Abstract
The AXIN1 gene has been implicated in caudal duplication anomalies. Its coding region was sequenced in both members of a monozygotic (MZ) twin pair discordant for a caudal duplication anomaly, but no mutation was found. Using bisulfite sequencing, we examined methylation at the promoter region of the AXIN1 gene in these twins and in twin and age-matched singleton controls. Methylation of the promoter region in peripheral blood mononucleated cells was variable among individuals, including MZ pairs. In the MZ pair discordant for the caudal duplication, this region of the affected twin was significantly more methylated than that of the unaffected twin (P<.0001), which was significantly more methylated than those of the controls (P=.02). We have confirmed that this CpG island does function as a promoter in vitro and that its activity is inversely proportional to the extent of methylation. This finding raises the possibility that hypermethylation of the AXIN1 promoter, by mechanisms as yet undetermined, is associated with the malformation. This case may be paradigmatic for some cases of MZ discordance.
The high rate of discordance in MZ pairs for most complex diseases is a continuing puzzle. Vague statements that “the environment” is responsible for this discordance are seldom backed up by data, despite often huge investments in searching for the etiologic agents1; smoking as a risk factor for lung cancer and heart disease is a rare exception.2 In recent years, there has been increasing interest in the possibility that stochastically or environmentally triggered differences in the epigenetic status of key genes may be responsible for some MZ discordance and, indeed, for much of the burden of complex disease.3,4 An obstacle to the testing of this hypothesis is that, for most complex traits, we still have very few identified genes whose methylation status can be investigated and, where we do, the pertinent tissue may be inaccessible for methylation studies on living subjects. Until such genes for common diseases become known, therefore, it is worth investigating cases of MZ discordance for rare diseases in which the causal gene(s) are known or strongly suspected.
In 2002, a pair of female MZ twins were described as discordant for caudal duplication anomaly (MIM 607864), in which one twin had a duplication of the distal spine and other organs as well as a tumor in the lumbosacral region and spina bifida.5 Caudal duplication anomaly is sporadic and can involve multiple congenital anomalies, but the hallmark is duplication of organs in the caudal region, such as the distal spine.6 This spinal duplication is similar to that seen in AxinFu mice, which carry a mutation at the Axin locus.7 Highly penetrant mice display bifurcating tails as a result of caudal duplication in the distal region. Axin encodes an inhibitor of the Wnt-signaling pathway and has been shown to regulate embryonic axis formation in mouse and in Xenopus.8 Suppression of wild-type Axin in Xenopus embryos results in the duplication of the body axis. The role of Axin in caudal duplication anomaly in humans is not known, but it remains the strongest candidate. Sequence analysis of the AXIN1 (MIM 603816) coding region in the twin with caudal duplication syndrome revealed no pathogenic mutations, although a rare missense variant was noticed in both twins and in the father.5 Clearly, this missense variant alone cannot account for the MZ discordance. We thought it possible that AXIN1 inactivation may have occurred via an epigenetic process.
In mammals, methylation of DNA occurs at cytosine residues, mainly at CpG dinucleotides. There are stretches of DNA, CpG islands, that are rich in CpG and are often found at promoter regions of genes. CpG islands are normally unmethylated, but there is now evidence that a small proportion of these are methylated in some tissues.9,10 Aberrant de novo methylation of normally unmethylated CpG islands in humans and mice is associated with transcriptional silencing.11 For example, the penetrance and severity of the abnormal tail phenotype in inbred—and, therefore, isogenic—AxinFu mice correlate with specific patterns of DNA methylation at the Axin locus.12 Here, we report the careful examination of cytosine methylation at the promoter and intergenic regions of human AXIN1 in the MZ twin pair discordant for caudal duplication anomaly, in their parents, and in several controls.
The probands were reported to be monochorionic and probably monoamniotic (H. Kroes, personal communication). Peripheral blood mononucleated cell (PBMC) DNA was collected from them at age 7 mo. Full blood counts taken at the same time from both were unremarkable. Methylation profiles may change slightly with age,13 so, for age-matched controls, we obtained PBMC DNA from two female hospital patients, the first aged 5 mo with a congenital malformation of the heart and trachea, and the second aged 11 mo with viral myocarditis. PBMC DNA from a further seven adult controls was also examined.
