Abstract
Arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C) is a disorder characterized by fibrofatty replacement of cardiac myocytes that typically manifests in the right ventricle. It is inherited as an autosomal dominant disease with reduced penetrance, although autosomal recessive forms of the disease also occur. We identified four probands with ARVD/C caused by mutations in DSG2, which encodes desmoglein-2, a component of the cardiac desmosome. No association between mutations in this gene and human disease has been reported elsewhere. One of these probands has compound-heterozygous mutations in DSG2, and the remaining three have isolated heterozygous missense mutations, each disrupting known functional components of desmoglein-2. We report that mutations in DSG2 contribute to the development of ARVD/C.
Arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C [MIM #107970]) is a heritable form of cardiomyopathy characterized by right-ventricular dysfunction and life-threatening ventricular arrhythmias. The diagnosis is made on the basis of criteria proposed by an expert consensus panel in 1994.1 Both dominant and recessive forms of inheritance occur.2,3 Mutations in genes encoding several components of the cardiac desmosome have been associated with ARVD/C. This was first recognized in the autosomal recessive condition Naxos syndrome (MIM #601214), in which affected individuals develop right-ventricular cardiomyopathy, palmoplantar keratoderma, and woolly hair due to homozygous mutations in the gene encoding junctional plakoglobin (JUP).3,4 Similarly, recessive mutations in DSP encoding desmoplakin have been described in Carvajal syndrome (MIM #605676), which consists of palmoplantar keratoderma, woolly hair, and arrhythmogenic cardiomyopathy.5 Djabali and colleagues reported additional locus heterogeneity for Naxos syndrome and described exclusion of DSP, JUP, and several other components of the desmosome in two families with Naxos syndrome.6
Dominantly inherited ARVD/C in a single family was first shown to be caused by desmoplakin (DSP) mutations that are predicted to disrupt binding of desmoplakin to plakoglobin.2 More recently, mutations in DSP were reported in four families with ARVD/C and a separate family segregating arrhythmogenic left ventricular cardiomyopathy.7,8 On the basis of impaired cardiac development in mice with deletions for Pkp2, which encode plakophilin-2,9 as well as involvement of other desmosomal elements in ARVD/C, Gerull and colleagues screened 120 probands with ARVD/C for mutations in PKP2.10 Remarkably, they found that 32 of these individuals harbored heterozygous mutations in this gene, for a 27% prevalence among patients with ARVD/C. We subsequently confirmed high prevalence of PKP2 mutations in a separate large cohort of patients with ARVD/C and reported that those with PKP2 mutations present with arrhythmia earlier than do patients with ARVD/C who do not have the PKP2 mutation.11
Other genes encoding nondesmosomal proteins have been implicated in ARVD/C. Mutations in RYR2, which encodes the cardiac ryanodine receptor, have been described in ARVD2, a subtype in which ventricular arrhythmias are particularly effort induced and structural right-ventricular involvement may be concealed.12,13 In addition, mutations in RYR2 result in catecholaminergic polymorphic ventricular tachycardia.14 Beffagna and colleagues recently described mutations in the 5′ and 3′ UTRs of the gene encoding transforming growth factor β3 (TGFB3) in association with ARVD/C.15 Transient transfection with luciferase reporter constructs harboring these alterations in the 5′ and 3′ UTRs suggests increased TGFB3 mRNA abundance, which implicates this cytokine in the pathogenesis of ARVD/C.
The desmogleins are desmosomal cadherins and, together with the desmocollins, form the two essential transmembrane components of the desmosome.16 Members of the desmoglein family (DSG1–4) each have four extracellular cadherin domains and a transmembrane domain. DSG2 is expressed in many tissues, including the myocardium.17 Because of the association between mutations in three components of the cardiac desmosome and ARVD/C, we analyzed probands with this disorder for mutations in DSG2, which encodes desmoglein-2.
