Abstract
Cowden syndrome (CS) and Bannayan-Riley-Ruvalcaba syndrome (BRRS) are autosomal dominant hamartoma syndromes. Germline PTEN mutations have been associated with 85% of CS cases and 65% of BRRS cases and also with other disorders, which are collectively referred to as the “PTEN hamartoma tumor syndrome.” The human PTEN gene has been previously found to express two naturally occurring splice variants (SVs). Recently, we identified eight novel naturally occurring PTEN SVs that result in different downstream signaling effects: SV3a, SV3b, SV3c (inclusion of various lengths of intron 3 3′ of exon 3), SV5a, SV5b, SV5c, SV5d (inclusion of various lengths of intron 5 3′ of exon 5), and SVΔEx6 (deletion of exon 6). We therefore sought to characterize the relative expression of 5′, middle, and 3′ full-length PTEN mRNA (FL-PTEN) and also of these eight PTEN SVs in 85 (65 female and 20 male) patients with CS/BRRS (with or without PTEN mutations) compared with 27 controls, using a SYBR green quantitative polymerase chain reaction method. Significantly reduced FL-PTEN levels were found in the probands, compared with those of controls (P<.01). Apart from FL-PTEN, SV3a is the most consistently relatively underexpressed in patients compared with controls. The patients showed relative underexpression of SV3a and SV3b and overexpression of SV5b (P=.005, P=.02, and P=.04, respectively). Indeed, there appears to be an SV expressional genotype-phenotype correlation in which the SV expressional profiles are distinct among CS, CS-like, and BRRS. The reduced FL-PTEN transcript expression, associated with differential expression of PTEN SVs, regardless of PTEN mutation status, supports the concept that modulation of PTEN inactivation may also occur at the transcription level influencing the specific phenotypes seen in these syndromes.
Cowden syndrome (CS [MIM 158350]) and BannayanRiley-Ruvalcaba syndrome (BRRS [MIM 153480]) are autosomal dominant disorders characterized by hamartomas and an increased risk of breast, thyroid, and uterine cancers. Germline PTEN mutations have been associated with 85% of CS cases and 65% of BRRS cases1–3 as well as other disorders, which are collectively referred to as the “PTEN hamartoma tumor syndrome” (PHTS).4 PTEN (phosphatase, tensin homologue, deleted on chromosome 10) encodes a tumor suppressor, which mediates cell-cycle arrest and apoptosis.5 It is a dual-specificity phosphatase that dephosphorylates both lipid and protein substrates.6 Its major lipid phosphatase activity acts on phosphatidylinositol(3,4,5)triphosphate, yielding phosphatidylinositol(4,5)biphosphate.7 Inactivation of PTEN allows constitutive and unregulated activation of the Akt/protein kinase B and mitogen-activated protein kinase (MAPK) signaling pathways, resulting in uncontrolled proliferation.8,9 In addition to its growth-suppressive function, PTEN dephosphorylates focal adhesion kinase, resulting in inhibition of cell spreading and migration.10
RNA splicing is essential for the generation of protein diversity and can also have regulatory functions. During splicing, alternative usage of splice donor or acceptor sites can lead to various exons and introns being skipped or retained, creating a diverse array of mRNAs from a single pre-mRNA, a process referred to as “alternative RNA splicing.”11 More than half of all human genes generate multiple mRNAs by alternative splicing, and ∼80% of those result in changes in the encoded protein.12 For the majority of alternative splice events, however, their functional significance is still unclear.13
The human PTEN gene has been found to express splice variants involving deletions from the normal coding sequence but also the inclusion of intronic segments.14 Because these variants were identified in normal and cancer tissues, the significance of these variants in carcinogenesis has become a controversy.15 Patterns of alternative splicing can be tissue, stimulus, or disease specific, and thus the presence or levels of specific alternatively spliced mRNAs may provide useful biomarkers, even without knowledge of their activities.13 A large number of cancer-related genes exhibiting alternative splicing have been characterized, such as BRCA1, MDM2, and WT1.13,16 These splice variants could either be causative of disease; be involved in disease development; influence the extent of tumor invasion, the status of lymph node involvement, and distant metastasis; or act as surrogate markers.13,17
Recently, we identified eight novel naturally occurring PTEN splice variants (SVs) that result in different downstream signaling effects.18 We therefore sought to characterize the relative expression levels of these eight PTEN SVs in a large series of germline PTEN mutation-positive and -negative patients with PHTS spanning various phenotypes compared with the levels in normal controls. We also quantified the expression of full-length (FL) PTEN mRNA, using three sets of primer pairs encompassing the 5′ end, middle, and 3′ end of PTEN. In addition to increasing our fundamental understanding of the impact of various splicing alterations on modulation of the protean manifestations of CS/BRRS, we hoped to identify specific splice variants that could help identify individuals with CS/BRRS phenotypes regardless of mutation status.
Material and Methods
Human Subjects and Samples
Eighty-five subjects (65 female and 20 male) were enrolled in a PTEN research study by referral from U.S. and international centers. All samples were acquired with informed consent in accordance with protocols approved by the human subjects protection committees of the respective institutions.
