Abstract
Cutis laxa is a condition characterized by redundant, pendulous, and inelastic skin. We identified a patient with recessive inheritance of a missense mutation (169G→A; E57K) in the Fibulin-4 gene. She had multiple bone fractures at birth and was diagnosed with cutis laxa, vascular tortuosity, ascending aortic aneurysm, developmental emphysema, inguinal and diaphragmatic hernia, joint laxity, and pectus excavatum by age 2 years. Her skin showed markedly underdeveloped elastic fibers, and the extracellular matrix laid down by her skin fibroblasts contained dramatically reduced amounts of fibulin-4. We conclude that fibulin-4 is necessary for elastic fiber formation and connective tissue development.
Cutis laxa is an acquired or inherited skin disease characterized by pendulous, redundant, and inelastic skin. Inherited forms of this disease show considerable locus heterogeneity. The cutis laxa genes discovered to date are all important for elastic fiber formation. X-linked cutis laxa, or occipital horn syndrome (OHS [MIM 304150]), is caused by mutations in the ATP7A Cu2+ transporter.1 Cu2+ is an essential cofactor of lysyl oxidases, a family of enzymes necessary for cross-linking fibrillar collagens and elastin. Autosomal dominant cutis laxa (ADCL [MIM 123700]) is caused by mutations in the elastin gene,2–6 which encodes the main structural component of elastic fibers. Finally, a severe autosomal recessive form of cutis laxa (ARCL [MIM 219100]) has been shown to be caused by a fibulin-5 mutation.7
Fibulins are a recently recognized family of extracellular matrix proteins with six known members. These molecules contain series of calcium-binding epidermal growth factor (cbEGF) modules and a C-terminal fibulin domain.8 Fibulins interact with a multitude of extracellular matrix proteins, including components of basement membranes and elastic fibers.9 Previous studies have demonstrated that Fibulin-5 gene mutations cause cutis laxa7 and have found reduced expression of fibulin-2 in patients with cutis laxa.10 Therefore, we hypothesized that mutations in other members of the fibulin gene family may cause cutis laxa as well and began a systematic mutational evaluation of fibulins. We report here a fibulin-4 mutation associated with a new recessive cutis laxa syndrome characterized by severe systemic connective-tissue abnormalities.
The patient, parents, and control individuals participated in this study after giving informed consent. This study was approved by the Human Studies Committee of Washington University School of Medicine. Tissue samples were collected via skin biopsies. Normal control tissue samples were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. Other investigators may have received specimens from the same subjects. Control DNA was obtained from healthy volunteers of diverse ethnic backgrounds. DNA was isolated from peripheral blood or dermal fibroblasts by isolation of nuclei, followed by proteinase K digestion and phenol extraction.11
Each exon of the Fibulin-4 gene (FBLN4) was amplified with 80–100 bp of flanking intronic sequences (table 1) in GeneAmp 9700 PCR machines (Applied Biosystems). Amplimers were subjected to direct sequence analysis on both strands by use of the BigDye terminator cycle sequencing chemistry (Applied Biosystems). A control DNA panel was screened for variants identified in the proband, by use of denaturing high-performance liquid chromatography.
Table 1.
Oligonucleotides for the Amplification of FBLN4 Exons[Note]
|
Primer(5′→3′) |
|||
| Exon | Forward | Reverse |
Product Size (bp) |
| 1 | TACAGGGAGTTGAGGTGCCG | ATTCCTGGAAGGGAACCAGAAG | 111 |
| 2 | TGAAGAGCCCGACAGCTACA | TGAAGAGCCCGACAGCTACA | 468 |
| 3 | TTAGCTTGTTGAGCAGAGTGGG | TTAAGACGCCTGGTGAGAGGA | 396 |
| 4 | TCAGTGTTGCTGGGGATGTT | TGAAGCCTGGCTTGAATGG | 426 |
| 5 | TGGTGAAGCCTTGAGTCCCA | ATGGAGAACTGTGTGGCAGG | 552 |
| 6 | TTCTTGACCAGCCCAGGTAGA | ATGGAGAACTGTGTGGCAGGA | 284 |
| 7 | TTCTTCCAGTTCTGGGCAGAGA | TCATCAATGTCTGTGCCAGGG | 438 |
| 8 | TCTGCCAAGGTACAGGTTGC | ATTCCCATCATCCCTCAGGG | 333 |
| 9 | ATCTATTCAGACCAGCGAGTGCC | TGTGCATGTCAGTTGAGGGTTG | 401 |
| 10 | TAGCACACAGGCTGACACAA | TCAGCCACCAGGACTTTCTT | 320 |
Note.— Annealing temperature was 53°C for all amplifications.