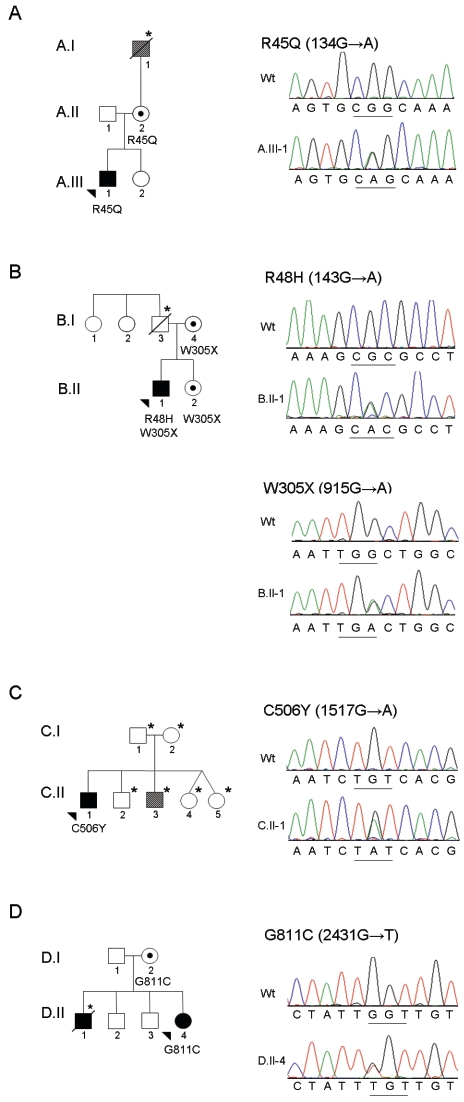
Thirty-three cases of ARVD/C were identified in which no mutation was present in PKP2 or DSP, genes previously associated with the classic form of ARVD/C. Written informed consent for genetic analysis was obtained from all participating individuals after approval of this protocol by the Johns Hopkins Institutional Review Board. Amplification and sequencing of the exonic and adjacent intronic sequence for the entire DSG2 gene was performed (see table A1 for primer sequences). Four individuals with ARVD/C were identified with mutations in DSG2 (table 1 and fig. 1). Each mutation disrupts a highly conserved amino acid within a functional domain of desmoglein-2 (fig. 2). Each mutation is absent in a control population of at least 110 ethnically matched individuals (220 control chromosomes).
Table 1. .
DSG2 Mutations
| Proband | Exon | Nucleotide | Amino Acid |
ARVD/C Criteriaa |
| A.III-1 | 3 | 134G→A | R45Q | Structural or functional right ventricular abnormality*, histopathologic right ventricular fibrofatty replacement*, ECG repolarization abnormality†, and diagnostic arrhythmias† |
| B.II-1 | 3 | 143 G→A | R48H | Structural or functional right ventricular abnormality*, ECG depolarization abnormality*, ECG repolarization abnormality†, and diagnostic arrhythmias† |
| B.II-1 | 8 | 915 G→A | W305X | |
| C.II-1 | 11 | 1517 G→A | C506Y | Structural or functional right ventricular abnormality†, ECG repolarization abnormality†, ECG depolarization abnormality†, and diagnostic arrhythmias† |
| D.II-4 | 15 | 2431 G→T | G811C | Family history*, ECG repolarization abnormality†, and structural or functional right ventricular abnormality† |
An asterisk (*) indicates that a major criterion is filled in the category; a dagger (†) indicates that a minor criterion is filled in the category.
Figure 1. .
Pedigrees and chromatograms demonstrating mutations in DSG2 for four probands with ARVD/C. In each pair of chromatograms, the top panel is from an unaffected control and the bottom panel is from the affected individual. A, Family A: individual A.III-1 with heterozygous R45Q (134G→A) mutation. B, Family B: individual B.II-1 with heterozygous R48H (143G→A) mutation and heterozygous W305X (915G→A) mutation. C, Family C: individual C.II-1 with heterozygous C506Y (1517G→A) mutation. D, Family D: individual D.II-4 with heterozygous G811C (2431G→T) mutation. Sequence analyses were performed bidirectionally with an ABI 3730 DNA Analyzer (Applied Biosystems). Chromatograms were analyzed using Sequencher 4.1 software for mutational analysis and with MacVector 7.2.2 software for ClustalW alignment. For pedigrees, squares represent males, circles represent females, blackened symbols indicate affected status, unblackened symbols indicate unaffected status, striped symbols indicate subdiagnostic features of ARVD/C, arrowheads indicate probands, dots indicate mutation carrier(s), and an asterisk (*) indicates that no DNA was available for sequencing.
