To the Editor: In the January issue of the American Journal of Human Genetics, Grupe and colleagues1 published evidence suggesting genetic association between SNP rs498055 on chromosome 10q24, located in a putative homologue of ribosomal protein S3a (RPS3A [MIM 180478]), and risk for Alzheimer disease (AD [MIM 104300]) in four of six independent case-control samples. The authors reached this conclusion after testing nearly 1,400 SNPs, using an exploratory case-control sample, followed by assessments of a number of independent data sets of different size, origin, and ascertainment. Although three of the replication samples showed significant risk effects for the G allele of rs498055, this effect was not confirmed in two smaller series of neuropathologically confirmed AD cases and controls. None of the other 68 “hits” uncovered in the first pass received the same degree of consistent replication as did rs498055. Overall, the effect of the putative risk allele was modest (yielding odds ratios [ORs] between ∼1.3 and 1.4) and—according to the authors’ conclusion—likely reflects linkage disequilibrium (LD) with another genetic variant nearby. Here, we have set out to independently assess the association between rs498055 and AD risk in two large and carefully characterized samples of AD-affected families comprising nearly 1,900 subjects from 654 pedigrees. However, in contrast to the findings of Grupe and colleagues, we observed no evidence of association between rs498055 and AD in any of our analyses.
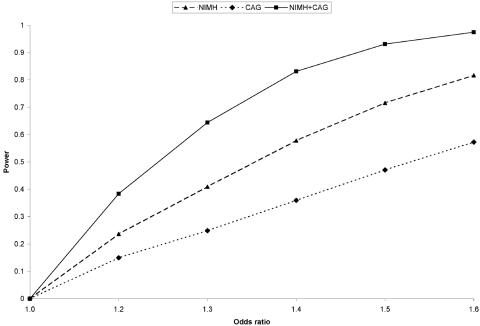
Using high-efficiency fluorescence polarization (HEFP) technology, we genotyped this SNP in two family-based AD samples: (1) 1,439 subjects from 437 multiplex AD-affected families recruited as part of the National Institute of Mental Health (NIMH) Genetics Initiative AD Study Sample (average age at onset [±SD] of affected individuals was 72.4±7.7 years) and (2) 489 subjects from 217 independent families, mostly consisting of discordant sibships, recruited as part of the Consortium on Alzheimer’s Genetics (CAG) (average age at onset was 71.2±9.1 years). These samples, as well as the genotyping procedures, are described in detail in the work of Bertram et al.2; PCR and HEFP primer sequences for rs498055 are available on request. Average genotyping efficiency across both samples was 98.4%, with a genotyping error rate <1% (on the basis of ∼10% duplicated samples). Power analyses (fig. 1) in the combined sample showed that, at a disease-allele frequency of 0.47 (i.e., the average frequency of the G allele in U.S. controls reported by Grupe et al.) and α=.05, power was 64% for an OR of 1.3 and was 83% for an OR of 1.4 (see fig. 1 for more details). Naturally, power was lower for the two samples considered separately, but it was still 40%–60% for the NIMH sample alone, comparable to the power of the replication samples in the study by Grupe et al.
Figure 1. .
Power to detect a range of effect sizes in the family samples analyzed. Power estimates were done with PBAT (v3.1).12 Estimates are based on approximation and are calculated for an additive disease model based on parameters published by Grupe et al.1 (i.e., disease-allele frequency of 0.47 and OR 1.2–1.6), with the exception of disease prevalence, which was set to 10%. Although the precise prevalence of AD is unknown and difficult to estimate, power does not change appreciably when prevalence is varied from 5% to 15% (data not shown). ORs are for heterozygous carriers of the disease allele versus homozygous noncarriers. Families were modeled after the observed pedigree structure for each sample, with both parents set as “missing.” Note that PBAT can currently handle a maximum offspring number of only four; however, 68 (16%) of NIMH pedigrees actually have more than four genotyped and phenotyped offspring, so that the power for “NIMH” and “NIMH+CAG” is likely to be underestimated (see the PBAT Web site for more details).
In contrast to the findings of Grupe et al., we did not observe any significant evidence of association between rs498055 and AD risk, neither in the two samples individually nor after combining both data sets (table 1), overall or when stratified by age at onset (with age 65 years as cutoff) or apolipoprotein E (APOE [MIM 107741]) ɛ4-carrier status. Interestingly, and in contrast to the overtransmission of the G allele noted by Grupe et al., in our two samples, this allele was generally undertransmitted to affected individuals, which approached statistical significance in two of our stratified analyses in the combined sample (P=.09 in “late-onset” families, and P=.06 in “APOE ɛ4-positive” families [table 1]). Finally, we also tested for association between rs498055 and age at onset of AD (age at last examination of unaffected individuals), using FBAT-LOGRANK, FBAT-Wilcoxon, and FBAT-Flemington-Harrington3,4 in the unstratified samples. However, none of these tests showed even marginally significant P values (data not shown). Since quantitative trait analyses are expected to be more powerful than analyses of binary traits if the underlying association is true,3 these results strengthen our overall negative conclusion.