MZ twinning is itself a duplication event, so we were keen to ensure that any abnormalities of methylation of the AXIN1 gene were specific to the malformation and not inherent to MZ twinning. We obtained PBMC DNA from nine MZ pairs aged 12–14 years at the time of collection, including eight sets of males and one set of females. These pairs were selected from a larger study14 as being probably dichorionic because the mother had reported two separate placentas at birth. It has been suggested that monochorionic pairs exchange hematopoietic stem cells in utero, making cotwins’ blood cell profiles (and perhaps methylation status) more similar than they might otherwise be. The extent of this alleged problem has never been quantified, but, to the extent that it exists, the monochorionicity of our proband pair would make our search for methylation differences between cotwins conservative. Monozygosity of all pairs, including the proband pair, was confirmed by typing them with the use of the ABI Profiler Plus set of nine highly polymorphic microsatellite markers and the amelogenin sex marker, yielding a probability of dizygosity, given concordance of <10−4.
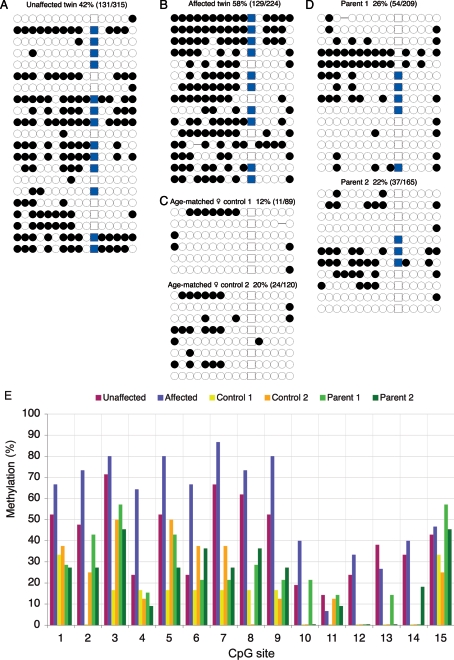
AXIN1 is positioned near the telomere of human chromosome 16 and has a CpG island at the 5′ end (the putative promoter).15 This CpG island surrounds exon 1 (Ensembl Build 35 [accession numbers 341973-343464]). In intron 2, there is a solitary long terminal repeat (LTR) that exhibits enhancer activity.16 We used bisulfite sequencing17 to examine the pattern of cytosine methylation at three distinct regions of AXIN1: the 5′ CpG island, intron 1, and intron 2-LTR (for PCR primers and conditions, see table A1). The AXIN1 intron 1 (8 CpGs) and intron 2-LTR (17 CpGs) regions were found to be hypermethylated in PBMC DNA from all individuals tested, with no significant difference between the affected MZ twin and controls. The average percentages of methylated CpGs were, for intron 1, 98% (n=11) for the affected twin and 100% (n=7) for the unaffected twin and, for intron 2-LTR, 80% (n=31) for the affected twin and 88% (n=29) for the unaffected twin. Methylation status of the AXIN1 5′ CpG island (15 CpGs) was determined for the proband family and for two age-matched controls (fig. 1). This region, in contrast to most of the unmethylated CpG islands in the human genome, is methylated to varying degrees both within and among individuals. The precise pattern of methylation varies considerably from clone to clone in all individuals, including controls. The proportion of the 15 CpGs that are methylated also varies considerably from clone to clone in all individuals (fig. 2). Although whole blood contains a mix of nucleated cell types, the majority are neutrophils, and the complexity of the clonal patterns cannot be explained by this mix alone.
Figure 1. .
AXIN1 CpG island methylation in the MZ twin pair (A, unaffected; B, affected) discordant for caudal duplication anomaly, their parents (D), and age- and sex-matched controls (C). An open circle represents an unmethylated CpG, a filled circle represents a methylated CpG, and each row of circles represents the methylation state of all 15 CpGs on a single allele. The square represents a SNP (G→T [dbSNP rs12928797]) commonly found in the samples analyzed. Overall percentage of methylation was calculated by determining the proportion of methylated CpGs relative to all CpGs. A detailed analysis of the methylation state at each CpG site is also shown (E).
