Abstract
Alagille syndrome (AGS) is caused by mutations in the gene for the Notch signaling pathway ligand Jagged1 (JAG1), which are found in 94% of patients. To identify the cause of disease in patients without JAG1 mutations, we screened 11 JAG1 mutation-negative probands with AGS for alterations in the gene for the Notch2 receptor (NOTCH2). We found NOTCH2 mutations segregating in two families and identified five affected individuals. Renal manifestations, a minor feature in AGS, were present in all the affected individuals. This demonstrates that AGS is a heterogeneous disorder and implicates NOTCH2 mutations in human disease.
Alagille syndrome (AGS [MIM 118450]) is a dominant, multisystem disorder defined clinically by hepatic bile duct paucity and cholestasis in association with cardiac, skeletal, and ophthalmologic manifestations. There are characteristic facial features and less-frequent clinical involvement of the renal and vascular systems.1,2 Expressivity is known to be highly variable. AGS is caused by mutations in the gene encoding Jagged1 (JAG1), a ligand in the Notch signaling pathway.3,4 Notch signaling is involved in cell fate determination and is essential for normal embryonic development. At least five ligands and four Notch receptors are expressed in humans, and many genes have been identified that function downstream of Notch.5 Mutations in JAG1 were identified in 94% of individuals with a clinically confirmed diagnosis of AGS.6 Failure to identify mutations in the remaining patients could have been because the mutations were located in noncoding regions not screened by present techniques or were in another gene. Data from the mouse have implicated the Notch2 gene in the etiology of clinical features associated with AGS. Although the Jagged1 knockout heterozygote mouse did not mimic the AGS phenotype,7 a Jagged1/Notch2 double heterozygote was found to have liver, cardiac, ocular, and renal manifestations similar to those seen in patients with AGS.8 Additionally, the spatial and temporal expression pattern of Notch2 in tissues involved in AGS makes it an excellent candidate to be the receptor interacting with Jagged1.9,10 This led us to screen a cohort of JAG1 mutation-negative patients with AGS for alterations in NOTCH2.
Eleven probands were screened for the coding region (34 exons) of NOTCH2. All individuals were enrolled in an institutional review board–approved protocol at The Children’s Hospital of Philadelphia, after informed consent was obtained. Screening was accomplished by direct sequencing of purified genomic DNA after PCR amplification. The first four exons of NOTCH2 (located within chromosome band 1p12) have a high degree of homology with a distinct but related gene, N2N, which is located at 1q21.11 We therefore designed primers for the first four exons of NOTCH2 so that the 3′ end sat on a nucleotide unique to NOTCH2, to avoid amplification of N2N. Primers are listed in table 1.
Table 1. .
NOTCH2 Mutation-Screening Primers
| Primer Sequence(5′→3′) |
||
| Exon(s) | Forward | Reverse |
| 1 | CAC ACG AGG CTG CTT CGT | CGG CGA TGT CCA AAC TCT T |