For RNA and protein analyses, fibroblasts were cultured to confluency at passages 4–10 in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 25 mM HEPES, l-glutamine, and antibiotics. Confluent cultures were used to extract RNA by use of TRI Reagent (Molecular Research Center). First-strand cDNA was synthesized from total RNA samples (1 μg each) by use of SuperScript III reverse transcriptase (Invitrogen). Standard PCR reactions used AmpliTaqGold polymerase (Applied Biosystems) under conditions recommended by the manufacturer, with annealing temperature of 57°C. Oligonucleotide primers were designed for exon 1 (5′-TGCTACTGTTGCTCTTGGGATCA-3′) and exon 5 (5′-AAGGAGCCAGGCAGGTTCA-3′), to amplify a region containing mutation E57K. A single amplification product was obtained from both the patient and the healthy controls, which suggests a lack of alternative splicing or mutation-induced aberrant splicing in this region of the mRNA. RT-PCR products were subjected to DNA sequencing, to confirm the presence of the mutation at the mRNA level.
For immunoblotting, first, the media fraction was collected. To extract extracellular matrix, the cell layers were washed using 10 ml of prewarmed PBS 2–3 times, and 2 ml of 10-mM EDTA (pH 8.0) in PBS was added, followed by shaking at room temperature for 5 min. The EDTA extract was centrifuged at 2,000 g for 10 min. Subsequently, cell lysates were prepared by incubating the cell layer in 4 M urea, 1 M sodium borate, and 0.15 M NaCl for 20 min at 4°C with agitation, followed by scraping the cell layers and centrifugation. The concentration of each fraction was determined by the Bradford assay (BioRad), and a volume corresponding to 25 μg of protein was taken. Proteins were concentrated by incubating each fraction with 10 μl of StrataClean resin (Stratagene), and the resin with bound protein was resuspended in 15 μl of 2× Laemmli loading buffer and was boiled for 5 min before loading onto SDS-PAGE. After electrophoresis, the proteins were transferred onto polyvinylidene fluoride membranes. The blots were blocked using 5% fat-free milk in Tris-buffered saline Tween (TBST [150 mM NaCl; 20 mM Tris-HCl, pH 7.5; and 0.1% Tween 20) and were incubated with a monoclonal anti–fibulin-4 antibody (1:100 [clone 7B9 isotype IgG2a]). The blot was washed using TBST, and horseradish-peroxidase–conjugated secondary antibody (1:10,000 [catalog number 674281, ICN Biomedical]) was added to the blot in fresh blocking solution. Antibody binding was detected using a SuperSignal West Pico Chemiluminescent Kit (Pierce). After fibulin-4 detection, blots were stripped and reprobed for β-actin (1:5,000 [Oncogene]). Immunoblotting experiments were repeated multiple times and with the use of cells cultured under different conditions (e.g., in serum-containing and serum-free media, with and without transforming growth factor–β1 treatment), and similar results were obtained. Fibulin-4 was undetectable in conditioned media of both patient and control fibroblasts. Thus, media blots are not shown.