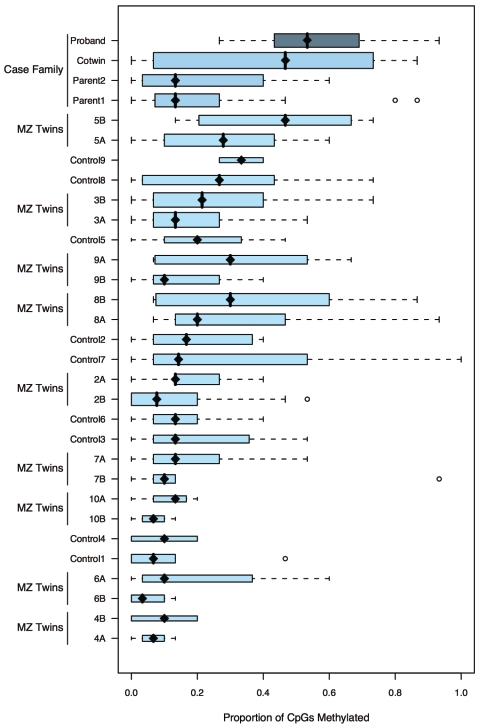
Figure 2. .
Box-and-whisker plot of proportion-methylated CpG sites in the AXIN1 promoter per clone for each individual. Individuals are ordered by familial mean methylation level, with the index family at the top. Length of bar is interquartile range, and bar width is proportional to the number of clones analyzed for that individual. Solid diamonds are medians, and dashed lines extend to the observations 1.5× the interquartile range from the median, with open circles representing more-extreme values.
We modeled methylation at 15 CpG sites in twins and controls, using a binomial generalized linear mixed model, as implemented in the lme4 package18 in the R statistical programming language.19 This approach allows the appropriate modeling of within-family and within-individual correlation in the binary (absent or present) methylation status across the different CpG sites. The multilevel random effects were at the level of the family (19 families), individual (4 individuals in the proband family [twins and their parents], 2 in each of the other 9 MZ pairs, and 9 single individuals, for a total of 31 individuals), and clone (3–17 distinct clones per individual, since, in the case of duplicate methylation signatures from the same PCR, one was discarded in case it represented clonal bias; 30 of 311 clones were so discarded). The outcome of the analysis is essentially the same with or without this filtering process. To reduce the impact of any bias, multiple PCRs were undertaken for each sample. Disease status was treated as a fixed effect and its significance was assessed using a likelihood-ratio test. Because of the presence of a SNP within the amplified region of the proband pair, we were able to see that both parental alleles were represented in the data set and that they each showed a range of methylation states (see fig. 1A and 1B).
The total level of methylation in the affected twin was significantly higher than that in the unaffected twin (P<.0001). The unaffected twin was also significantly more methylated than were controls (P=.02). The methylation levels of MZ twins differ from one another but are more similar to each other than they are to unrelated individuals. We estimated the intraclass correlation for total methylation at all 15 sites to be 0.76 (P=.02) for all 10 MZ pairs. Excluding the proband pair, the correlation was still 0.52. The MZ twin correlation in methylation status at each of the 15 sites varied between 0.01 and 0.86, but, with only 10 pairs available, this is still in keeping with homogeneity, so we cannot say whether there is differential genetic control of methylation between sites. The results suggest that the relatively high methylation levels that we observe at the AXIN1 CpG island in the affected and unaffected MZ twins are unlikely to be associated with twinning itself, since the MZ twin controls do not have significantly higher levels of methylation than the singleton controls (P=.73) (fig. 2).
For eight of these individuals (the proband pair, their parents, and four other unaffected singles), we had buccal DNA in addition to PBMC DNA. Buccal epithelia, unlike PBMC, is ectodermal in origin. Methylation of CpG sites in buccal DNAs was absent from all sites in all individuals, including the proband pair and their parents (data not shown), so the variable hypermethylation at the AXIN1 5′ CpG island is tissue specific.