Table 1. .
Association Analyses of rs498055 in Two Independent Family Samples[Note]
|
FBAT Statistic Result |
||||
| Samplea | G Allele Frequency | No. of Informative Families | Z Scoreb | P |
| All Families: | ||||
| NIMH | .519 | 123 | −1.096 | .27 |
| CAG | .504 | 84 | −.194 | .85 |
| Combined | .514 | 207 | −1.001 | .32 |
| Families with late-onset disease: | ||||
| NIMH | .537 | 81 | −1.541 | .12 |
| CAG | .531 | 66 | −.808 | .42 |
| Combined | .535 | 147 | −1.703 | .09 |
| APOE ɛ4-positive families: | ||||
| NIMH | .494 | 107 | −1.488 | .14 |
| CAG | .515 | 31 | −1.380 | .17 |
| Combined | .499 | 138 | −1.929 | .06 |
Note.— Association tests were performed using FBAT (v1.5.5) with an additive transmission model, the empirical variance function, and an equal-weight offset correction for affected and unaffected individuals (see the FBAT Web site for more details).
Families were classified as “late onset” when all sampled affected individuals had age at onset of >65 years and were classified as “APOE ɛ4 positive” when at least one affected individual per family carried the ɛ4 allele. The smaller strata of remaining families (i.e., those displaying an earlier age at onset or those in which none of the affected individuals carried an ɛ4 allele) also failed to show evidence of significant association (data not shown).
For the G allele of rs498055, which was reported as the putative risk allele by Grupe et al.1 (positive values indicate overtransmission to affected individuals). Note that the direction of transmission is consistent for both family samples analyzed here and is opposite to that seen in the previous publication.1
Our study is the first to independently assess the potential association between rs498055 and AD that had emerged from a semisystematic screen of 1,397 SNPs on chromosome 10.1 The fact that we failed to replicate the previous findings is noteworthy for several reasons. First, rs498055 is located within the chromosome 10q24 linkage peak reported elsewhere for this collection of NIMH families.5 Thus, our sample should be particularly well suited to detect disease associations underlying this linkage signal. Second, our study is the first to analyze this SNP with use of family-based methodologies in which affected subjects are compared with related unaffected subjects from the same family. Results from such analyses are more robust to bias due to population admixture or other sources of skewed genotype distributions in cases or controls; this is of particular note, given the differences in allele frequencies reported for the two control populations from the St. Louis area in the work of Grupe et al. (see below). Despite the strengths of our approach, it is possible that we have missed a putative risk effect at rs498055 because of insufficient power, especially when aiming to detect minor effects with ORs of ⩽1.3 (fig. 1). However, the differences between our findings and those of Grupe et al. are unlikely to result from lack of power alone, since we see under- rather than overtransmission of the G allele in both samples. It is unclear whether these discrepancies are caused by chance or by differential patterns of LD across the various samples.
The long arm of chromosome 10 has been a focus of work for many AD genetics laboratories since the discovery of significant linkage with AD phenotypes by three independent groups, including ours (AD6 [MIM 605526]).5–7 These publications were followed by two additional studies suggesting the presence of an AD risk and/or age-at-onset–modifying gene on this chromosome.8,9 Although nearly 30 positional candidate genes have since been assessed as potential AD risk factors underlying these linkage signals and several positive association findings have been published, no gene has received consistent support from independent follow-up studies,10 and none shows evidence of conclusive and significant summary effects in systematic meta-analyses of all published and available genotype data (AlzGene).
Unfortunately, the present failure to replicate the promising results of Grupe and colleagues is consistent with this overall pattern. There are several possible reasons for the differences between our findings and theirs. First, the difference might be due to chance, because the initial finding is a false-positive result. The 69 hits among 1,397 SNPs in the exploratory data set is close to the expected value by chance alone, as is the confirmation of 5 of these 69 signals in at least one of the two direct follow-up samples. However, we agree with the authors that a significant overrepresentation of the same allele in three of five confirmation samples, as observed for rs498055, may be unlikely to occur by chance alone. Second, the difference might have arisen by chance because our finding is a false-negative result. Although this is possible, it should be noted that our sample is as large or larger than many replication samples in the field. In addition, the difference is unlikely to result from insufficient power alone, because the putative risk allele, if anything, is undertransmitted in our samples. Third, the differences may relate to our use of family-based methods, which are more robust to bias due to population admixture. Although the degree to which admixture may lead to spurious association findings in case-control samples is controversial,11 the issue is a concern here, given the marked difference in allele frequencies across the two independent Washington University control samples (47% for the case-control sample—similar to the other U.S. control sample—and 44% for the controls used in comparison with the linkage sample), a difference substantial enough that the allelic association between rs498055 and AD in the linkage sample (49.8% risk-allele frequency) would not have been significant had the other Washington University control set (or the University of California–San Diego controls) been used. Finally, the differences across studies may be due to differences in patterns of LD across the various samples, which are impossible to assess as long as the precise nature of the putative risk allele at this locus remains unknown.