For the purposes of this study, a series of PHTS-affected individuals with germline PTEN mutations, a series of CS/BRRS-affected individuals without mutations, and a series of normal controls were required. Among the 85 patients in this study, 22 have no detectable germline PTEN mutations, and 63 have various germline PTEN alterations throughout the gene, comprising 43 with pathogenic mutations and 20 with variants (table 1). The 27 normal controls were defined as having no documented illness, no CS/BRRS phenotype or CS/BRRS features, and no PTEN mutation. The 63 subjects who were PTEN alteration positive were included in this study regardless of clinical syndrome name. Most (28) had classic CS and BRRS, 26 had CS-like, and 1 had BRRS-like. Of the 63 patients with PTEN germline pathogenic mutations, 8 could not be classified as having CS/CS-like or BRRS/BRRS-like, because each had only hamartomatous/hyperplastic polyps, breast hamartomas, breast cancer, or breast cancer plus glycogenic acanthosis as the sole clinical manifestation. Three patients had no reported clinical findings but had been found to carry the same mutation (R130X) carried by a sibling, and one patient had no specific clinical information available. The 22 patients who were PTEN mutation negative were all classified as having classic CS (by the International Cowden Consortium operational diagnostic criteria)4 or BRRS.
Table 1. .
Germline PTEN Mutation or Variant and Phenotype of the Patients in the Study
| Group and Sample | Location | Mutation or Variant | Phenotype |
| Sequence variant: | |||
| SCP12* | Promoter region | −903 G/A | CS-like |
| SCP19* | Promoter region | −903 G/A | CS |
| SCP20* | Promoter region | −903 G/A | CS |
| SCP36* | Promoter region | −903 G/A | Breast cancer and glycogenic acanthosis |
| SCP38* | Promoter region | −903 G/A | CS-like |
| SCP39* | Promoter region | −903 G/A | CS-like |
| SCP43* | Promoter region | −903 G/A | CS-like |
| SCP51* | Promoter region | −903 G/A | CS-like |
| SCP57* | Promoter region | −903 G/A | Breast hamartomas |
| SCP58* | Promoter region | −903 G/A | CS-like |
| SCP63* | Promoter region | −903 G/A | CS-like |
| SCP14* | Promoter region | −1026 C/A | CS |
| SCP42* | Promoter region | −1026 C/A | CS-like |
| SCP60* | Promoter region | −1026 C/A | CS-like |
| SCP64* | Promoter region | −1026 C/A | CS |
| SCP56* | Promoter region | −1026 C/A | CS-like |
| SCP23* | Promoter region | −1026 C/A | CS-like |
| SCP26* | Promoter region | −1026 C/A | CS |
| SCP25* | Promoter region | −1084 C/T | CS |
| SCP28* | Promoter region | −1084 C/T | CS-like |
| SCP4* | Exon 6 | L193L (CTG193CTA) | CS |
| SCP66* | Exon 4 | T78T (ACC78ACT) | CS-like |
| SCP59* | Exon 2 | G44G (GGC44GGT) | CS-like |
| Germline PTEN mutation positive: | |||
| SCP45* | Promoter region | −843 C/T | CS-like |
| SCP67* | Promoter region | −843 C/T | CS-like |
| SCP72* | Promoter region | −970 T/G | CS-like |
| SCP13* | Promoter region | −1142 C/T | CS |
| SCP24* | IVS1 | IVS1 +35 C/T | Juvenile polyps |
| SCP2* | Exon 1 | T26P (ACC26CCC) | CS-like |
| SCP32* | Exon 1 | R15S (AGA15AGT) | CS-like |
| SCP40* | Exon 1 | Y16X (TAT16TAA), Q17X (CAA17TAA) | BRRS |
| SCP47* | Exon 1 | Y16X (TAT16TAA), Q17X (CAA17TAA) | CS |
| SCP71* | Exon 1 | I32N (ATT32AAT) | CS/BRRS overlap |
| SCP46* | IVS2 | IVS2 −38 ins G | BRRS |
| SCP16* | IVS2 | IVS2 −31 G/T | CS |
| SCP11* | Exon 3 | Y68X (TAC68TAA) | BRRS |
| SCP15* | Exon 5 | 445 ins A, IVS4 −29 insT | CS-like |
| SCP6* | Exon 5 | 445 ins A | CS-like |
| SCP35* | Exon 5 | 445 ins A | CS-like |
| SCP27* | Exon 5 | G132V (GGT132GTT) | CS |
| SCP18* | Exon 5 | 315 del T | BRRS |
| SCP21* | Exon 5 | R130Q (CGA130CAA) | BRRS |
| SCP37* | Exon 5 | L152P (CTA152CCA) | CS-like |
| SCP54* | Exon 5 | C136Y (TGT136TAT) | CS |
| SCP1* | Exon 5 | R130X (CGA130TGA) | No signs reported |
| SCP3* | Exon 5 | R130X (CGA130TGA) | CS |
| SCP5* | Exon 5 | R130X (CGA130TGA) | CS |
| SCP7* | Exon 5 | R130X (CGA130TGA) | No signs reported |
