Abstract
Many lines of evidence implicate mitochondria in phenotypic variation: (a) rare mutations in mitochondrial proteins cause metabolic, neurological, and muscular disorders; (b) alterations in oxidative phosphorylation are characteristic of type 2 diabetes, Parkinson disease, Huntington disease, and other diseases; and (c) common missense variants in the mitochondrial genome (mtDNA) have been implicated as having been subject to natural selection for adaptation to cold climates and contributing to “energy deficiency” diseases today. To test the hypothesis that common mtDNA variation influences human physiology and disease, we identified all 144 variants with frequency >1% in Europeans from >900 publicly available European mtDNA sequences and selected 64 tagging single-nucleotide polymorphisms that efficiently capture all common variation (except the hypervariable D-loop). Next, we evaluated the complete set of common mtDNA variants for association with type 2 diabetes in a sample of 3,304 diabetics and 3,304 matched nondiabetic individuals. Association of mtDNA variants with other metabolic traits (body mass index, measures of insulin secretion and action, blood pressure, and cholesterol) was also tested in subsets of this sample. We did not find a significant association of common mtDNA variants with these metabolic phenotypes. Moreover, we failed to identify any physiological effect of alleles that were previously proposed to have been adaptive for energy metabolism in human evolution. More generally, this comprehensive association-testing framework can readily be applied to other diseases for which mitochondrial dysfunction has been implicated.
Mitochondria play a central role in energy metabolism, are composed of >700 known proteins,1 and are essential for generating ATP and for regulating apoptosis.2 The human mitochondrial oxidative phosphorylation (OXPHOS) machinery, which synthesizes most intracellular ATP, consists of five complexes with 85 known protein subunits. Thirteen OXPHOS subunits, 2 rRNA genes, and 22 tRNA genes are encoded by the 16.6-kb mitochondrial genome (mtDNA).
Rare mutations in both nuclear-encoded OXPHOS genes and in mtDNA result in disease syndromes with neurological, muscular, or metabolic manifestations, proving that defects in mitochondrial OXPHOS can play a causal role in human disease. Mutations in nuclear-encoded components of OXPHOS complexes have been identified in many early-onset diseases, such as Leigh syndrome (MIM 256000) and cardioencephalomyopathy (MIM 604377).2,3 Nuclear genes that encode OXPHOS assembly factors, influence mtDNA maintenance or translation, modify mitochondrial tRNA, or encode biosynthetic enzymes may be mutated in rare mitochondrial diseases.4,5 In addition, >250 mtDNA point mutations and deletions have been linked to human disease, and several of these include glucose defects or diabetes mellitus as component phenotypes—for example, mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS [MIM 540000]), Kearns-Sayre syndrome (KSS [MIM 530000]), and maternally inherited diabetes and deafness syndrome (MIDD [MIM 520000]).6,7 A mutation in the mitochondrial Leu tRNA gene (A3243G) causes MIDD,8 and a novel substitution in a highly conserved region of the mitochondrial Ile tRNA gene (T4291C) causes hypertension, hypercholesterolemia, and hypomagnesemia (MIM 500005),9 suggesting that mutations in mtDNA can cause diabetes and other metabolic defects.
Alterations in the function of OXPHOS have been recognized consistently in type 2 diabetes (MIM 125853). Reduced activity of OXPHOS enzymes and fewer, smaller mitochondria are seen by transmission electron microscopy in skeletal muscle from diabetics compared with nondiabetics.10–12 From global gene-expression–profiling studies, we and others identified a subset of OXPHOS genes that are co-coordinately downregulated in muscles from individuals with type 2 diabetes compared with healthy control individuals.13,14 In addition, functional in vivo spectroscopy studies demonstrate that OXPHOS activity and rates of ATP synthesis are lower in the insulin-resistant offspring of diabetic individuals and in elderly insulin-resistant people.15–17
Population genetics analysis has suggested a functional role for common variants in mtDNA. On the basis of differential conservation of missense variants in different mtDNA lineages, it was argued that positive selection influenced common mtDNA variation. Moreover, it was hypothesized that mtDNA SNPs favorable for selective adaptation to cold Northern climates during human evolution predispose to energy metabolism diseases today.2,18,19
These many lines of evidence suggest a “mitochondrial” hypothesis of disease. Specifically, inherited defects in mitochondria may play a causal role in type 2 diabetes20,21 and neurodegenerative diseases, such as Alzheimer (MIM 104300), Parkinson (MIM 168600), and Huntington (MIM 143100) diseases.22,23 Population genetics analysis specifically suggests that common mtDNA variants may be functional and relevant to disease. However, no comprehensive test, even of common variation in mtDNA, has yet been performed. Several previous reports did not consistently reproduce associations of common mtDNA variants with diabetes or metabolic traits.24–29 However, these studies tested only a subset of mtDNA variation in relatively modestly sized samples of <2,000 individuals.