Figure 2. .
Location and conservation of mutated residues. A, Location of mutations along schematic representation of pre-pro-desmoglein-2. SS = signal peptide sequence; Pro = propeptide; TM = transmembrane domain; IA = intracellular anchor; IPL = intracellular proline-rich linker; RUD = repeated-unit domains; DTD = desmoglein-specific terminal domain (adapted from the work of Getsios et al.18). B, ClustalW sequence alignment of human DSG2 to orthologues and to other human desmogleins (DSG1, DSG3, and DSG4) and desmocollins (DSC2, DSC3,and DSC4). Identical residues are shaded in black; conserved residues are shaded in gray. Mutations identified in the present study are shown at the top of each alignment. The furin-recognition motif and plakoglobin binding domain are indicated.
Two of these individuals have mutations in the furin-cleavage site of the desmoglein-2 proprotein. Individual A.III-1 has a G→A transition at nt 134, which results in the substitution of a conserved arginine with glutamine (R45Q). Individual B.II-1 has a G→A transition at nt 143, which results in the substitution of a conserved arginine with histidine (R48H). These arginines occur as the first and fourth amino acids within the R-X-K-R furin-cleavage motif (fig. 2). ClustalW alignment demonstrates that these arginines are absolutely conserved in all species and subtypes of desmoglein, as is the lysine residue.
Newly synthesized desmogleins are initially inactive because of an NH2-terminal propeptide sequence, which is then posttranslationally cleaved by subtilisin-like proprotein convertases (PCs), to create mature, active proteins. These endoproteases cleave C-terminal to the recognition motif Arg-X-Lys-Arg (RXKR), which is highly conserved throughout all cadherin molecules. Coexpression of the PC furin with human desmoglein-1 or desmoglein-3 proproteins results in efficient propeptide processing.19 Mutation of the tetrapeptide recognition site or selective inhibition of PCs prevents endoproteolytic processing of cadherin propeptides and abrogates their adhesive function.20,21 Therefore, we predict that the R45Q and R48H mutations abolish furin cleavage of pro-desmoglein-2, thereby disrupting production of mature, functional protein.
Individual C.II-1 has a mutation (C506Y) in the extracellular anchor (EA) domain, immediately distal to the fourth extracellular cadherin (EC) domain, which ends at residue 502. Cysteine→tyrosine substitution confers loss of a sulfur residue that likely participates in disulfide bonding in the nonreducing extracellular environment. This cysteine is conserved in all species of desmoglein-2 (fig. 2).
Individual D.II-4 has a mutation (G811C) in the intracellular cadherin-typical sequence (ICS) of desmoglein-2. This region is responsible for binding desmoglein-2 to plakoglobin, as determined elsewhere by mutagenesis analyses of residues 794–824.22–24 All species and subtypes of desmogleins have glycine at this position (fig. 2).
In addition to the R48H mutation in the furin-cleavage domain, individual B.II-1 has a G→A transition at np 915 that results in a premature termination codon (PTC) in exon 8 (W305X). His 77-year-old unaffected mother (individual B.I-4) shares the W305X mutation but does not have the R48H mutation. She underwent screening echocardiography, electrocardiography (ECG), exercise stress testing, and several 24-h ambulatory Holter monitors. Results were notable for normal right and left ventricular size and function, normal ECG depolarization and repolarization, atrial fibrillation, and lack of significant ventricular arrhythmias. Because of the absence of any features of ARVD/C, endomyocardial biopsy was not performed. The proband’s father (individual B.I-3) died—at age 74 years from diabetes mellitus and sepsis—prior to providing a DNA sample or consent for analysis. The father’s surviving siblings (individuals B.I-1 and B.I-2) do not have the R48H mutation. Although the proband may have inherited this mutation from his father, the mutation may also be a de novo mutation. RT-PCR analysis, with use of mRNA derived from the proband’s lymphocytes, indicates that the R48H mutation occurs on the paternal allele (fig. 3).
Figure 3. .