| SCP8* | Exon 5 | R130X (CGA130TGA) | No signs reported |
| SCP10* | Exon 6 | C211X (TGC211TGA) | CS |
| SCP53* | Exon 6 | C211X (TGC211TGA) | CS |
| SCP52* | Exon 6 | 549 ins A | BRRS |
| SCP55* | Exon 6 | 604–610 del 7 (ACTATTC) | BRRS like |
| SCP9* | Exon 7 | 706–8 GAC/TTGT | CS |
| SCP49* | Exon 7 | 683 del A | No information |
| SCP61* | Exon 7 | Q245X (CAG245TAG) | CS |
| SCP68* | Exon 7 | K260R (AAA260AGA) | Glycogenic ancanthosis |
| SCP33* | Exon 8 | 972 del T | CS |
| SCP65* | Various | IVS3 −39 A/G, IVS4 −29 ins T, IVS7 +1 del G | CS |
| SCP41* | Various | −1084 C/T and −9 C/G | CS-like |
| SCP62* | Various | −1084 C/T and −9 C/G | CS-like |
| SCP22 | Various | −903 G/A and L112V (CTA112GTA) | CS-like |
| SCP17* | Various | −903 G/A and R130Q (CGA130CAA) | CS |
RNA Extraction
Lymphocyte pellets were separated from total peripheral blood of patients and normal controls by use of red blood cell lysis buffer (89.9 g NH4Cl, 10.0 g KHCO3, and 2.0 ml 0.5-M EDTA) and were resuspended in PBS that was subsequently aspirated after centrifugation. The pellet could then be immediately processed for RNA extraction or stored at −80°C until further use. Total RNA was extracted using Versagene (Gentra Systems) total RNA purification kit, according to the manufacturer’s instructions.
mRNA Expression of FL-PTEN and Its Splice Variants
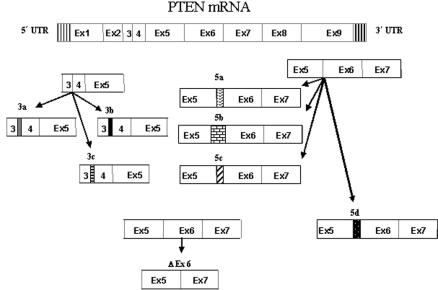
We studied the patients and the normal controls for expression of eight novel SVs of PTEN identified in our lab—namely, SV3a, SV3b, SV3c, SV5a, SV5b, SV5c, SV5d, and SVΔEx6.18 With the exception of SVΔEx6, all variants consisted of insertion of intronic regions following the end of the exons (fig. 1 and table 2). For example, SV3a was generated by alternative splicing of the end of exon 3 (genomic sequence 83526) to a splicing acceptor site within the third intron (intron C), which thus inserted 29 bp (85517–85545) of the genomic sequence (1,991 bp downstream of the end of exon 3). The other SVs were generated similarly, with inclusions of various lengths of intron 3 3′ of exon 3 (SV3a, 3b, and 3c) or inclusions of intron 5 3′ of exon 5 (SV5a, 5b, 5c, and 5d) (table 2). SVΔEx6 was generated by the exclusion of the entire exon 6. To compare the expression of these SVs with FL-PTEN mRNA, we also measured, in all the patients and controls, three regions representing the 5′ end, middle, and 3′ end of the gene: exons 1 and 2 (1-2); exons 3, 4, and 5 (3-4-5); and exons 7 and 8 (7-8).
Figure 1. .
Diagram showing splice variants of PTEN: SV3a, 3b, 3c, 5a, 5b, 5c, 5d, and ΔEx6. With the exception of SVΔEx6, all variants consist of insertions of intronic regions (represented by the bars with different patterns) following the 3′ end of the exons. SVΔEx6 was generated by the deletion of the entire exon 6. See table 2 for more details. Ex = exon.
Table 2. .
Size and Position of the Intronic Insertions or Exon Deletions That Generated the Splice Variants[Note]
| Splice Variant | Description | Change |
| SV3a | IVS3+1991 to 2019 | Ins 29 bp |
| SV3b | IVS3+1991 to 2042 | Ins 52 bp |
| SV3c | IVS3+4660 to 4699 | Ins 40 bp |
| SV5a | IVS5+3617 to 3654 | Ins 38 bp |
| SV5b | IVS5+1377 to 1703 | Ins 327 bp |
| SV5c | IVS5+9356 to 9514 | Ins 158 bp |
| SV5d | IVS5+3028 to 3088 | Ins 60 bp |
| SVΔEx6 | Exon 6: 2422 to 2563 | Del 142 bp |
Note.— For example, SV3a was generated by the insertion of 29 bp (from intron 3 sequence) at 1,991 bp following the 3′ end of exon 3 and ending at 2,019 bp following the 3′ end of exon 3. SVΔEx6 was the only one generated by a deletion of the entire exon 6, from nucleotide 2422 to 2563.
cDNA Synthesis Reaction
One microgram of RNA extracted from the patients and normal controls was treated with DNase at room temperature by use of 2 U amplification-grade DNase (Invitrogen). DNase in the reaction mix was inactivated by EDTA and heating at 65°C for 5 min. cDNA was synthesized using Superscript Reverse Transcriptase II (Invitrogen) and random primers (Invitrogen), according to the manufacturer’s instructions.