| 2 | AAA CAC AGA GAA ATA AGA GCA TC | CAT CCA CAT CCT TCC ATC |
| 3 | CCT GCC AGG ACT CAA AAG GA | TAT CTG CTG AAG GTA GGG AAC |
| 4 | CAT CTC CTA TTT CTG TGG C | ACT TCC CTT TTT CCT TGG |
| 5 | GGA ACA GAG CAG GTC GTT TC | CAG TCT GCC TCT GGT TAC CA |
| 6 | CCT TTG AGA CTC AAG AGC CC | GAG AGG GCA GAG TCC TGA AT |
| 7 | TCC CCA AGT GAG AGA CAT | ATG GCT ATG CTG AAA GGT |
| 8 | CTA TAA CCA TCA GAT GCT CT | ATA CTG AAT GCT CTA CCA AG |
| 9 | CTC GCA AAG GAT GTA GTT AT | CCC TGC TCT CTT CCC TGT |
| 10 | GTG CCT CTG ATT CCC TGA | AAA CTT ACT GCT GCC AAA A |
| 11 | TAT CTG CCT TGG TCT TTG AG | TGG GTG GGG TGG AAC TTA |
| 12 | CTG TGG ACT GCA TTT TGA GA | GGC AGC AAA TAA ACA CAT AA |
| 13 | GGG TCT ATT TCC TAA GCA TT | CTA CCA AAT CTT CAG CAA A |
| 14 | CAC CAA GTC CTG TAT TTC AC | GAG AAA AGG AAA CTA AGA AG |
| 15 | TGT AGT TGG AGG GGC ATT AG | TAA AGG CAG GAA GGA CAA CT |
| 16 | TGC CCA AGG TTA CAC AGT TA | CCC CGA AGA CAA GGA CAA TG |
| 17 | TCC TGC GTA GAT TTT TGT GG | CAC TGG GCT CTC CTC TGC TC |
| 18 | ATA AAT GAG GGA GTT GGC A | GAT GAC TGT GGA CTG GGA T |
| 19 | GGT ATT TGC CTC TGT ATG AT | TTC TTG GTT AGC CTC TTT TC |
| 20 | TAC CTC GGA ACA CTG AAT C | CCT TCC CTC TTA TTC CAC A |
| 21 | ATT CCA CTT TCA GGG TTT A | AGA GAG GCT GAA AGA AAC A |
| 22 | AAA GAG GAG TAG GTA AAT GC | AGA GAT GGG AGA ATA AAA TG |
| 23 | TGC CCT GCC CAA CTA CAA G | GCT TCA CTT GGG TCT GGG A |
| 24 | TAG TTC CCA TCC CAG CAA AGT T | CCA CAC ATC AGC CCT ATT TTC A |
| 25 | CTG TTT TCC TAC CAA TGC C | GAG CAA GAA CGA GAA ACT GA |
| 26 | CAA ATG AAA AAG CAA ATA GAG C | GGC AAA AGG TTA TGG ATT CTG G |
| 27–28 | CTT CTC CCT TGG CAT CC | ACG CTC TGA CTG TGG TTT C |
| 29 | ATG GTG GAA AGT GTT GAA AA | AGG ATA AGG AAG AGA TGT GG |
| 30 | GAT TGA TAG GGA GCA TTG TT | AGG GGT TTA TGA CTG GAC A |
| 31 | AAC TAC CTT CTC CTT CTT GA | TAC CTG CCC TAA TCT CTC A |
| 32 | TCA TTA TTT TCC TTC ATC CA | AAC TCT ATT CCC CTC GTC AG |
| 33 | CAG AGG AAA AAG AAG AGA TT | GAG AAA GAG TTA CAC CAA GC |
| 34A | TCA CAG ATA CAC ATT CAG TAC ATT | CTC TCA GAC AGT TGG ACC TT |
| 34B | CAA TGT GAC CCC AAG CCC | GGA GCC AGG ACC ATA CCA |
| 34C | AGT CCC AGT CCC AGC AGA TT | GAC CTT CAT TTG TTC CTC AGC A |
| 32–34 (cDNA) | GAG GGA ATG GTG GCA GAA | GGC AAG GTT AGG GAG GCT |
Mutations were identified in two probands. Proband 1 had cholestatic liver disease, cardiac disease (peripheral pulmonic stenosis and a small atrial septal defect), characteristic facial features, and severe infantile renal disease (small kidneys with cysts bilaterally, renal tubular acidosis, and renal insufficiency) (fig. 1a). He died of cardiopulmonary arrest at age 2 years. His mother had valvular and peripheral pulmonic stenosis, characteristic facial features, and dysplastic kidneys and proteinuria that resulted in renal failure and a kidney transplant. A NOTCH2 mutation of the splice acceptor of exon 33 (c.5930−1G→A) was identified in the proband and his mother (fig. 1b). Maternal grandparents and three of the mother’s siblings were also tested, and they did not carry the mutation, indicating it was a de novo change in the proband’s mother.