Skin tissue in paraffin blocks were cut to 5-μm-thick sections. Sections were deparaffinized in Histo-Clear (National Diagnostics) and were rehydrated. All histological sections were stained with hematoxylin-eosin and Hart’s elastic stain. For Hart’s elastin stain, sections were immersed in 0.25% potassium permanganate solution for 5 min. Slides were then cleared in 5% oxalic acid and were soaked in resorcin-fuchsin solution overnight. After being washed in water, sections were counterstained with tartrazine (yellow), were dehydrated in ethanol, were cleared in xylene, and were mounted. Sections were viewed under bright-field optics by use of an Olympus Bx60 microscope. Images were acquired through a charge-coupled device camera (Carl Zesis AxioCam HR), were digitized via the Carl Zesis AxioVision 3.1 Image Processing System, and were archived in a desktop personal computer.
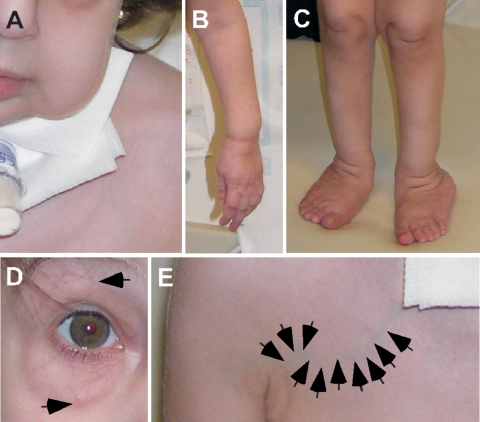
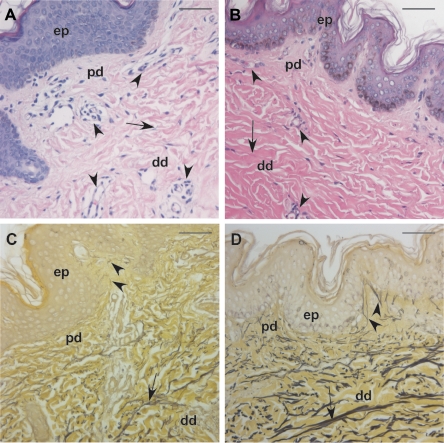
The patient was born to unaffected parents who indicated they were nonconsanguineous and had no history of cutis laxa in their respective families. Both parents were of Iraqi descent. The patient was born with multiple fractures at gestational age 36 wk, after an uneventful pregnancy, although oligohydramnios was reported. An examination at age 9 mo revealed generalized cutis laxa with soft, velvety, and transparent skin. Additional observations included hypotonia, emphysema, generalized arterial tortuosity, inguinal and diaphragmatic hernia, joint laxity, and pectus excavatum. At age 10 mo, she underwent diaphragmatic plication. Surgical observations included both arterial and venous tortuosity, emphysematous and hyperexpanded lungs, and an esophagus not fixed to the posterior chest. The diaphragm contained very little muscle and mainly consisted of a pleuroperitoneal membrane. Subsequent breathing difficulties required the placement of a tracheal tube (fig. 1A), and, at age 2 years, she received a diagnosis of an aortic root aneurysm. Clinical photographs taken at age 2 years confirmed generalized cutis laxa (fig. 1A–1C) and vascular tortuosity (fig. 1D and 1E). Histological evaluation of a skin biopsy specimen taken at the same time revealed smaller-than-normal collagen bundles (fig. 2A and 2B), increased vascularization of the upper dermis, and severely underdeveloped elastic fibers compared with specimens from age-matched healthy controls (fig. 2C and 2D).
Figure 1.
Generalized cutis laxa and vascular tortuosity in the proband. Clinical photographs of the patient show sagging cheeks and tracheal tube (A), redundant skin in the extremities (B and C), and tortuous vessels in the periorbital area (arrows in D). In panel E, arrows point to a tortuous vein in the right upper chest area.
Figure 2.