SYBR Green Quantitative PCR
Before the relative quantification procedure, the cDNA samples were amplified using HotstarTaq master mix (Qiagen) and primers for PTEN exons and splice variants (table 3). PCR was performed in a Peltier Thermal Cycler-200 (MJ Research, Bio-Rad) by use of specific conditions for each primer pair, optimized to ensure that a single band of the appropriate length was amplified (table 3). Products were analyzed on a 2% agarose gel stained with ethidium bromide.
Table 3. .
Primers and PCR Conditions Used to Amplify the PTEN Transcript or Its Splice Variants[Note]
|
PTEN Targets and Primers |
Sequence | Annealing Temperature (°C) |
No. of Cycles |
| 1-2: | 55 | 40 | |
| 1F | CAGCCATCATCAAAGAGATCG | ||
| 2R | TTGTTCCTGTATACGCCTTCAA | ||
| 3-4-5: | 59 | 40 | |
| 3F | TGGATTCAAAGCATAAAAACCA | ||
| 4–5R | AAAAGGATATTGTGCAACTCTGC | ||
| 7-8: | 59 | 40 | |
| 7F | TCCACAAACAGAACAAGATG | ||
| 8R | CTGGTCCTGGTATGAAGAAT | ||
| SV3a: | 59 | 48 | |
| 1F | CAGCCATCATCAAAGAGATCG | ||
| IVS3a R | CTTTCAGCACAATTAACTTCTCT | ||
| SV3b: | 59 | 44 | |
| 1F | CAGCCATCATCAAAGAGATCG | ||
| IVS3b R | CTGTGTGACCTTGTTCAACTCA | ||
| SV3c: | 60 | 48 | |
| 1F | CAGCCATCATCAAAGAGATCG | ||
| IVS3c R | GCAGTACCCTGGTAACTCCAA | ||
| SV5a: | 60 | 48 | |
| 4F | TCTTTGTGCTGAAAGACATT | ||
| IVS5a R | AGCCTTCTCTTGGATTTAATTTGGACTT | ||
| SV5b: | 61.2 | 48 | |
| 4F | TCTTTGTGCTGAAAGACATT | ||
| IVS5b R | CGCCTCGGCCTCCCAAAGT | ||
| SV5c: | 59 | 38 | |
| 4F | TCTTTGTGCTGAAAGACATT | ||
| IVS5c R | GGCCTCTACAAGGTCAGGATCAT | ||
| SV5d: | 60 | 48 | |
| 4F | TCTTTGTGCTGAAAGACATT | ||
| IVS5d R | GCCTTCTCTTGGATTTAATTTGG | ||
| SVΔEx6: | 59 | 42 | |
| 5–7F | AGGACCAGAGACAAAAAGATC | ||
| 8R | CTGGTCCTGGTATGAAGAAT |
Note.— The first three pairs define exonic regions and, hence, FL-PTEN, whereas the other primer pairs define the splice variants studied. Primer3 was used for design.
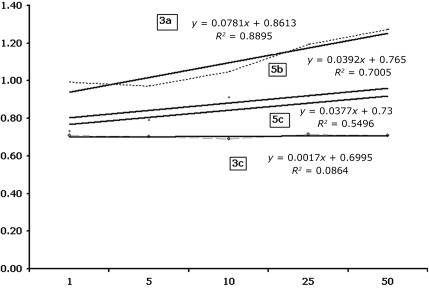
Relative quantification of FL-PTEN mRNA and its splice variants was performed using a SYBR green quantitative PCR method for comparison between patients and normal controls. Compared with β-actin, the GAPDH gene primer pair showed similar amplification efficiency to FL-PTEN and its splice variants in the cDNA dilution standardization experiment (fig. 2), and thus GAPDH was chosen as the control gene. Five serial dilutions of cDNA were amplified using gene-specific primers. The ΔCT was calculated for each cDNA dilution. The data were fit using linear regression analysis. If the absolute value of the slope is close to zero, the efficiencies of the target and reference genes are similar, and the ΔΔCT calculation for the relative quantification of the target may be used.19 The reactions were performed using SYBR Green PCR MasterMix (Bio-Rad) in the iCycler apparatus (Bio-Rad Laboratories). Amplification reactions, in a final volume of 25 μl, included 12.5 μl of SYBR Green PCR MasterMix (Bio-Rad), ∼0.3 μg of cDNA, and 10 mM of each primer (forward and reverse). For the quantification of expression of FL-PTEN mRNA transcript, three sets of primer pairs were used that represented the beginning, middle, and end of the gene: exons 1 and 2 (1-2); exons 3, 4, and 5 (3-4-5); and exons 7 and 8 (7-8). For the splice variants with intronic insertions, the forward primer was in the exon preceding the inserted intron sequence, and the reverse primer was in the inserted intronic sequence (table 3). For SVΔEx6, the forward primer included the ending and beginning sequence of exons 5 and 7, respectively, and the reverse primer was within exon 8. In all cases, we performed 80 cycles of melting, to verify the absence of nonspecific products. All the reactions were performed in triplicate, and the comparative CT method was used for the quantification of the expression for each segment, by use of GAPDH as a normalization control gene. All the reactions were done with a negative control to ensure that we had no contamination. One normal control sample was chosen to be used as a standard control for all the reactions with the patient samples. After each reaction, the products were analyzed on a 2% agarose gel stained with ethidium bromide, to rule out the presence of any extra amplified bands.