Compound heterozygous mutations in DSG2 for individual B.II-1. Total RNA was isolated from Epstein-Barr virus–transformed lymphoblasts from individual B.II-1 (Trizol [Invitrogen]; RNeasy clean-up [Qiagen]), and cDNA was generated by reverse transcription (Superscript III 1st-strand cDNA synthesis kit [Invitrogen]). PCR products amplified with primers spanning the proband’s two mutations were cloned into the pCR2.1 plasmid (TOPO-TA cloning kit [Invitrogen]), and several individual clones were sequenced using both M13 forward and reverse primers. Representative chromatograms from two clones are displayed, which demonstrate that the R48H and W305X mutations lie on different alleles.
Three of the four probands have single, heterozygous missense mutations in DSG2. A fourth proband (B.II-1) has both a missense mutation on one allele of DSG2 and a nonsense mutation on the other. This raises the question of whether mutations in DSG2 cause disease by a recessive or dominant mode of inheritance. If inheritance is recessive, the three singly heterozygous probands could harbor mutations on a second, as-yet-unidentified disease-causing gene. This could explain why individual B.I-4 shares the R45Q mutation but has no evidence of ARVD/C on cardiac magnetic resonance imaging, ECG, Holter monitoring, or signal-averaged ECG. Alternatively, these findings could result from dominant inheritance with low penetrance, as has been reported for other genetic forms of ARVD/C.25 Complete loss of desmoglein-2 in mice results in early embryonic lethality.26 The presence of compound heterozygous mutations in individual B.II-1 suggests either partial function or a functional redundancy in humans that is not present in mice.
Among these probands, none has woolly hair or keratoderma. As a child, after receiving penicillin, individual B.II-1 developed a disseminated cutaneous exanthem with extensive mucosal involvement, which was diagnosed as Stevens Johnson syndrome (SJS). Other than a severe postoperative wound infection, he has had no other cutaneous diseases. The occurrence of SJS in the proband with compound heterozygosity for mutations in desmoglein-2 suggests that this desmosomal protein may be involved in the pathogenesis of SJS. Several members of the desmoglein family have been associated with other diseases of the skin. Stanley et al. demonstrated that desmoglein-1 is the antigen target in pemphigus foliaceus, an autoimmune disorder affecting skin.27 Similarly, desmoglein-3 and desmoglein-4 have been identified as antigen targets in pemphigus vulgaris.28,29 Mutation in DSG1 has also been described in the dominantly inherited skin disease striate palmoplantar keratoderma type I.30 Mutations in DSG4 result in localized autosomal recessive hypotrichosis.29 Finally, cleavage of desmoglein-1 by staphylococcal exfoliative toxins has been shown to result in staphylococcal scalded skin syndrome by facilitating the spread of Staphylococcus aureus under the stratum corneum.31
Results of clinical screening for ARVD/C and DSG2 genetic testing on those family members who consented to these analyses are available in table A2. Incomplete penetrance is evident, as described in other forms of ARVD/C.25 The absence of phenotypic manifestations of ARVD/C in the mother and sister of individual B.II-1, who share the single W305X mutation, could be due to incomplete penetrance, or this mutation may be insufficient to result in ARVD/C in isolation. Because the mutation creates a PTC, mutant transcripts are predicted to be rapidly degraded by the nonsense-mediated mRNA decay (NMD) pathway.32 This would then suggest that haploinsufficiency for desmoglein-2 is not the mechanism for disease. Even if the mutant transcript escapes NMD, the resulting truncated protein lacks a transmembrane domain (aa 612–634) and would likely be secreted extracellularly, which would prevent a potentially deleterious dominant negative effect.33 In this model, the phenotypic manifestations of heterozygous DSG2 mutation carriers would be same as for those with compound heterozygous mutation, with one mutation causing a PTC prior to the transmembrane domain (TM). However, additional individuals harboring mutations that prevent transmembrane anchoring of desmoglein-2 are required for definitive conclusions regarding the role of haploinsufficiency.
The mechanism whereby mutations affecting components of the cardiac desmosome result in ARVD/C remains in question. Some have suggested that either a lack of desmosomal protein or incorporation of mutant protein will disrupt cell-cell adhesion, predisposing regions of greatest stretch to fibrofatty degeneration.10,34 Others have invoked a pathogenic role for viral infection, perhaps worsened by a mutant cardiac desmosome.35,36 Finally, alteration in cytokines such as transforming growth factor β3 may also contribute to disease pathogenesis.15
In conclusion, we report that mutations in DSG2 contribute to the development of ARVD/C in the absence of mutations in PKP2 or DSP. This provides further evidence that disruption of the cardiac desmosome is important in the pathogenesis of this condition. Since sudden cardiac death is a prominent manifestation of ARVD/C, recognition of those at highest risk of developing the condition may be improved by genetic screening within affected families.