The goal of the present study was to perform a comprehensive test of the hypothesis that mtDNA variants influence type 2 diabetes and metabolic traits. Specifically, we sought to (1) inventory all SNPs with frequency >1% in Europeans, (2) select tagging SNPs (tSNPs) to capture this variation efficiently, (3) test all common variants and haplogroups for association with type 2 diabetes and a range of metabolic phenotypes (although sample sizes for some traits were limited), and (4) assess studywide significance of our findings by permutation testing.
Material and Methods
Alignment of Sequences
We aligned all human mtDNA coding-region sequences from GenBank (719 sequences) and 536 sequences from Mitokor30 and identified 3,240 variant sites. We excluded ∼0.8 kb of the hypervariable mtDNA D-loop promoter region from the study, since this region is best addressed by direct resequencing in case-control samples because of its high mutation rate.
Subjects and Samples
The sample consisted of 6,608 white subjects from Scandinavia (Sweden and Botnia, hereafter referred to as the “Swedish” and “Scandinavian” samples, respectively), Canada, Poland, and the United States; metabolic phenotype information was available for a subset of the subjects (table 1).31,32 The Scandinavian and Canadian diabetic case-control samples were matched for sex, age, region, and BMI (calculated as weight in kilograms divided by the square of height in meters), whereas the Polish and U.S. case-control samples were matched for sex, age, and geographical region but not tightly for BMI. BMI, blood pressure, and cholesterol values were available for diabetic and nondiabetic subjects from all five populations, and insulinogenic index and homeostasis model assessment of insulin resistance (HOMA-IR) values were obtained for nondiabetic Scandinavian and Swedish subjects (from a 2-h oral glucose tolerance test [OGTT]). The study of human subjects was approved by the institutional review boards at parent institutions for all samples and at the Broad Institute for Harvard and MIT. Genomic DNA was extracted from blood samples and lymphoblastoid cell lines.
Table 1. .
Clinical Characteristics of the Diabetes Case-Control Study Samples and Quantitative Metabolic Traits in Subjects Used to Study Genotype-Phenotype Correlations[Note]
| Sample or Traita | n | No. of Males; Females | Age (years) |
BMI | Fasting Plasma Glucose (mmol/liter) |
HbA1cb (%) |
Plasma Glucose by 2-h OGTT (mmol/liter) |
Trait Value |
| Case-control sample: | ||||||||
| Scandinavian: | ||||||||
| DM/severe IGT | 459 | 247; 212 | 60.9 ± 10.2 | 28.2 ± 4.6 | 9.8 ± 3.4 | … | 15.0 ± 5.3 | … |
| NGT | 459 | 247; 212 | 59.6 ± 10.4 | 26.3 ± 3.6 | 6.2 ± 1.8 | … | 6.8 ± 2.8 | … |
| Swedish: | ||||||||
| DM/severe IGT | 505 | 264; 241 | 66.3 ± 11.8 | 27.6 ± 4.1 | 9.8 ± 3.4 | 6.5 ± 1.5 | … | … |
| NGT | 505 | 264; 241 | 66.6 ± 11.6 | 27.7 ± 4.1 | 6.2 ± 1.8 | ND | ND | … |
| Canadian: | ||||||||
| DM | 123 | 67; 56 | 53.5 ± 7.9 | 29.2 ± 4.4 | 6.4 ± 1.8 | … | 12.8 ± 2.1 | … |
| NGT | 123 | 67; 56 | 52.1 ± 7.9 | 28.6 ± 4.1 | 5.1 ± 0.6 | … | 6.1 ± 1.1 | … |
| U.S.c: | ||||||||
| DM | 1,214 | 641; 573 | 62.6 ± 11.0 | 33.0 ± 6.9 | 9.8 ± 3.0 | 8.0 ± 3.1 | … | … |
| NGT | 1,214 | 641; 573 | 60.9 ± 9.7 | 27.4 ± 5.2 | 5.1 ± 0.9 | ND | ND | … |
| Polishc: | ||||||||
| DM | 1,003 | 420; 583 | 61.8 ± 9.6 | 29.6 ± 4.8 | 8.9 ± 4.0 | 7.9 ± 1.3 | … | … |
| NGT | 1,003 | 420; 583 | 58.7 ± 7.2 | 26.1 ± 3.6 | 4.8 ± 1.2 | ND | ND | … |
| Trait: | ||||||||
| Ins index | 342 | 181; 161 | 58.5 ± 10.1 | 26.2 ± 3.6 | … | … | … | 22.6 ± 31.3 |
| HOMA-IR | 399 | 210; 189 | 59.2 ± 10.3 | 26.3 ± 3.7 | … | … | … | 2.1 ± 1.3 |
| Cholesterol | 1,274 | 682; 592 | 62.5 ± 11.1 | 27.4 ± 4.2 | … | … | … | 6.0 ± 1.2 mmol |
| Systolic BP | 2,047 | 1,090; 957 | 62.2 ± 11.7 | 27.7 ± 4.2 | … | … | … | 144 ± 22 mm Hg |
| Diastolic BP | 2,047 | 1,090; 957 | 62.2 ± 11.7 | 27.7 ± 4.2 | … | … | … | 83 ± 10 mm Hg |
Note.— Data are means ± SD. ND = not determined.