Figure 2. .
Plot of the log cDNA dilution versus ΔCT. The plots show the efficiency of amplification of the target sequences (SV3a, 3c, 5b, and 5c) and internal control (GAPDH) examined using SYBR green quantitative PCR. Five serial dilutions (X-axis) of cDNA were amplified using gene-specific primers. The ΔCT was calculated for each cDNA dilution. The data were fit using linear regression analysis.
To assure the segments were being correctly amplified, sequencing of randomly chosen real-time PCR products was performed using dGTP technology and the ABI 3730 analyzer (Applied Biosystems, Perkin-Elmer) according to the manufacturer’s recommendation. The Sequencher software package (version 4.2 [GeneCodes]) was used for sequence analysis.
Results
To address the hypothesis that differential expression of naturally occurring PTEN SVs will occur in patients with CS/BRRS compared with controls, we performed relative quantitation of SVs, FL-PTEN transcript, and the housekeeping GAPDH in 85 patients (63 with germline PTEN variation, comprising 43 with proven pathogenic mutations and a further 20 with variants of unknown significance [VUS]) and 27 normal controls, in a blinded fashion. After molecular analyses were complete, phenotype and mutation status were unblinded for statistical analysis (table 1).
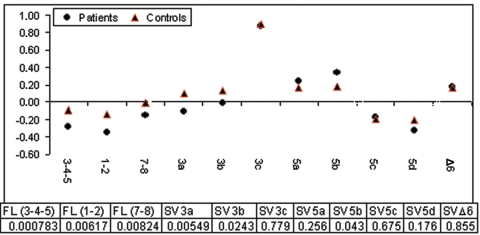
FL-PTEN transcript and all the naturally occurring splice variants were present in both patients and controls and showed significant differences in levels of expression in each of the groups studied (figs. 3 and 4 and table 4). Among the SVs, in both patients and controls, the highest level of relative expression was seen for SV3c, and the lowest relative expression was seen for SV5d (fig. 3). The group comprising all patients showed relative underexpression of FL-PTEN transcript, most accurately reflected in the exon 7–8 amplicon, compared with normal controls (figs. 3 and 4). Interestingly, differential expression of specific SVs was evident in the patient group compared with controls (figs. 3 and 4). SV3a and SV3b were relatively underexpressed in patients compared with controls (P=.005 and P=.002, respectively; figs. 3 and 4). In contrast, SV5b was slightly overexpressed in the patients compared with normal controls (P=.04; fig. 4).
Figure 3. .
Log-transformed expression values (Y-axis) of FL-PTEN (1-2, 3-4-5, and 7-8) and different splice variants (SV3a, 3b, 3c, 5a, 5b, 5c, 5d, and ΔEx6) for controls (red triangles) and patients (black circles). Table 4 gives the P values (from t test). See text and figure 4 for more details.
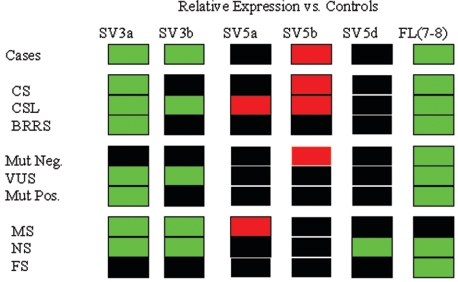
Figure 4. .
Two-color “heat map” representing relative expression of SV3a, SV3b, SV5a, SV5b, SV5d, and FL (7-8) transcripts in all cases, phenotypic subgroups, and mutation-status subgroups compared with normal controls. Green boxes represent significant (P<.05) relative underexpression; red boxes, significant overexpression; and black boxes, no difference in relative expression compared with controls. Mut Neg. = mutation negative; Mut Pos. = mutation positive; NS = nonsense mutation; MS = missense mutation; FS = frameshifting mutation.
Table 4. .
P Values (from t Test) from the Comparisons of Relative Expression of SVs between Controls and Patients Grouped According to Clinical Phenotype, Mutation Status, and Mutation Type[Note]
| Transcript or SV |
Patients | CS | CSL | BRRS | VUS | PTEN+ | PTEN– | MS | NS | FS |
| FL (3-4-5) | .000783 | .00118 | .00109 | .0203 | .0094 | .0003 | .0249 | .0094 | .000175 | .00259 |
| FL (1-2) | .00617 | .00228 | .00434 | .275 | .1310 | .0092 | .0014 | .1310 | .00138 | .0592 |
| FL (7-8) | .00824 | .0477 | .0043 | .018 | .0064 | .0178 | .1550 | .0064 | .00217 | .296 |
| SV3a | .00549 | .00122 | .0208 | .0224 | .0002 | .0193 | .1690 | .0002 | .00121 | .0998 |
| SV3b | .0243 | .0576 | .0395 | .0898 | .0049 | .0649 | .2560 | .0049 | .029 | .279 |
| SV3c | .779 | .957 | .985 | .649 | .3710 | .8650 | .6560 | .3710 | .105 | .878 |
| SV5a | .256 | .42 | .0275 | .346 | .4890 | .3170 | .1720 | .4890 | .498 | .419 |
| SV5b | .043 | .00329 | .0489 | .661 | .4040 | .0567 | .0170 | .4040 | .198 | .402 |
| SV5c | .675 | .884 | .302 | .726 | .5790 | .5160 | .3280 | .5790 | .806 | .302 |
| SV5d | .176 | .233 | .265 | .863 | .1620 | .2950 | .5660 | .1620 | .0198 | .653 |
| SVΔEx6 | .855 | .868 | .359 | .603 | .5910 | .4700 | .9870 | .5910 | .386 | .253 |
Note.— PTEN+ = PTEN mutation positive, PTEN– = PTEN mutation negative; MS = missense mutations; NS = nonsense mutations; FS = frameshift mutations.