Acknowledgments
We thank the patients and their families for participation in this study. This work was supported by grants from the W. W. Smith Charitable Trust (to D.P.J.) and the D. W. Reynolds Foundation. We also acknowledge the Johns Hopkins ARVD Program, which is supported by the Bogle Foundation, the Campanella family, and the Wilmerding Endowments.
Appendix
Table A1. .
Oligonucleotide Primer Sequences[Note]
| Primer Sequence(5′→3′) |
||
| Region | Sense | Antisense |
| Exon 1 | ACCCAAGGACGTCACGGTCCC | CCAAGAGGATTTTCCGAAGCC |
| Exon 2 | AGGAGTCAGTATGGATCCAGG | ATTCAGCACCTCGTCATGGAC |
| Exon 3 | TAGACAATGAAGCCTCATAGG | CAATGATGCTGCATCTTCCGG |
| Exon 4 | ATCACTTCTTAGGCTTTTGGC | |
| Exon 5 | AGGAAAGCAGTTCTGAACTGC | |
| Exon 6 | CTAATGTGTTCTAATTGGTTGGACC | TTGGCTCACTGCAACTTCTGCC |
| Exon 7 | TTCTGCAAAAGCTCTGACTGC | |
| Exon 8 | GAATTTGAACCATAGTGTGACC | |
| Exon 9 | AGTTGGACTATTCAGTGCTGC | TTTGGGGAACTATAATGCTGG |
| Exon 10 | TGACATGCAGTAAAGAGAGGG | CAAAGTGCTGGGATTACAGGC |
| Exon 11 | AAACATCTTCATCAACCTCTGG | TTCCAGTGCATCTTTGTGAACG |
| Exon 12 | CAGCAATGAAAGAACATTTGTGG | CAGTTGTTTCCCTATTCACCC |
| Exon 13 | GTGAAGACAAGTCCAGGAAGG | CAAAGGCACATGAGTGAAATCC |
| Exon 14 | AGCTTATACCTTCCTATGCCC | AGTCTCATTTGGATCCAAGGC |
| Exon 15 | TTGTGTTTCCCTGATGGTTCC | CTGTAAGGCTCATGAAAAATCAGG |
| cDNA | TGCTTTAACGTTGGAAGTGG | TTGATGGGAATGGGTGTAGG |
Note.— Intronic sense and antisense oligonucleotide primers used for sequencing the 15 exons of human DSG2 are shown. Primers used to amplify the coding region containing both the R48H mutation and the W305X mutation are shown (cDNA). Reference human sequence was obtained from Ensembl.
Table A2. .
Clinical and Genetic Findings in Study Families
| Finding or Result |
|||||||||
| Subjects | DSG2 Mutation | Agea (years) |
Family Historyb | Right Ventricle Structure Abnormality |
Depolarization Abnormality | Repolarization Abnormality | Arrhythmia | Histopathology | Criteriac (Major/Minor) |
| Family A: | |||||||||
| I-1 | Not tested | 62* | Minor | Not screened | Not screened | Not screened | Not screened | Not screened | 0/1 |
| II-1 | Not tested | 46 | Minor | None | None | None | None | Not screened | 0/1 |
| II-2 | R45Q | 46 | Minor | None | None | None | None | Not screened | 0/1 |
| III-1 | R45Q | 19 | Proband | Major | None | Minor | Minor | major | 2/2 |
| III-2 | Not tested | 16 | Minor | None | None | None | None | Not screened | 0/1 |
| Family B: | |||||||||
| I-1 | None | 81 | Minor | Not screened | Not screened | Not screened | Not screened | Not screened | NA |
| I-2 | None | 80 | Minor | Not screened | Not screened | Not screened | Not screened | Not screened | NA |
| I-3 | Not tested | 74* | Minor | None | None | None | None | Not screened | 0/1 |
| I-4 | W305X | 77 | Minor | None | None | None | None | Not screened | 0/1 |
| II-1 | W305X R48H | 42 | Proband | Major | Major | Minor | Minor | Not screened | 2/2 |
| II-2 | W305X | 39 | Minor | None | None | None | Not screened | Not screened | 0/1 |
| Family C: | |||||||||
| I-1 | Not tested | 73 | Minor | None | None | None | None | Not screened | 0/1 |
| I-2 | Not tested | 69 | Minor | None | None | None | None | Not screened | 0/1 |
| II-1 | C506Y | 45 | Proband | None | Minor | Minor | Minor | Not screened | 0/4 |
| II-2 | Not tested | 44 | Minor | None | None | None | None | Not screened | 0/1 |
| II-3 | Not tested | 43 | Minor | Minor | None | None | None | Not screened | 0/2 |
| II-4 | Not tested | 39 | Minor | None | None | None | None | Not screened | 0/1 |