Clearly, additional analyses of sufficiently powered and independent samples are needed to assert whether rs498055, or a polymorphism in LD with it, makes a relevant contribution to AD risk. At least in the two family samples investigated here—one of which shows linkage to the same chromosomal interval as rs498055—this SNP is not a major determinant of AD risk.
Acknowledgments
The authors thank all families for participating in this study. This work was sponsored by grants from the NIMH, the National Institute on Aging (NIA) (at the Alzheimer Disease Research Center), and the Alzheimer Association. L.B. is supported by NIA grant 1R01 AG023667-01 and by a Young Investigator Award from the National Alliance for Research on Schizophrenia and Depression.
Web Resources
The URLs for data presented herein are as follows:
- AlzGene, http://www.alzgene.org/ (Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi R. The AlzGene Database–Published AD Candidate Genes; Alzheimer Research Forum; last accessed 3/1/06)
- FBAT, http://www.biostat.harvard.edu/~fbat/fbat.htm
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RPS3A, AD, APOE, and AD6)
- PBAT, http://www.biostat.harvard.edu/~clange/default.htm
References
- 1.Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, Smemo S, Kauwe JSK, et al (2006) A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet 78:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, Mullin K, Menon R, Sampson AJ, Hsiao MY, Elliott KJ, Velicelebi G, Moscarillo T, Hyman BT, Wagner SL, Becker KD, Blacker D, Tanzi RE (2005) Family-based association between Alzheimer’s disease and variants in UBQLN1. N Engl J Med 352:884–894 10.1056/NEJMoa042765 [DOI] [PubMed] [Google Scholar]
- 3.Lange C, DeMeo DL, Laird NM (2002) Power and design considerations for a general class of family-based association tests: quantitative traits. Am J Hum Genet 71:1330–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang H, Harrington D, Raby BA, Bertram L, Blacker D, Weiss ST, Lange C (2006) Family-based association test for time-to-onset data with time-dependent differences between the hazard functions. Genet Epidemiol 30:124–132 10.1002/gepi.20132 [DOI] [PubMed] [Google Scholar]
- 5.Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis MG, Go RC, Vekrellis K, Selkoe DJ, Saunders AJ, Tanzi RE (2000) Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science 290:2302–2303 10.1126/science.290.5500.2302 [DOI] [PubMed] [Google Scholar]
- 6.Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG (2000) Linkage of plasma Aβ42 to a quantitative locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Science 290:2303–2304 10.1126/science.290.5500.2303 [DOI] [PubMed] [Google Scholar]
- 7.Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R, Hamshere M, Abraham R, Tunstall N, Rice F, Carty S, Lillystone S, Kehoe P, Rudrasingham V, Jones L, Lovestone S, Perez-Tur J, Williams J, Owen MJ, Hardy J, Goate AM (2000) Susceptibility locus for Alzheimer’s disease on chromosome 10. Science 290:2304–2305 10.1126/science.290.5500.2304 [DOI] [PubMed] [Google Scholar]
- 8.Li Y-J, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, et al (2002) Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet 70:985–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JH, Mayeux R, Mayo D, Mo J, Santana V, Williamson J, Flaquer A, Ciappa A, Rondon H, Estevez P, Lantigua R, Kawarai T, Toulina A, Medrano M, Torres M, Stern Y, Tycko B, Rogaeva E, St George-Hyslop P, Knowles JA (2004) Fine mapping of 10q and 18q for familial Alzheimer’s disease in Caribbean Hispanics. Mol Psychiatry 9:1042–1051 10.1038/sj.mp.4001538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertram L, Tanzi RE (2004) Alzheimer’s disease: one disorder, too many genes? Hum Mol Genet Spec No 1 13:R135–R141 10.1093/hmg/ddh077 [DOI] [PubMed] [Google Scholar]
- 11.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72:1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM (2004) PBAT: tools for family-based association studies. Am J Hum Genet 74:367–369 [DOI] [PMC free article] [PubMed] [Google Scholar]