The patients were then subdivided according to the three major phenotypic groups (30 with CS, 39 with CSL, and 7 with BRRS) and were analyzed for relative expression of FL transcript and the various SVs (fig. 4). Just as with the entire patient group as a whole, FL transcript and SV3a expression levels were reduced in each of the phenotypic groups—CS (P=.001), CSL (P=.02), and BRRS (P=.02)—compared with controls. The CS group and CSL group, but not the BRRS group, each showed a higher expression of SV5b (P=.003 and .048, respectively; fig. 4). Interestingly, SV3b expression, which was found to be reduced in the entire patient group, showed a relative underexpression in only the CSL group (P=.0395; fig. 4), not in the CS or BRRS group. Further, SV5a was significantly overexpressed in only the CSL group compared with controls (P=.0275; fig. 4). This observation contrasts with the entire patient group, which did not show differential expression of SV5a.
For further analysis, the 85 patients were regrouped into three by germline PTEN status—namely, the PTEN mutation-positive group (n=43), PTEN mutation-negative group (n=22), and VUS group (n=20). Each of the three groups showed relative underexpression of FL transcript compared with controls (P<.02; fig. 4). Interestingly, the mutation-positive and VUS groups but not the mutation-negative group showed significantly lower expression levels of SV3a (P<.02) compared with controls (fig. 4). Therefore, decreased expression of SV3a seen in patients was not specific for any of three major phenotypes but was an important marker for those with promoter alterations that, theoretically, would not interfere with PTEN function. Furthermore, the VUS group but not the mutation-positive or mutation-negative group had a significantly lower expression level of SV3b (P<.01). Of note, PTEN mutation-negative patients had a significantly higher SV5b expression level (P<.02) compared with controls, a difference not seen in either the mutation-positive or the VUS groups.
We analyzed SV expression levels by dividing the patients who had PTEN mutations according to mutation type (missense, nonsense, frameshift, etc.). Patients with germline truncating mutations (nonsense and frameshift) but not those with missense mutations had reduced levels of FL transcript, as expected (fig. 4). We found that patients with missense mutations had lower expression levels of SV3a and SV3b (P<.01) but higher SV5a expression levels (P<.05; fig. 4) than those of controls. Therefore, our data together show that the relative underexpression of variant SV3b was more specific to patients with CSL when promoter alterations (VUS) were present. Patients with nonsense mutations had lower expression levels of SV3a, SV3b, and SV5d compared with those of controls (P<.02; fig. 4). Interestingly, this differential SV expression was not noted among patients with frameshifting mutations (microinsertions or deletions) who had decreased FL-PTEN only.
Discussion
There has been considerable interest in alternatively spliced transcripts and their role in normal development and pathological conditions.20–22 Because of their presence in normal tissues, it is natural to suppose that they might have important biological functions. In addition, increasing evidence suggests that alternative RNA processing is involved in human cancer during transformation and/or acquisition of metastatic potential.17,21 Different SV profiles could also have implications in the risk of developing cancer as well as potentially influence prognosis and response to therapies.11,20 The use of alternative splice sites might result in the inappropriate expression of different protein isoforms that ultimately would affect cellular physiology and consequently contribute to tumorigenesis.
While somatic alternative splicing and SVs have been studied extensively in neoplasia, few studies, if any, have systematically analyzed splice variation in the germline of those with heritable cancer syndromes. We have found eight novel naturally occurring splice variants transcribed from PTEN (fig. 1). Of the eight SVs, five show differential expressional levels in CS/BRRS cases compared with normal controls (fig. 4). The observation that FL-PTEN transcript is underexpressed in cases compared with normal controls, except for those with missense mutations (this last group included all promoter mutations), serves as a good internal experimental control. The fact that the frameshift mutation group, compared with the control group, showed relative underexpression of only FL transcript, with no differences in expression of any SV (fig. 4), also serves as an experimental control. Consistent with the literature, frameshifting mutations result in transcripts that undergo rapid nonsense-mediated degradation.23 Thus, the observation of underexpression of only FL transcript in our patients with germline frameshift mutations, which likely reflects mainly the remaining wild-type allele, is consistent with our hypothesis. In the context of our observation of differential SV expression shown in figure 4, this particular observation may give circumstantial evidence that the differential SV expression seen in the other subgroups (fig. 4) reflects alternative splice usage from the affected allele.