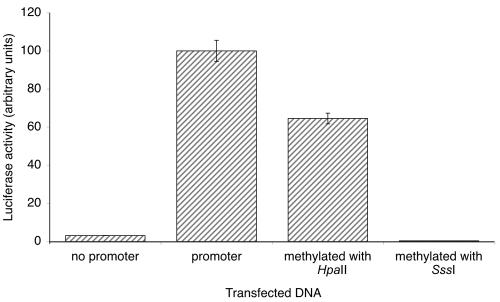
To confirm that this CpG island functions as a promoter, we cloned a fragment corresponding to nucleotides −416 to +114 relative to the start site of transcription, upstream of a luciferase reporter gene, and we performed transient transfections into the human cell line HEK293. The fragment did function as a promoter, and in vitro methylation of the construct decreased expression; partial methylation (16% of CpGs) resulted in a reduction of expression, and complete methylation resulted in no expression (see fig. 3). Similar results were obtained after transfection into HeLa cells (data not shown).
Figure 3. .
The degree of methylation of the AXIN1 promoter correlates with its activity in HEK 293 cells. A fragment of the AXIN1 locus, corresponding to nucleotides −416 to +114 relative to the start point of transcription (as determined by Evidence Viewer), was amplified by PCR from human genomic DNA (Promega). For a more detailed description of the methods used, see the main text. A similar trend was also seen in HeLa cells (data not shown). Each data point is the average of six replicates. Error bars represent two SDs.
The primers used to generate this insert were as follows. The upstream primer contained a BglII site followed by the AXIN1 sequence, CCAGTCTAGATCTCCACGCTCCTCACTTTATCCT. The downstream primer contained a HindIII site followed by the AXIN1 sequence, CCAGTCAAGCTTGAGCCCGGCCCTACTCAC. The insert was cloned into pGL3-Basic (Promega). The resulting construct was methylated in vitro by either HpaII methylase (New England Biolabs) or SssI methylase (New England Biolabs). Methylation was verified by digestion with the methylation-sensitive restriction enzyme, HpaII (New England Biolabs). pRL-Luc was cotransfected, to control for transfection efficiency. Methylation of 16% of the CpG dinucleotides in the AXIN1 insert by HpaII correlated with a 36% reduction in luciferase activity, whereas methylation of 100% of the CpG dinucleotides by SssI was associated with a complete silencing of the luciferase gene.
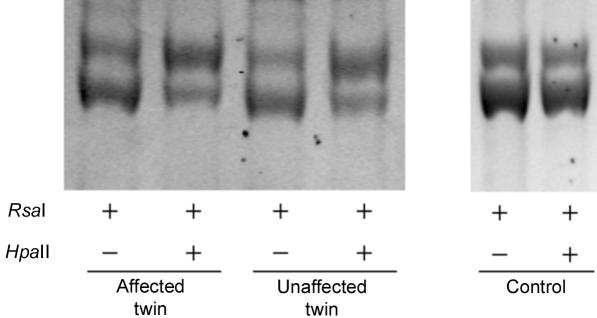
Since the proband pair are female, it is possible that unequal skewing of X inactivation resulting in expression differences in X-linked genes and in any downstream autosomal genes could underlie the phenotypic discordance in the proband twin pair. Analysis of methylation, by use of the human androgen gene assay,20 showed some evidence for skewing in both the proband and her cotwin in PBMC DNA, but the pattern was identical in both (fig. 4). This finding is consistent with a previous study21 that showed that most monochorionic MZ twins display highly similar X-inactivation patterns, and it rules out skewed X inactivation as an explanation for the discordance in this instance.
Figure 4. .
Quantification of X-chromosome inactivation in PBMCs by PCR amplification of the polymorphic human androgen-receptor (HUMARA) locus after digestion with RsaI or RsaI and HpaII. Although the pair both display some skewing towards the larger allele, the pattern is highly consistent in both twins.
We were curious to discover whether other neighboring CpG promoters were also methylated in PBMC in the proband. The α-globin locus (HBA1 [MIM 141800] and HBA2 [MIM 141850]) lies 170 kb downstream of the AXIN1 gene. Bisulfite analysis of 16 CpGs in the HBA1 CpG island promoter showed overall hypomethylation (0%–17%) in all individuals tested, including the proband, the unaffected cotwin, the two parents, and a pair of control MZ twins (data not shown). These results are consistent with previous reports that the α-globin promoters are relatively unmethylated in PBMCs of normal individuals22 and suggest that the hypermethylation observed in the proband is specific to the AXIN1 CpG island.