Skin pathology in the proband. A and B, Hematoxylin-eosin staining of samples from the patient (A) and an age- and sex-matched control (B) revealed increased vascularization (arrowheads) and reduced collagen bundle size (arrows) in the dermis of the patient. C and D, Hart’s elastin staining showed severely underdeveloped elastic fibers in both the papillary dermis (pd) (arrowheads) and the deep dermis (dd) (arrows) of the patient (C) compared with a matched control (D). There is no apparent pathology in the epidermis (ep). Magnification bars = 50 μm.
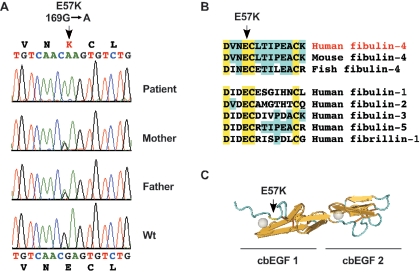
Screening of FBLN4 revealed a homozygous missense mutation (169G→A; E57K) in the patient (fig. 3A). Both of her parents were heterozygous, which suggests autosomal recessive inheritance. Mutation E57K was not found in 123 unrelated healthy controls. Multiple alignment showed that the E57 residue was completely conserved in cbEGF modules (fig. 3B), consistent with this residue being essential for calcium binding (fig. 3C).
Figure 3.
Recessive inheritance of E57K mutation in fibulin-4. A, DNA sequence analysis of FBLN4 revealed a homozygous 169G→A (E57K) mutation in the patient. Both parents were heterozygous, and a healthy control individual showed wild-type sequence (Wt). B, Multiple alignment of the peptide sequences of fibulins and fibrillin-1 shows the conservation of the E57 residue in all cbEGF-like modules. C, Three-dimensional structure of a pair of cbEGF modules, based on nuclear magnetic resonance studies of fibrillin-1 peptides,12 shows that the residue corresponding to E57 is part of the calcium-binding site.
To investigate the effect of this mutation on fibulin-4 expression, we first conducted RT-PCR experiments, using mRNA isolated from skin fibroblasts obtained from the patient and healthy controls. FBLN4 mRNA expression in the patient was comparable to normal expression, and sequencing of RT-PCR products confirmed the presence of the E57K mutation in the patient (data not shown). Immunoblot analysis of fibulin-4 in fibroblasts cultured under a variety of conditions showed normal abundance of fibulin-4 in patient cell lysates, but extracellular matrix extracts for patient fibroblast cultures showed severely reduced fibulin-4 content (fig. 4).
Figure 4.
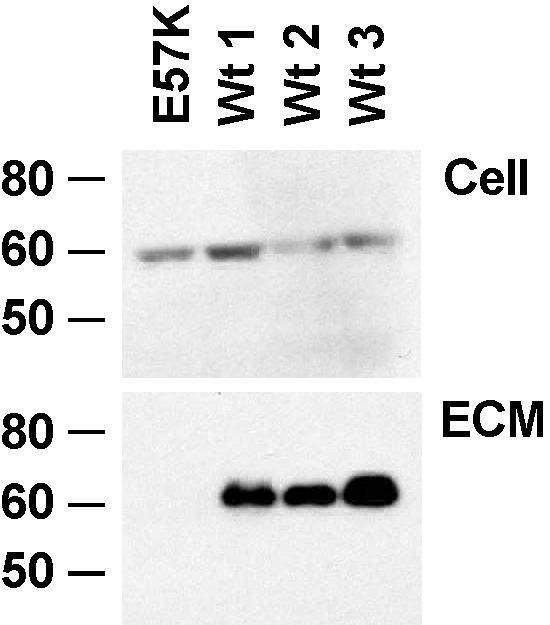
Deficient matrix incorporation of K57 fibulin-4. Immunoblot analysis of fibulin-4 expression in fibroblasts from the patient (E57K) and three healthy controls (Wt 1–Wt 3). Cell lysates (Cell) show approximately equal expression of fibulin-4 in the patient and controls. However, EDTA extract of the extracellular matrix (ECM) shows no fibulin-4 in the patient sample.