Inspection of figure 4 reveals virtually unique SV relative expression profiles between cases and controls and among the various groups of cases. Except for the frameshift mutation group and the mutation-negative group, which had similar profiles, every group had a unique profile (fig. 4). Apart from FL transcript, SV3a is the most consistently relatively underexpressed in cases and among all groups compared with controls. The cases show relative underexpression of SV3a and SV3b and overexpression of SV5b. It is interesting to note that the SV5b overexpression profile is contributed by mainly patients with CS who comprise the majority of cases and who would also have the highest likelihood of being mutation positive. Indeed, there appears to be an SV expressional genotype-phenotype correlation in which the SV expressional profiles are distinct among CS, CS-like, and BRRS cases.
Germline VUS in cancer susceptibility genes are a diagnostic dilemma in clinical practice. Here, we demonstrate that the relative SV expressional profile in the VUS group did not mirror that in the controls but, instead, showed relative underexpression of SV3a and SV3b in addition to FL transcript. This only underscores our lack of understanding of the complex downstream consequences of VUS. However, our observations may suggest that SV typing may help resolve the diagnostic dilemma, if our data can be independently confirmed. Germline mutation-negative cases in any heritable syndrome represent a challenge in the practice of clinical cancer genetics and genetic counseling as well. Our study shows that PTEN mutation-negative CS/BRRS cases have a relative SV expressional profile similar to that of the frameshift mutation group, for which only FL transcript, but no SV, was underexpressed compared with normal controls. While it could be argued that there is a variant in the promoter leading to relative underexpression of PTEN, our mutation-negative cases were not found to have any variants in the PTEN promoter. However, it is altogether possible that PTEN has an as-yet-unrecognized alternative promoter(s), and germline variants/mutations there may contribute to our observed profile. Nonetheless, again, relative transcript quantitation may prove to be a useful diagnostic adjunct if our data can be independently replicated.
While we have demonstrated that FL-PTEN transcript and its SVs show distinct relative expressional profiles in CS/BRRS, the mechanism by which these SVs lead to or modulate the phenotype is currently unknown. We can only postulate that SVs could play a role in CS/BRRS pathogenesis in at least two non–mutually exclusive ways. First, SVs may play an indirect role by mediating, either by facilitating or interfering with, FL-PTEN transcription and/or translation. And it is this fine balance among SV3a, SV3b, SV5a, SV5b, and SV5d that modulates the rate or quantity of FL transcription or translation. Second, SVs may actually be translated into protein and exert their effect in the signaling pathways downstream of PTEN. In a recent study, we functionally analyzed the PTEN SVs by measuring PTEN’s downstream readouts—that is, the effect that expression of these SVs has on phosphorylation of Akt and cyclin D1 promoter activity. We have demonstrated that SV5b protein increases cyclin D1 promoter activity in vitro, which is the opposite of PTEN protein action.18 This corroborates our observation here that SV5b is overexpressed in cases, particularly CS and CS-like, and in mutation-positive patients (fig. 4). We also have shown that overexpression of SV5a resulted in slightly decreased P-Akt levels and decreased cyclin D1 promoter activity as measured by the reporter firefly luciferase.18 Therefore, SV5a functions in a manner similar to PTEN.18 Superficially, these functional data appear contradictory to our current observations that SV5a is relatively overexpressed in CS-like cases and the missense mutation group. However, the CS-like phenotype is milder (i.e., missing phenotypic criteria) than classic CS.24 Hence, we may speculate that SV5a, which mimics PTEN downstream action, may partially compensate for altered PTEN function and thus modulate phenotype. Similarly, we postulate that missense mutations result in PTEN protein that can act in a dominant negative manner (as has been shown, at least in vitro9), and so relative overexpression of SV5a would worsen the dominant negative effect in cases with germline missense mutations.
In conclusion, our data favor the concept that reduced FL-PTEN transcript expression, associated with differential expression of PTEN SVs, regardless of PTEN mutation status, may modulate its action and therefore lead to different phenotypes. This influence can occur at the transcription or translation level of PTEN, producing partially active transcripts that could act in the same way as the primary protein or in opposition to the wild-type allele, having, in this latter case, a dominant negative action. The study of these variants with their characterization and quantification is particularly important in PTEN mutation-negative patients and patients with VUS, given that we could demonstrate that these groups have distinct profiles compared with those of normal controls. If confirmed by other studies, we might be able to use these novel data to help establish the diagnosis of CS/BRRS regardless of the mutation status. The complete comprehension of the mechanism involved in the alternative splicing, its correlation with normal or mutant PTEN, and the differential downstream signaling not only may add to the fundamental understanding of PTEN’s complex pathways in health and disease but also may help us understand the resultant phenotype of the different syndromes associated with this gene.
Acknowledgments
We are grateful to Frank Weber and to Kevin Zbuk for critical review of the manuscript and for helpful discussions. This work was supported in part by the American Cancer Society (RSG-02-151-01-CCE [to C.E.]). M.S.S. is an International Scholar of The Endocrine Society. C.E. is a recipient of the Doris Duke Distinguished Clinical Scientist Award.