It is commonly held that, in normal cells, CpG islands are unmethylated, except when they are located on the inactive X chromosome of females or in the differentially methylated regions (DMRs) of imprinted genes. Hypermethylation of normally unmethylated CpG islands is associated with transcriptional silencing. Our analysis of the AXIN1 CpG island in MZ twins discordant for caudal duplication anomaly identified the affected twin as the individual with the highest methylation state, which may explain the disease phenotype. However, some degree of AXIN1 CpG island methylation is observed even in unaffected individuals. AXIN1 is not an X-linked gene and does not show the characteristic allele-specific methylation patterns of an imprinted gene. Presumably, it represents a recently identified class of variably methylated CpG islands in the human genome, which are not DMRs of imprinted genes and are not on the inactive X chromosome.23 It is interesting that these elements have been shown to map to telomere-proximal positions on several different chromosomes,23 since AXIN1 lies at the telomere of the short arm of human chromosome 16.15
The variability in methylation at the AXIN1 CpG island, both within an individual and between individuals, is interesting. Yamada and colleagues used methylation-sensitive restriction enzyme digestion to identify 8 CpG islands (of 149 analyzed on chromosome 21q) that exhibited so-called incomplete methylation, where several CpGs on the same allele are differentially methylated,9 similar to those reported in this article. All eight were located at promoter or 5′-UTR gene regions, but differences in methylation between individuals were not addressed.9 Interindividual variation in methylation state, though not necessarily at CpG islands, was reported at the major histocompatibility complex region of human chromosome 6 and, similar to our results, this variation was not observed in every tissue.24 The consistent hypomethylation of the 5′ CpG island of AXIN1 in buccal tissue suggests that the methylation occurred after differentiation of the three germ layers, since buccal tissue is ectodermal in origin, whereas blood is mesodermal. It is worth noting that mesoderm gives rise to the notochord, which defines the body’s axis and eventually becomes the vertebral column. Ideally, we would examine methylation in the vertebral column, but results from a tissue (blood) derived from the same embryonic tissue layer are more likely to reflect the relevant epigenetic state than results from a tissue (buccal epithelium) that is not.
To the best of our knowledge, this study of human AXIN1 is the first detailed analysis, by bisulfite sequencing, of a CpG island promoter that exhibits variable methylation both within a tissue and among individuals and, in particular, between genetically identical individuals. Such alleles are referred to as “metastable epialleles” in mice and plants.3 On the basis of what we have learned about their behavior in other organisms,3,25–27 it is likely that they will be sensitive both to genetic background and to environment, properties we should expect in agents of sporadic and complex disease. Although, in this instance, a candidate-gene approach was appropriate, in many cases where no obvious candidates are available, newly developed genomewide approaches13,28 are likely to be fruitful.
Although we favor the hypothesis that the hypermethylation at AXIN1 in the proband is a purely epigenetic event, a genetic rearrangement in cis is almost impossible to rule out, since we know that such events can affect the epigenetic state over long distances.29 It is also possible that blood supply differences in utero or other postzygotic events30,31 could be underlying causes of both the phenotypic discordance and the increased methylation levels. We are aware of this and other limitations of our study but believe nevertheless that MZ twins provide the best opportunity to understand the relationship between epigenotype and phenotype in humans, and we hope that our findings make a useful contribution to an inherently difficult field.
Acknowledgments
This work was supported in part by grants from the National Health and Medical Research Council of Australia, the Australian Research Council, and the GenomEUtwin project, which is supported by the European Union (contract number QLRT-2001-01254). We also thank Professor Ron Trent, University of Sydney, for providing some of the control samples, and we thank the proband twins and their parents for their generous cooperation. We are also grateful to A. Verger and M. Crossley for their assistance with the transient transfection experiments. This project was approved by the human research ethics committees of University of Sydney (approval number X03-0129), Royal Children’s Hospital (2005/023), and Queensland Institute of Medical Research (P879).