| II-5 | Not tested | 39 | Minor | None | None | None | None | Not screened | 0/1 |
| Family D: | |||||||||
| I-1 | None | 73 | Major | None | None | None | Not screened | 1/0 | |
| I-2 | G811C | 73 | Major | Not screened | Not screened | Not screened | Not screened | Not screened | NA |
| II-1 | Not tested | 32* | Proband | On autopsy | Not screened | Not screened | Not screened | On autopsy | Autopsyd |
| II-2 | None | 45 | Major | None | None | None | None | Normal | 1/0 |
| II-3 | None | 44 | Major | None | None | None | None | Not screened | 1/0 |
| II-4 | G811C | 40 | Major | Minor | None | Minor | None | Not screened | 1/2 |
An asterisk (*) indicates age at death.
Minor = satisfies minor criterion in this category; major = satisfies major criterion in this category.
To establish a diagnosis of ARVD/C, the subject must meet two major criteria, one major and two minor criteria, or four minor criteria in different categories. NA = not available.
The diagnosis was established during postmortem autopsy rather than by use of the clinical criteria.
Web Resources
The URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/Homo_sapiens/index.html
- Johns Hopkins ARVD Program, http://www.arvd.com/
- Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/ (for ARVD/C, Naxos syndrome, and Carvajal syndrome)
References
- 1.McKenna WJ, Thiene G, Nava A, Fontaliran F, Blomstrom-Lundqvist C, Fontaine G, Camerini F, for the Task Force of the Working Group on Myocardial and Pericardial Disease of the European Society of Cardiology and of the Scientific Council on Cardiomyopathies of the International Society and Federation of Cardiology (1994) Diagnosis of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Br Heart J 71:215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, Towbin JA, Danieli GA (2002) Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet 71:1200–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman M, Baboonian C, Jeffery S, McKenna WJ (2000) Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet 355:2119–2124 10.1016/S0140-6736(00)02379-5 [DOI] [PubMed] [Google Scholar]
- 4.Protonotarios N, Tsatsopoulou A, Patsourakos P, Alexopoulos D, Gezerlis P, Simitsis S, Scampardonis G (1986) Cardiac abnormalities in familial palmoplantar keratosis. Br Heart J 56:321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norgett EE, Hatsell SJ, Carvajal-Huerta L, Ruiz Cabezas J-C, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP (2000) Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet 9:2761–2766 10.1093/hmg/9.18.2761 [DOI] [PubMed] [Google Scholar]
- 6.Djabali K, Martinez-Mir A, Horev L, Christiano AM, Zlotogorski A (2002) Evidence for extensive locus heterogeneity in Naxos disease. J Invest Dermatol 118:557–560 10.1046/j.0022-202x.2001.01627.x [DOI] [PubMed] [Google Scholar]
- 7.Bauce B, Basso C, Rampazzo A, Beffagna G, Daliento L, Frigo G, Malacrida S, Settimo L, Danieli G, Thiene G, Nava A (2005) Clinical profile of four families with arrhythmogenic right ventricular cardiomyopathy caused by dominant desmoplakin mutations. Eur Heart J 26:1666–1675 10.1093/eurheartj/ehi341 [DOI] [PubMed] [Google Scholar]
- 8.Norman M, Simpson M, Mogensen J, Shaw A, Hughes S, Syrris P, Sen-Chowdhry S, Rowland E, Crosby A, McKenna WJ (2005) Novel mutation in desmoplakin causes arrhythmogenic left ventricular cardiomyopathy. Circulation 112:636–642 10.1161/CIRCULATIONAHA.104.532234 [DOI] [PubMed] [Google Scholar]