Our results demonstrate for the first time, to our knowledge, that homozygous mutations in FBLN4 cause a severe, recessive connective-tissue disease including cutis laxa, bone fragility, vascular tortuosity and aneurysm, developmental emphysema, and diaphragmatic and inguinal hernia. Evidence that the mutation E57K is disease causing includes conservation of this residue in cbEGF-like domains, where it is part of the calcium-binding consensus sequence. Lack of this mutation in 123 healthy controls provides further supporting evidence. Moreover, mutant fibroblasts contained an undetectable amount of fibulin-4, showing that mutation E57K impairs the secretion, matrix binding, or stability of this protein. Additional evidence supporting our conclusion is provided by the observation that the same substitution at a homologous residue in fibrillin-1 (E1073K) causes severe, neonatal Marfan syndrome.13,14
Reduced amount of elastic fibers in the skin and the clinical observations of cutis laxa, emphysema, and aortic aneurysm in our patient implicate fibulin-4 in elastic fiber formation. Similar clinical and pathologic observations were made in autosomal dominant cutis laxa caused by elastin gene mutations.5,6 A recent report of targeted inactivation of fibulin-4 in mice uncovered a neonatal lethal phenotype very similar to our observations in a human: defective elastic fiber formation, vascular tortuosity, aneurysms, rupture, and developmental emphysema.15 Thus, the role of fibulin-4 in elastic fiber formation is evolutionarily conserved in mammals. However, additional observations in our study, such as vascular tortuosity and bone fragility, indicate that fibulin-4 may have additional, pleiotropic functions in vascular patterning and collagen biosynthesis. Indeed, the recent discovery of fibulin-4 expression in chondrocytes supports the notion that this protein is important for skeletal function.16
Interestingly, a missense mutation in the Fibulin-5 gene was also reported to cause recessive cutis laxa and severe lung disease associated with abnormal elastic fiber formation.7 Of note, systemic complications of fibulin-5 mutations included supravalvular aortic stenosis and obstructive vascular disease, and no collagen-related abnormalities were observed.7 Studies of animals with targeted disruption of the fibulin-5 gene confirmed the importance of fibulin-5 in elastic fiber formation17,18 and demonstrated the ability of fibulin-5 to bind both cell surface receptors and tropoelastin. These studies, together with the present report, suggest that both fibulin-4 and fibulin-5 play essential, nonoverlapping roles in elastogenesis.
Acknowledgments
This study was funded by National Institutes of Health grant HL073703 (to Z.U.) and by the Department of Pediatrics, Washington University. We are grateful to the patient and family members whose cooperation made this study possible.
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for OHS, ADCL, and ARCL)
References
- 1.Das S, Levinson B, Vulpe C, Whitney S, Gitschier J, Packman S (1995) Similar splicing mutations of the Menkes/mottled copper-transporting ATPase gene in occipital horn syndrome and the blotchy mouse. Am J Hum Genet 56:570–576 [PMC free article] [PubMed] [Google Scholar]
- 2.Tassabehji M, Metcalfe K, Hurst J, Ashcroft GS, Kielty C, Wilmot C, Donnai D, Read AP, Jones CJ (1998) An elastin gene mutation producing abnormal tropoelastin and abnormal elastic fibres in a patient with autosomal dominant cutis laxa. Hum Mol Genet 7:1021–1028 10.1093/hmg/7.6.1021 [DOI] [PubMed] [Google Scholar]