Appendix A
Table A1. .
Primers and Conditions Used for Amplification of AXIN1 and HBA1 Promoter Regions[Note]
| Region and Primer Name |
PCR Round | Primer Sequence |
| AXIN1 5′ CpG island: | ||
| ACGL1 | 1 and 2 | 5′-GGAGGTTTTGGTTTTTTAGAGAGYGGAG-3′ |
| ACGR1 | 1 | 5′-AAACCCTAACCATCCCTACCTACCRACC-3′ |
| ACGR1.5 | 2 | 5′-TCCTAAAAACCTACTTCCCTCAC-3′ |
| HBA15′ CpG island: | ||
| HBAL1 | 1 and 2 | 5′-TTTTATAGTTTAGAGAGAATTTATTATGG-3′ |
| HBAR1 | 1 | 5′-TAACCCTTAACCTAAACAAAACC-3′ |
| HBAR2 | 2 | 5′-AAAAAAAACAAAAACATCCTAC-3′ |
| AXIN1 intron 1: | ||
| AxIL1 | 1 | 5′-TGTTTATAATTTTAGTTATTTGGGAAGGT-3′ |
| AxIR1 | 1 | 5′-ACCCCTTATTTTTACTCACACTTCTATT-3′ |
| AxIL2 | 2 | 5′-ATAATTTTAGTTATTTGGGAAGGTTGAG-3′ |
| AxIR2 | 2 | 5′-AACTTACTTAAATCCACAAACCCTATTT-3′ |
| AXIN1 intron 2-LTR: | ||
| AxLTRL1 | 1 | 5′-GGATAAATATAGAAAAGGGTTAGGAATG-3′ |
| AxLTRR1 | 1 and 2 | 5′-ATAAACTAAAAAACTCCTCAAATACCAC-3′ |
| AxLTRL2 | 2 | 5′-TTTTTAAATTAAGTATGTGAAATTATTA-3′ |
Note.— A 263-bp fragment of the AXIN1 5′ CpG island and a 256-bp fragment of the HBA1 promoter regions were amplified using semi-nested bisulphite PCR. The PCR protocol involved 2 min at 94°C followed by 36 cycles for 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C, with a final extension of 6 min at 72°C. The PCR contained BSA (4 mg/mL) and, for the AXIN1 5′ CpG island region, Polymate additive (Bioline) as well. The resulting PCR fragments were subcloned into the p-GEM-T Easy Vector (Promega) and were sequenced.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for rs12928797)
- Ensembl Build 35, http://www.ensembl.org (accession numbers 341973-343464)
- Evidence Viewer, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for caudal duplication anomaly, AXIN1, HBA1, and HBA2)
References
- 1.Torrey EF, Bowler AE, Taylor EH, Gottesman II (1994) Schizophrenia and manic-depressive disorder: the biological roots of mental illness as revealed by the landmark study of identical twins. Basic Books, New York [Google Scholar]
- 2.Floderus B, Cederlof R, Friberg L (1988) Smoking and mortality: a 21-year follow-up based on the Swedish Twin Registry. Int J Epidemiol 17:332–340 [DOI] [PubMed] [Google Scholar]
- 3.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E (2002) Metastable epialleles in mammals. Trends Genet 18:348–351 10.1016/S0168-9525(02)02709-9 [DOI] [PubMed] [Google Scholar]
- 4.Petronis A, Gottesman II, Kan P, Kennedy JL, Basile VS, Paterson AD, Popendikyte V (2003) Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull 29:169–178 [DOI] [PubMed] [Google Scholar]
- 5.Kroes HY, Takahashi M, Zijlstra RJ, Baert JA, Kooi KA, Hofstra RM, van Essen AJ (2002) Two cases of the caudal duplication anomaly including a discordant monozygotic twin. Am J Med Genet 112:390–393 10.1002/ajmg.10594 [DOI] [PubMed] [Google Scholar]
- 6.Dominguez R, Rott J, Castillo M, Pittaluga RR, Corriere JN Jr (1993) Caudal duplication syndrome. Am J Dis Child 147:1048–1052 [DOI] [PubMed] [Google Scholar]
- 7.Vasicek TJ, Zeng L, Guan XJ, Zhang T, Costantini F, Tilghman SM (1997) Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics 147:777–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL 3rd, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F (1997) The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90:181–192 10.1016/S0092-8674(00)80324-4 [DOI] [PubMed] [Google Scholar]
- 9.Yamada Y, Watanabe H, Miura F, Soejima H, Uchiyama M, Iwasaka T, Mukai T, Sakaki Y, Ito T (2004) A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res 14:247–266 10.1101/gr.1351604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA (2005) Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci USA 102:3336–3341 10.1073/pnas.0408436102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issa JP (2004) CpG island methylator phenotype in cancer. Nat Rev Cancer 4:988–993 10.1038/nrc1507 [DOI] [PubMed] [Google Scholar]