- 9.Grossmann KS, Grund C, Huelsken J, Behrend M, Erdmann B, Franke WW, Birchmeier W (2004) Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J Cell Biol 167:149–160 10.1083/jcb.200402096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L (2004) Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet 36:1162–1164 10.1038/ng1461 [DOI] [PubMed] [Google Scholar]
- 11.Dalal D, Molin LH, Piccini JP, Tichnell C, James C, Bomma C, Prakasa K, Towbin JA, Marcus FI, Spevak PJ, Bluemke DA, Abraham T, Russell SD, Calkins H, Judge DP (2006) Clinical features of arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in plakophilin-2. Circulation 113:1641–1649 10.1161/CIRCULATIONAHA.105.568642 [DOI] [PubMed] [Google Scholar]
- 12.Rampazzo A, Nava A, Erne P, Eberhard M, Vian E, Slomp P, Tiso N, Thiene G, Danieli G (1995) A new locus for arrhythmogenic right ventricular cardiomyopathy (ARVD2) maps to chromosome 1q42-q43. Hum Mol Genet 4:2151–2154 [DOI] [PubMed] [Google Scholar]
- 13.Bauce B, Rampazzo A, Basso C, Bagattin A, Daliento L, Tiso N, Turrini P, Thiene G, Danieli GA, Nava A (2002) Screening for ryanodine receptor type 2 mutations in families with effort-induced polymorphic ventricular arrhythmias and sudden death: early diagnosis of asymptomatic carriers. J Am Coll Cardiol 40:341–349 10.1016/S0735-1097(02)01946-0 [DOI] [PubMed] [Google Scholar]
- 14.Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, Sorrentino V, Danieli GA (2001) Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103:196–200 [DOI] [PubMed] [Google Scholar]
- 15.Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, Bauce B, Carraro G, Thiene G, Towbin JA, Danieli GA, Rampazzo A (2005) Regulatory mutations in transforming growth factor-β3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res 65:366–373 10.1016/j.cardiores.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 16.Schwarz MA, Owaribe K, Kartenbeck J, Franke WW (1990) Desmosomes and hemidesmosomes: constitutive molecular components. Annu Rev Cell Biol 6:461–491 10.1146/annurev.cb.06.110190.002333 [DOI] [PubMed] [Google Scholar]
- 17.Schafer S, Koch PJ, Franke WW (1994) Identification of the ubiquitous human desmoglein, Dsg2, and the expression catalogue of the desmoglein subfamily of desmosomal cadherins. Exp Cell Res 211:391–399 10.1006/excr.1994.1103 [DOI] [PubMed] [Google Scholar]
- 18.Getsios S, Huen AC, Green KJ (2004) Working out the strength and flexibility of desmosomes. Nat Rev Mol Cell Biol 5:271–281 10.1038/nrm1356 [DOI] [PubMed] [Google Scholar]
- 19.Posthaus H, Dubois CM, Muller E (2003) Novel insights into cadherin processing by subtilisin-like convertases. FEBS Lett 536:203–208 10.1016/S0014-5793(02)03897-8 [DOI] [PubMed] [Google Scholar]
- 20.Ozawa M, Kemler R (1990) Correct proteolytic cleavage is required for the cell adhesive function of uvomorulin. J Cell Biol 111:1645–1650 10.1083/jcb.111.4.1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson ED, Thomas L, Hayflick JS, Thomas G (1993) Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed α-1-antitrypsin variant. J Biol Chem 268:24887–24891 [PubMed] [Google Scholar]
- 22.Chitaev NA, Averbakh AZ, Troyanovsky RB, Troyanovsky SM (1998) Molecular organization of the desmoglein-plakoglobin complex. J Cell Sci 111:1941–1949 [DOI] [PubMed] [Google Scholar]
- 23.Pai LM, Kirkpatrick C, Blanton J, Oda H, Takeichi M, Peifer M (1996) Drosophila α-catenin and E-cadherin bind to distinct regions of Drosophila armadillo. J Biol Chem 271:32411–32420 10.1074/jbc.271.50.32411 [DOI] [PubMed] [Google Scholar]