- 3.Zhang MC, He L, Giro M, Yong SL, Tiller GE, Davidson JM (1999) Cutis laxa arising from frameshift mutations in exon 30 of the elastin gene (ELN). J Biol Chem 274:981–986 10.1074/jbc.274.2.981 [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Revenga L, Iranzo P, Badenas C, Puig S, Carrio A, Mila M (2004) A novel elastin gene mutation resulting in an autosomal dominant form of cutis laxa. Arch Dermatol 140:1135–1139 10.1001/archderm.140.9.1135 [DOI] [PubMed] [Google Scholar]
- 5.Urban Z, Gao J, Pope FM, Davis EC (2005) Autosomal dominant cutis laxa with severe lung disease: synthesis and matrix deposition of mutant tropoelastin. J Invest Dermatol 124:1193–1199 10.1111/j.0022-202X.2005.23758.x [DOI] [PubMed] [Google Scholar]
- 6.Szabo Z, Crepeau MW, Mitchell AL, Stephan MJ, Puntel RA, Loke KY, Kirk RC, Urban Z (2006) Aortic aneurysmal disease and cutis laxa caused by defects in the elastin gene. J Med Genet 43:255–258 10.1136/jmg.2005.034157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeys B, Van Maldergem L, Mortier G, Coucke P, Gerniers S, Naeyaert JM, De Paepe A (2002) Homozygosity for a missense mutation in fibulin-5 (FBLN5) results in a severe form of cutis laxa. Hum Mol Genet 11:2113–2118 10.1093/hmg/11.18.2113 [DOI] [PubMed] [Google Scholar]
- 8.Argraves WS, Greene LM, Cooley MA, Gallagher WM (2003) Fibulins: physiological and disease perspectives. EMBO Rep 4:1127–1131 10.1038/sj.embor.7400033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timpl R, Sasaki T, Kostka G, Chu ML (2003) Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol 4:479–489 10.1038/nrm1130 [DOI] [PubMed] [Google Scholar]
- 10.Markova D, Zou Y, Ringpfeil F, Sasaki T, Kostka G, Timpl R, Uitto J, Chu ML (2003) Genetic heterogeneity of cutis laxa: a heterozygous tandem duplication within the fibulin-5 (FBLN5) gene. Am J Hum Genet 72:998–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrmann BG, Frischauf AM (1987) Isolation of genomic DNA. Methods Enzymol 152:180–183 [DOI] [PubMed] [Google Scholar]
- 12.Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, Handford PA (1996) Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85:597–605 10.1016/S0092-8674(00)81259-3 [DOI] [PubMed] [Google Scholar]
- 13.Nijbroek G, Sood S, McIntosh I, Francomano CA, Bull E, Pereira L, Ramirez F, Pyeritz RE, Dietz HC (1995) Fifteen novel FBN1 mutations causing Marfan syndrome detected by heteroduplex analysis of genomic amplicons. Am J Hum Genet 57:8–21 [PMC free article] [PubMed] [Google Scholar]
- 14.Putnam EA, Cho M, Zinn AB, Towbin JA, Byers PH, Milewicz DM (1996) Delineation of the Marfan phenotype associated with mutations in exons 23–32 of the FBN1 gene. Am J Med Genet 62:233–242 [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin PJ, Chen Q, Horiguchi M, Starcher BC, Stanton JB, Broekelmann TJ, Marmorstein AD, McKay B, Mecham R, Nakamura T, Marmorstein LY (2006) Targeted disruption of fibulin-4 abolishes elastogenesis and causes perinatal lethality in mice. Mol Cell Biol 26:1700–1709 10.1128/MCB.26.5.1700-1709.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang Y, Sekine T, Nakamura H, Imajoh-Ohmi S, Fukuda H, Yudoh K, Masuko-Hongo K, Nishioka K, Kato T (2006) Fibulin-4 is a target of autoimmunity predominantly in patients with osteoarthritis. J Immunol 176:3196–3204 [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T, Lozano PR, Ikeda Y, Iwanaga Y, Hinek A, Minamisawa S, Cheng CF, Kobuke K, Dalton N, Takada Y, Tashiro K, Ross J Jr, Honjo T, Chien KR (2002) Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature 415:171–175 10.1038/415171a [DOI] [PubMed] [Google Scholar]
- 18.Yanagisawa H, Davis EC, Starcher BC, Ouchi T, Yanagisawa M, Richardson JA, Olson EN (2002) Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 415:168–171 10.1038/415168a [DOI] [PubMed] [Google Scholar]