Figure 1. .
NOTCH2 mutation in family 1. a, Proband and his mother demonstrate features of AGS. Family members marked with an asterisk (*) were tested for the presence of the mutation but had only wild-type sequence. Individuals marked with a section symbol (§) had mutant sequences. b, Genomic DNA (gDNA) and cDNA sequencing demonstrates the presence and consequences of a mutation in the splice site of exon 33 in the proband and his mother. c, cDNA amplification and electrophoresis demonstrate the abnormally spliced product (arrow). d, Predicted protein product is represented below the diagram of the wild-type Notch2 protein (mutation marked by an asterisk). EGFR = EGF-like repeats.
To determine the effect of this mutation on splicing, we analyzed cDNA prepared from a lymphoblastoid cell line from the proband. Gel electrophoresis of an amplified portion of the proband’s NOTCH2 cDNA encompassing exons 32–34 confirmed the presence of an abnormal band, which, by sequencing, was shown to have the 98-bp exon 33 spliced out (fig. 1c). The transcript resulting from this mutation is predicted to have a premature termination codon within exon 34, and, since this is the last exon, the transcript is not predicted to undergo nonsense-mediated mRNA decay.12 The resulting protein is predicted to lack three of the seven ankyrin repeats and the ensuing 3′ sequence. (fig. 1d). The ankyrin repeat is found in a number of proteins and is responsible for mediating protein-protein interactions.13 These repeats in the intracellular domain of the Notch proteins interact with nuclear cofactors that serve to modulate Notch signaling, and the ankyrin repeats have been shown to be crucial for Notch activity.14
Proband 2 had cholestatic liver disease, which led to a liver transplant. She had cardiac disease (tetralogy of Fallot) and ocular findings (posterior embryotoxon). She demonstrated renal disease (tubular acidosis and dysplastic kidneys), and currently, at age 8 years, she is awaiting a renal transplant. Her mother has a history of asymptomatic hematuria and proteinuria. She came to medical attention at age 26 years with a mildly elevated urine protein level (155 mg/24 hr), which increased steadily over the next 10 years (584 mg/24 hr at age 36 years). Hypertension was diagnosed when she was age 36 years. Abdominal ultrasound indicated normal renal sizes. No cardiovascular or gastrointestinal abnormalities were present. The diagnosis was subnephrotic-range proteinuria with microscopic hematuria and no evidence of renal insufficiency. Examination by a dysmorphologist revealed the presence of facial features characteristic of AGS (fig. 2a). The proband’s maternal grandmother has advanced chronic renal insufficiency of undetermined etiology, which was first noted at age 59 years. Her renal insufficiency worsened until age 65 years, when she began peritoneal dialysis. An ultrasound of the kidneys showed a right atrophic kidney, which was thought to be congenital. Cardiac evaluation was negative for a murmur, and there was no history of liver disease. Adult-onset diabetes was diagnosed just before dialysis was begun, but this condition is well controlled with diet alone.
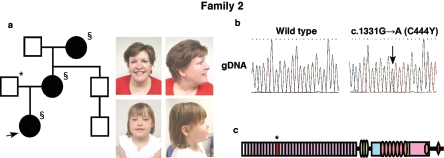
Figure 2. .
NOTCH2 mutation in family 2. a, Proband, her mother, and her grandmother had clinical features associated with AGS. The family member marked with an asterisk (*) was tested for the presence of the mutation but had only wild-type sequence. Individuals marked with a section symbol (§) had mutant sequences. b, Sequence analysis shows a mutation in exon 8 in all three affected individuals. c, The mutation (asterisk) is predicted to cause substitution of a tyrosine for a cysteine residue in EGF-like repeat 11 of Notch2.
