosier RN, Shears SB, Reynolds PR (2000) Targeted deletion of Minpp1 provides new insight into the activity of multiple inositol polyphosphate phosphatase in vivo. Mol Cell Biol 20:6496–6507 10.1128/MCB.20.17.6496-6507.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waite KA, Eng C (2002) Protean PTEN: form and function. Am J Hum Genet 70:829–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waite KA, Eng C (2003) BMP2 exposure results in decreased PTEN protein degradation and increased PTEN levels. Hum Mol Genet 12:679–684 10.1093/hmg/12.6.679 [DOI] [PubMed] [Google Scholar]
- 28.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, Mishina Y, Li L (2004) BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nat Genet 36:1117–1121 10.1038/ng1430 [DOI] [PubMed] [Google Scholar]
- 29.Schmickel RD (1986) Contiguous gene syndromes: a component of recognizable syndromes. J Pediatr 109:231–241 [DOI] [PubMed] [Google Scholar]
- 30.Brook-Carter PT, Peral B, Ward CJ, Thompson P, Hughes J, Maheshwar MM, Nellist M, Gamble V, Harris PC, Sampson JR (1994) Deletion of the TSC2 and PKD1 genes associated with severe infantile polycystic kidney disease—a contiguous gene syndrome. Nat Genet 8:328–332 10.1038/ng1294-328 [DOI] [PubMed] [Google Scholar]
- 31.Marsh DJ, Dahia PL, Coulon V, Zheng Z, Dorion-Bonnet F, Call KM, Little R, Lin AY, Eeles RA, Goldstein AM, Hodgson SV, Richardson AL, Robinson BG, Weber HC, Longy M, Eng C (1998) Allelic imbalance, including deletion of PTEN/MMACI, at the Cowden disease locus on 10q22-23, in hamartomas from patients with Cowden syndrome and germline PTEN mutation. Genes Chromosomes Cancer 21:61–69 [DOI] [PubMed] [Google Scholar]
- 32.Reifenberger J, Rauch L, Beckmann MW, Megahed M, Ruzicka T, Reifenberger G (2003) Cowden’s disease: clinical and molecular genetic findings in a patient with a novel PTEN germline mutation. Br J Dermatol 148:1040–1046 10.1046/j.1365-2133.2003.05322.x [DOI] [PubMed] [Google Scholar]
- 33.Zhou XP, Loukola A, Salovaara R, Nystrom-Lahti M, Peltomaki P, de la Chapelle A, Aaltonen LA, Eng C (2002) PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am J Pathol 161:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nassif NT, Lobo GP, Wu X, Henderson CJ, Morrison CD, Eng C, Jalaludin B, Segelov E (2004) PTEN mutations are common in sporadic microsatellite stable colorectal cancer. Oncogene 23:617–628 10.1038/sj.onc.1207059 [DOI] [PubMed] [Google Scholar]
- 35.Grotsky HW, Rickert RR, Smith WD, Newsome JF (1982) Familial juvenile polyposis coli: a clinical and pathologic study of a large kindred. Gastroenterology 82:494–501 [PubMed] [Google Scholar]
- 36.Arimura T, Hayashi T, Terada H, Lee SY, Zhou Q, Takahashi M, Ueda K, Nouchi T, Hohda S, Shibutani M, Hirose M, Chen J, Park JE, Yasunami M, Hayashi H, Kimura A (2004) A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J Biol Chem 279:6746–6752 10.1074/jbc.M311849200 [DOI] [PubMed] [Google Scholar]
- 37.Selcen D, Engel AG (2005) Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann Neurol 57:269–276 10.1002/ana.20376 [DOI] [PubMed] [Google Scholar]
- 38.Braghetta P, Ferrari A, De Gemmis P, Zanetti M, Volpin D, Bonaldo P, Bressan GM (2004) Overlapping, complementary and site-specific expression pattern of genes of the EMILIN/Multimerin family. Matrix Biol 22:549–556 10.1016/j.matbio.2003.10.005 [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y, Liu YE, Goldberg ID, Shi YE (2004) γ Synuclein, a novel heat-shock protein-associated chaperone, stimulates ligand-dependent estrogen receptor α signaling and mammary tumorigenesis. Cancer Res 64:4539–4546 10.1158/0008-5472.CAN-03-3650 [DOI] [PubMed] [Google Scholar]
- 40.Smith TJ, Peterson PE, Schmidt T, Fang J, Stanley CA (2001) Structures of bovine glutamate dehydrogenase complexes elucidate the mechanism of purine regulation. J Mol Biol 307:707–720 10.1006/jmbi.2001.4499 [DOI] [PubMed] [Google Scholar]
- 41.Stanley CA, Lieu YK, Hsu BY, Burlina AB, Greenberg CR, Hopwood NJ, Perlman K, Rich BH, Zammarchi E, Poncz M (1998) Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N Engl J Med 338:1352–1357 10.1056/NEJM199805073381904 [DOI] [PubMed] [Google Scholar]
- 42.Gimm O, Chi H, Dahia PL, Perren A, Hinze R, Komminoth P, Dralle H, Reynolds PR, Eng C (2001) Somatic mutation and germline variants of MINPP1, a phosphatase gene located in proximity to PTEN on 10q23.3, in follicular thyroid carcinomas. J Clin Endocrinol Metab 86:1801–1805 10.1210/jc.86.4.1801 [DOI] [PubMed] [Google Scholar]
- 43.ul Haque MF, King LM, Krakow D, Cantor RM, Rusiniak ME, Swank RT, Superti-Furga A, Haque S, Abbas H, Ahmad W, Ahmad M, Cohn DH (1998) Mutations in orthologous genes in human spondyloepimetaphyseal dysplasia and the brachymorphic mouse. Nat Genet 20:157–162 10.1038/2458 [DOI] [PubMed] [Google Scholar]
- 44.Comer KA, Dennis PA, Armstrong L, Catino JJ, Kastan MB, Kumar CC (1998) Human smooth muscle α-actin gene is a transcriptional target of the p53 tumor suppressor protein. Oncogene 16:1299–1308 10.1038/sj.onc.1201645 [DOI] [PubMed] [Google Scholar]
- 45.Kumar MS, Hendrix JA, Johnson AD, Owens GK (2003) Smooth muscle α-actin gene requires two E-boxes for proper expression in vivo and is a target of class I basic helix-loop-helix proteins. Circ Res 92:840–847 10.1161/01.RES.0000069031.55281.7C [DOI] [PubMed] [Google Scholar]
- 46.Chaponnier C, Gabbiani G (2004) Pathological situations characterized by altered actin isoform expression. J Pathol 204:386–395 10.1002/path.1635 [DOI] [PubMed] [Google Scholar]