- 24.Stappert J, Kemler R (1994) A short core region of E-cadherin is essential for catenin binding and is highly phosphorylated. Cell Adhes Commun 2:319–327 [DOI] [PubMed] [Google Scholar]
- 25.Syrris P, Ward D, Asimaki A, Sen-Chowdhry S, Ebrahim HY, Evans A, Hitomi N, Norman M, Pantazis A, Shaw AL, Elliott PM, McKenna WJ (2006) Clinical expression of plakophilin-2 mutations in familial arrhythmogenic right ventricular cardiomyopathy. Circulation 113:356–364 10.1161/CIRCULATIONAHA.105.561654 [DOI] [PubMed] [Google Scholar]
- 26.Eshkind L, Tian Q, Schmidt A, Franke WW, Windoffer R, Leube RE (2002) Loss of desmoglein 2 suggests essential functions for early embryonic development and proliferation of embryonal stem cells. Eur J Cell Biol 81:592–598 10.1078/0171-9335-00278 [DOI] [PubMed] [Google Scholar]
- 27.Stanley JR, Koulu L, Klaus-Kovtun V, Steinberg MS (1986) A monoclonal antibody to the desmosomal glycoprotein desmoglein I binds the same polypeptide as human autoantibodies in pemphigus foliaceus. J Immunol 136:1227–1230 [PubMed] [Google Scholar]
- 28.Amagai M, Klaus-Kovtun V, Stanley JR (1991) Autoantibodies against a novel epithelial cadherin in pemphigus vulgaris, a disease of cell adhesion. Cell 67:869–877 10.1016/0092-8674(91)90360-B [DOI] [PubMed] [Google Scholar]
- 29.Kljuic A, Bazzi H, Sundberg JP, Martinez-Mir A, O’Shaughnessy R, Mahoney MG, Levy M, Montagutelli X, Ahmad W, Aita VM, Gordon D, Uitto J, Whiting D, Ott J, Fischer S, Gilliam TC, Jahoda CA, Morris RJ, Panteleyev AA, Nguyen VT, Christiano AM (2003) Desmoglein 4 in hair follicle differentiation and epidermal adhesion: evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell 113:249–260 10.1016/S0092-8674(03)00273-3 [DOI] [PubMed] [Google Scholar]
- 30.Rickman L, Simrak D, Stevens HP, Hunt DM, King IA, Bryant SP, Eady RA, Leigh IM, Arnemann J, Magee AI, Kelsell DP, Buxton RS (1999) N-terminal deletion in a desmosomal cadherin causes the autosomal dominant skin disease striate palmoplantar keratoderma. Hum Mol Genet 8:971–976 10.1093/hmg/8.6.971 [DOI] [PubMed] [Google Scholar]
- 31.Hanakawa Y, Schechter NM, Lin C, Garza L, Li H, Yamaguchi T, Fudaba Y, Nishifuji K, Sugai M, Amagai M, Stanley JR (2002) Molecular mechanisms of blister formation in bullous impetigo and staphylococcal scalded skin syndrome. J Clin Invest 110:53–60 10.1172/JCI200215766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendell JT, Dietz HC (2001) When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell 107:411–414 10.1016/S0092-8674(01)00583-9 [DOI] [PubMed] [Google Scholar]
- 33.Huber O, Kemler R, Langosch D (1999) Mutations affecting transmembrane segment interactions impair adhesiveness of E-cadherin. J Cell Sci 112:4415–4423 [DOI] [PubMed] [Google Scholar]
- 34.Sen-Chowdhry S, Syrris P, McKenna WJ (2005) Genetics of right ventricular cardiomyopathy. J Cardiovasc Electrophysiol 16:927–935 10.1111/j.1540-8167.2005.40842.x [DOI] [PubMed] [Google Scholar]
- 35.Calabrese F, Basso C, Carturan E, Valente M, Thiene G (2006) Arrhythmogenic right ventricular cardiomyopathy/dysplasia: is there a role for viruses? Cardiovasc Pathol 15:11–17 10.1016/j.carpath.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 36.Bowles NE, Ni J, Marcus F, Towbin JA (2002) The detection of cardiotropic viruses in the myocardium of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol 39:892–895 10.1016/S0735-1097(02)01688-1 [DOI] [PubMed] [Google Scholar]