Screening of the NOTCH2 gene in the proband, her mother, and her grandmother identified a G→A change at position 1331 in the cDNA sequence (c.1331G→A) in exon 8, which causes a substitution of a tyrosine for a cysteine residue in the 11th epidermal growth factor (EGF)–like repeat (C444Y) (fig. 2b and 2c). The EGF-like repeat motif consists of six cysteine residues, forming three intramolecular disulfide bridges, which are crucial for conformational stability of the protein.15 Loss or gain of cysteine residues in the EGF-like repeats of other genes has been shown to cause disease. Examples include loss or gain of EGF-like cysteine residues in fibrillin, which results in Marfan syndrome,16 and Notch3, which results in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL).17 The Drosophila and mammalian Notch genes are highly conserved, and both contain 36 EGF-like repeats, with EGF repeats 11 and 12 required for ligand binding of the Drosophila Notch, providing further support for the significance of the observed mutation.18
Neither of the mutations we identified was found in 110 ethnically matched control individuals (220 chromosomes). These studies identified a number of polymorphisms in NOTCH2, which are listed in table 2.
Table 2. .
Polymorphisms Identified in NOTCH2
| Nucleotide Changea |
Exon or Intron (IVS) |
Amino Acid |
Frequencyb |
| c.−214C→G | Exon 1 | 7/138 | |
| c.15C→T | Exon 1 | Arg5 | 16/138 |
| c.874+10G→A | IVS 6 | 3/138 | |
| c.939C→T | Exon 6 | Gly507 | 2/138 |
| c.1396C→A | Exon 8 | Glu466Lys | 1/358 |
| c.1681+142A→C | IVS 11 | 1/138 | |
| c.1915+89C→T | IVS 12 | 5/138 | |
| c.1915+201G→C | IVS 12 | 6/138 | |
| c.2365+39T→G | IVS 15 | 17/138 | |
| c.2365+47T→A | IVS 15 | 18/138 | |
| c.2479+20G→C | IVS 16 | 46/138 | |
| c.2753−44C→T | IVS 18 | 20/138 | |
| c.3034T→C | Exon 19 | Leu1012 | 1/138 |
| c.3184−76G→A | IVS 20 | 19/138 | |
| c.3523−58G→A | IVS 22 | 25/138 | |
| c.3655+96G→T | IVS 23 | 26/138 | |
| c.3656−22G→A | IVS 23 | 20/138 | |
| c.3980A→G | Exon 24 | Asp1327Gly | 3/358 |
| c.4005+45A→G | IVS 25 | 16/138 | |
| c.4005+232C→G | IVS 25 | 1/138 | |
| c.4014C→T | Exon 25 | Ser1338 | 2/138 |
| c.4305G→A | Exon 25 | Arg1435 | 3/138 |
| c.4859+54G→C | IVS 27 | 1/138 | |
| c.4859+152C→T | IVS 27 | 20/138 | |
| c.4859+191G→A | IVS 27 | 3/138 | |
| c.5103A→G | Exon 28 | Lys1701 | 1/138 |
| c.5310+171G→A | IVS 30 | 24/138 | |
| c.5310+195T→G | IVS 30 | 3/138 | |
| c.6028−116C→T | IVS 34 | 12/138 | |
| c.6028−84G→T | IVS 34 | 2/138 | |
| c.6224G→A | Exon 34 | Val2075Met | 2/358 |
| c.6421C→T | Exon 34 | Leu2141 | 2/138 |
| c.7341T→A | Exon 34 | Gly2447 | 20/138 |
Numbering is based on NOTCH2 cDNA sequence (GenBank accession number NM_024408.2).
Frequency is presented as number of occurrences per number of chromosomes sequenced and refers to both patients and controls.