Web Resources
The URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CS and BRRS)
- Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi (for PCR primer design)
References
- 1.Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PLM, Zheng Z, Liaw D, et al (1998) Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet 7:507–515 10.1093/hmg/7.3.507 [DOI] [PubMed] [Google Scholar]
- 2.Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, Crowe C, Curtis MA, Dasouki M, Dunn T, Feit H, Geraghty MT, Graham JM Jr, Hodgson SV, Hunter A, Korf BR, Manchester D, Miesfeldt S, Murday VA, Nathanson KL, Parisi M, Pober B, Romano C, Eng C (1999) PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8:1461–1472 10.1093/hmg/8.8.1461 [DOI] [PubMed] [Google Scholar]
- 3.Zhou XP, Waite KA, Pilarski R, Hampel H, Fernandez MJ, Bos C, Dasouki M, Feldman GL, Greenberg LA, Ivanovich J, Matloff E, Patterson A, Pierpont ME, Russo D, Nassif NT, Eng C (2003) Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am J Hum Genet 73:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilarski R, Eng C (2004) Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet 41:323–326 10.1136/jmg.2004.018036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eng C (2003) PTEN: one gene, many syndromes. Hum Mutat 22:183–198 10.1002/humu.10257 [DOI] [PubMed] [Google Scholar]
- 6.Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, Parsons R, Tonks NK (1997) P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci USA 94:9052–9057 10.1073/pnas.94.17.9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waite KA, Eng C (2002) Protean PTEN: form and function. Am J Hum Genet 70:829–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng LP, Smith WM, Brown JL, Eng C (2001) PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet 10:605–616 10.1093/hmg/10.6.605 [DOI] [PubMed] [Google Scholar]
- 9.Weng L, Brown J, Eng C (2001) PTEN induces apoptosis and cell cycle arrest through phosphoinositol-3-kinase/Akt-dependent and -independent pathways. Hum Mol Genet 10:237–242 10.1093/hmg/10.3.237 [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Yu Q, He J, Zha X (2004) Study of the PTEN gene expression and FAK phosphorylation in human hepatocarcinoma tissues and cell lines. Mol Cell Biochem 262:25–33 10.1023/B:MCBI.0000038212.78008.7f [DOI] [PubMed] [Google Scholar]
- 11.Bracco L, Kearsey J (2003) The relevance of alternative RNA splicing to pharmacogenomics. Trends Biotechnol 21:346–353 10.1016/S0167-7799(03)00146-X [DOI] [PubMed] [Google Scholar]
- 12.Faustino NA, Cooper TA (2003) Pre-mRNA splicing and human disease. Genes Dev 17:419–437 10.1101/gad.1048803 [DOI] [PubMed] [Google Scholar]
- 13.Brinkman BM (2004) Splice variants as cancer biomarkers. Clin Biochem 37:584–594 10.1016/j.clinbiochem.2004.05.015 [DOI] [PubMed] [Google Scholar]
- 14.Sharrard RM, Maitland JN (2000) Alternative splicing of the human PTEN/MMAC1/TEP1 gene. Biochim Biophys Acta 1494:282–285 [DOI] [PubMed] [Google Scholar]
- 15.Wang NM, Chang JG (1999) Are aberrant transcripts of FHIT, TSG101, and PTEN/MMAC1 oncogenesis related? Int J Mol Med 3:491–495 [DOI] [PubMed] [Google Scholar]
- 16.Bartel F, Harris LC, Wurl P, Taubert H (2004) MDM2 and its splice variant messenger RNAs: expression in tumors and down-regulation using antisense oligonucleotides. Mol Cancer Res 2:29–35 [PubMed] [Google Scholar]
- 17.Philips AV, Cooper TA (2000) RNA processing and human disease. Cell Mol Life Sci 57:235–249 10.1007/PL00000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal S, Eng C (2006) Differential expression of novel naturally occurring splice variants of PTEN and their functional consequences in Cowden syndrome and sporadic breast cancer. Hum Mol Genet 15:777–787 10.1093/hmg/ddi492 [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgent TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and 2-ΔΔCT method. Methods 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 20.Poola I, Speirs V (2001) Expression of alternatively spliced estrogen receptor alpha mRNAs is increased in breast cancer tissues. J Steroid Biochem Mol Biol 78:459–469 10.1016/S0960-0760(01)00118-2 [DOI] [PubMed] [Google Scholar]
- 21.Hsieh HF, Yu JC, Ho LI, Chiu SC, Harn HJ (1999) Molecular studies into the role of CD44 variants in metastasis in gastric cancer. Mol Pathol 52:25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulis ML, Parsons M (2003) PTEN: from pathology to biology. Trends Cell Biol 13:478–483 10.1016/S0962-8924(03)00175-2 [DOI] [PubMed] [Google Scholar]
- 23.Agrawal S, Pilarski R, Eng C (2005) Different splicing defects lead to differential effects downstream of the lipid and protein phosphatase activities of PTEN. Hum Mol Genet 14:2459–2468 10.1093/hmg/ddi246 [DOI] [PubMed] [Google Scholar]
- 24.Eng C (2000) Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet 37:828–830 10.1136/jmg.37.11.828 [DOI] [PMC free article] [PubMed] [Google Scholar]