- 12.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E (2003) Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA 100:2538–2543 10.1073/pnas.0436776100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102:10604–10609 10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu G, Evans DM, Duffy DL, Montgomery GW, Medland SE, Gillespie NA, Ewen KR, Jewell M, Liew YW, Hayward NK, Sturm RA, Trent JM, Martin NG (2004) A genome scan for eye color in 502 twin families: most variation is due to a QTL on chromosome 15q. Twin Res 7:197–210 10.1375/136905204323016186 [DOI] [PubMed] [Google Scholar]
- 15.Daniels RJ, Peden JF, Lloyd C, Horsley SW, Clark K, Tufarelli C, Kearney L, Buckle VJ, Doggett NA, Flint J, Higgs DR (2001) Sequence, structure and pathology of the fully annotated terminal 2 Mb of the short arm of human chromosome 16. Hum Mol Genet 10:339–352 10.1093/hmg/10.4.339 [DOI] [PubMed] [Google Scholar]
- 16.Ling J, Pi W, Bollag R, Zeng S, Keskintepe M, Saliman H, Krantz S, Whitney B, Tuan D (2002) The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol 76:2410–2423 10.1128/jvi.76.5.2410-2423.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark SJ, Harrison J, Paul CL, Frommer M (1994) High-sensitivity mapping of methylated cytosines. Nucleic Acids Res 22:2990–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates D, Sarkar D (2005) lme4: linear mixed-effects models using S4 classes. v 0.995-2. University of Wisconsin, Madison [Google Scholar]
- 19.R Development Core Team (2005) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 20.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51:1229–1239 [PMC free article] [PubMed] [Google Scholar]
- 21.Monteiro J, Derom C, Vlietinck R, Kohn N, Lesser M, Gregersen PK (1998) Commitment to X inactivation precedes the twinning event in monochorionic MZ twins. Am J Hum Genet 63:339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR (1997) The relationship between chromosome structure and function at a human telomeric region. Nat Genet 15:252–257 10.1038/ng0397-252 [DOI] [PubMed] [Google Scholar]
- 23.Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, Feinberg AP (2002) A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res 12:543–554 10.1101/gr.224102. Article published online before print in March 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakyan VK, Hildmann T, Novik KL, Lewin J, Tost J, Cox AV, Andrews TD, Howe KL, Otto T, Olek A, Fischer J, Gut IG, Berlin K, Beck S (2004) DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol 2:e405 10.1371/journal.pbio.0020405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff GL (1978) Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics 88:529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooney CA, Dave AA, Wolff GL (2002) Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 132:2393S–2400S [DOI] [PubMed] [Google Scholar]
- 27.Waterland RA, Jirtle RL (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300 10.1128/MCB.23.15.5293-5300.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37:853–862 10.1038/ng1598 [DOI] [PubMed] [Google Scholar]
- 29.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34:157–165 10.1038/ng1157 [DOI] [PubMed] [Google Scholar]
- 30.Machin GA (1996) Some causes of genotypic and phenotypic discordance in monozygotic twin pairs. Am J Med Genet 61:216–228 [DOI] [PubMed] [Google Scholar]
- 31.Martin N, Boomsma D, Machin G (1997) A twin-pronged attack on complex traits. Nat Genet 17:387–392 10.1038/ng1297-387 [DOI] [PubMed] [Google Scholar]