Data presented here indicate that AGS is a genetically heterogeneous disorder caused by mutations in the Notch signaling pathway ligand gene, JAG1, or the gene for its receptor, NOTCH2. However, at this time, the vast majority of patients with AGS have mutations in the JAG1 gene, with only a small subset showing NOTCH2 mutations. This may be a matter of ascertainment bias, in that more patients with NOTCH2 mutations may be found who do not meet the full criteria we used to select patients with AGS. It is also possible that a single NOTCH2 mutation does not, by itself, cause AGS, but perhaps there are polymorphisms in JAG1 or other Notch signaling pathway genes that are necessary to cause the phenotype. We analyzed the complete JAG1 gene in the two probands with NOTCH2 mutations, and both had multiple JAG1 polymorphisms, although all of these were common and did not result in an amino acid alteration. Further work is needed to understand why NOTCH2 mutations are relatively rare in the AGS population.
To our knowledge, this is the first report of mutations in the NOTCH2 gene causing human disease. Germline mutations in two other Notch receptor genes have been associated with human disease: congenital cardiac disease, in patients with NOTCH1 mutations,19 and CADASIL, in patients with NOTCH3 mutations.17 Both probands we identified with NOTCH2 mutations clearly meet the diagnostic criteria for AGS, which require the presence of three of five clinical features (cholestasis, cardiac disease, ocular abnormality, skeletal abnormality, and characteristic facial features). It is of interest that the renal disease in both probands was severe. In addition, renal disease was present in all three mildly affected relatives who also carried the NOTCH2 mutation. Renal disease has been reported in 40%–70% of patients with a clinical diagnosis of AGS.1,2 Renal tubular acidosis and small kidneys are the most common findings, although severe renal phenotypes, including renal failure, have been described.20 Microscopic examination of the kidneys of 26 children with AGS revealed glomerular lesions of varying severity in 18 children and mild changes in the remaining 8 children.21 In our cohort, there were 59 JAG1 mutation-positive parents. Of the 59, 3 had renal anomalies. These included two parents with renal failure (one complicated by the presence of diabetes) and one parent with a deformity of the left ureter. There is evidence from mouse studies that functional Notch2 is required for normal kidney development, because mice homozygous for a hypomorphic Notch2 mutation died perinatally secondary to defects in glomerular development.22 This work raises the possibility that AGS caused by mutations in NOTCH2 will be found to have a phenotypic profile different from that of AGS caused by mutations in JAG1.
Acknowledgments
We are grateful to Drs. Bernard Kaplan, Jennifer Morrissette, and Elizabeth Rand and to Rob Bauer for thoughtful review and recommendations. We thank the physicians and families who provided samples and clinical information for these studies. This work was supported by National Institute for Diabetes and Digestive Diseases grants NIDDK-DK53104 and U54RR019455 and by the Fred and Suzanne Biesecker Center at The Children’s Hospital of Philadelphia.
Web Resources
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for NOTCH2 cDNA [accession number NM_024408.2])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AGS) [PubMed]
References
- 1.Alagille D, Estrada A, Hadchouel M, Gautier M, Odievre M, Dommergues JP (1987) Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): review of 80 cases. J Pediatr 110:195–200 10.1016/S0022-3476(87)80153-1 [DOI] [PubMed] [Google Scholar]
- 2.Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA (1999) Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology 29:822–829 10.1002/hep.510290331 [DOI] [PubMed] [Google Scholar]
- 3.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont MEM, Rand EB, Piccoli DA, Hood L, Spinner NB (1997) Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet 16:243–251 10.1038/ng0797-243 [DOI] [PubMed] [Google Scholar]
- 4.Oda T, Elkahlous AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PM, Spinner NB, Collins FS, Chandrasekharappa SC (1997) Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet 16:235–242 10.1038/ng0797-235 [DOI] [PubMed] [Google Scholar]
- 5.Gridley T (2003) Notch signaling and inherited disease syndromes. Hum Mol Genet Review Issue 1 12:R9–R13 10.1093/hmg/ddg052 [DOI] [PubMed] [Google Scholar]
- 6.Warthen DM, Moore EC, Kamath BM, Morrissette JJD, Sanchez P, Piccoli DA, Krantz ID, Spinner NB (2006) Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat 27:436–443 10.1002/humu.20310 [DOI] [PubMed] [Google Scholar]
- 7.Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T (1999) Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet 8:723–730 10.1093/hmg/8.5.723 [DOI] [PubMed] [Google Scholar]
- 8.McCright B, Lozier J, Gridley T (2002) A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 129:1075–1082 [DOI] [PubMed] [Google Scholar]
- 9.Loomes KM, Taichman DB, Glover CL, Williams PT, Markowitz JE, Piccoli DA, Baldwin HS, Oakey RJ (2002) Characterization of Notch receptor expression in the developing mammalian heart and liver. Am J Med Genet 112:181–189 10.1002/ajmg.10592 [DOI] [PubMed] [Google Scholar]
- 10.Crosnier C, Attie-Bitach T, Encha-Razavi E, Audollent S, Soudy F, Hadchouel M, Meunier-Rotival M, Vekemans M (2000) JAGGED1 gene expression during human embryogenesis elucidates the wide phenotypic spectrum of Alagille syndrome. Hepatology 32:574–581 10.1053/jhep.2000.16600 [DOI] [PubMed] [Google Scholar]
- 11.Duan Z, Li FQ, Wechsler J, Meade-White K, Williams K, Benson KF, Horwitz M (2004) A novel notch protein, N2N, targeted by neutrophil elastase and implicated in hereditary neutropenia. Mol Cell Biol 24:58–70 10.1128/MCB.24.1.58-70.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5:89–97 10.1038/nrm1310 [DOI] [PubMed] [Google Scholar]
- 13.Lubman OY, Korolev SV, Kopan R (2004) Anchoring Notch genetics and biochemistry: structural analysis of the ankyrin domain sheds light on existing data. Molecular Cell 13:619–626 10.1016/S1097-2765(04)00120-0 [DOI] [PubMed] [Google Scholar]
- 14.Kurooka H, Honjo T (2000) Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem 275:17211–17220 10.1074/jbc.M000909200 [DOI] [PubMed] [Google Scholar]
- 15.Campbell ID, Bork P (1993) Epidermal growth factor-like modules. Curr Opin Struct Biol 3:385–392 10.1016/S0959-440X(05)80111-3 [DOI] [Google Scholar]
- 16.Dietz HC, Saraiva JM, Pyeritz RE, Cutting GR, Francomano CA (1992) Clustering of fibrillin (FBN1) missense mutations in Marfan syndrome patients at cysteine residues in EGF-like domains. Hum Mutat 1:366–374 10.1002/humu.1380010504 [DOI] [PubMed] [Google Scholar]
- 17.Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C, Cruaud C, Maciazek J, Weissenbach J, Bousser MG, Bach JF, Tournier-Lasserve E (1997) Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet 350:1511–1515 10.1016/S0140-6736(97)08083-5 [DOI] [PubMed] [Google Scholar]
- 18.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S (1991) Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67:687–699 10.1016/0092-8674(91)90064-6 [DOI] [PubMed] [Google Scholar]
- 19.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D (2005) Mutations in NOTCH1 cause aortic valve disease. Nature 437:270–274 10.1038/nature03940 [DOI] [PubMed] [Google Scholar]
- 20.Harendza S, Hubner CA, Glaser C, Burdelski M, Thaiss F, Hansmann I, Gal A, Stahl RAK (2005) Renal failure and hypertension in Alagille syndrome with a novel JAG1 mutation. J Nephrol 18:312–317 [PubMed] [Google Scholar]
- 21.Habib R, Dommergues JP, Gubler MC, Hadchouel M, Gautier M, Odievre M, Alagille D (1987) Glomerular mesangiolipidosis in Alagille syndrome (arteriohepatic dysplasia). Pediatr Nephrol 1:455–464 10.1007/BF00849254 [DOI] [PubMed] [Google Scholar]
- 22.McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T (2001) Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491–502 [DOI] [PubMed] [Google Scholar]