BP = blood pressure; DM = diabetes mellitus, type 2; IGT = impaired glucose tolerance; Ins = insulinogenic; NGT = normal glucose tolerance.
HbA1c = hemoglobin A1c.
Sample from Genomics Collaborative Inc.
Genotyping
Genotyping was performed by primer extension of multiplex products with detection by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy with use of a Sequenom platform.33 Genotyping for the 64 tSNPs was >98%; maternal inheritance was confirmed for tSNPs by genotyping in 117 CEPH trios (not shown). Two SNPs (mt14470 and mt15884) were triallelic in this population. Duplicate genotypes were obtained for ∼20% of the sample with 22 SNPs and were highly concordant (0.1% discordant genotypes of 29,462 duplicate comparisons). For 69 samples (1.1% of samples genotyped), we observed spectral peaks for both the major and minor allele for 1–3 SNPs per sample. In total, 21 SNPs had heterozygous calls in at least one individual. These observations are consistent with either DNA contamination or heteroplasmy (more than one mtDNA haplotype arising from somatic/maternal germ line mutations or from the paternal mtDNA contribution). Signals for both alleles were seen in individuals from all populations, and no significant difference in the number of cases and controls with such genotypes was observed. For analysis, these genotypes were treated as missing data.
Results
Previous studies have tested subsets of mtDNA variants (from 10–32 sites) for association with a variety of traits,24–27,34–49 typically focusing on the nine canonical haplogroups.50 We began by cataloging all common sites (frequency >1%) in 15,770 bp of mtDNA (excluding the coding region) and by evaluating how well the complete inventory is captured by these canonical haplogroups in mtDNAs of European origin. Alignment of 928 European coding-region mtDNA sequences (>15,770 bp) identified 2,349 variable sites; 144 sites had a frequency >1% in European individuals (1 site per 110 bp), including 37 nonsynonymous variants, 81 synonymous variants, 25 rRNA/tRNA variants, and 1 noncoding variant (table 2). Fifty variants with frequency >5% were identified. The common variants (frequency >1%) explained 72% of the “heterozygosity” in the European population; this is notably less than in the nuclear genome, where ∼90% of heterozygosity is due to common variants.51–53
Table 2. .
Common European mtDNA Variants with Frequency >1%, by Gene of Origin
| Genea | OXPHOS Complex | Putatively Functional | Missense Mutations |
Synonymous Mutations |
Total |
| ND1 | 1 | … | 5 | 5 | 10 |
| ND2 | 1 | … | 6 | 11 | 17 |
| ND3 | 1 | … | 2 | 1 | 3 |
| ND4L | 1 | … | … | 1 | 1 |
| ND4 | 1 | … | … | 15 | 15 |
| ND5 | 1 | … | 6 | 15 | 21 |
| ND6 | 1 | … | 1 | 5 | 6 |
| Cyt B | 3 | … | 9 | 5 | 14 |
| COX1 | 4 | … | … | 10 | 10 |
| COXII | 4 | … | … | 4 | 4 |
| COXIII | 4 | … | 3 | 4 | 7 |
| ATP6 | 5 | … | 5 | 5 | 10 |
| 12SrRNA | … | 6 | … | … | 6 |
| 16SrRNA | … | 7 | … | … | 7 |
| tRNA Gln | … | 1 | … | … | 1 |
| tRNA Ser | … | 1 | … | … | 1 |
| tRNA Gly | … | 2 | … | … | 2 |
| tRNA Arg | … | 1 | … | … | 1 |
| tRNA Ser2 | … | 1 | … | … | 1 |
| tRNA Leu2 | … | 1 | … | … | 1 |
| tRNA Glu | … | 1 | … | … | 1 |
| tRNA Thr | … | 4 | … | … | 4 |
| Noncoding | … | … | … | … | 1 |
| Total | … | 25 | 37 | 81 | 144 |
One ATP6 missense variant is also a synonymous ATP8 variant; no common variants in 14 tRNA genes.
Evaluation of previous sets of tSNPs used for disease association studies revealed that ∼30% of variants (frequency ⩾1%) are captured with r2⩾0.8 (table 3). Recently,27 more-complete coverage of mtDNA variation was reported using 32 mtDNA SNPs; still, these SNPs capture less than half of the sites (42%) at r2⩾0.8. Thus, previous studies might have missed a true association simply because not all variants were captured by the tSNPs used.
Table 3. .
Estimated Fraction of mtDNA Coding-Region Variation Captured by Previous Association Studies Compared with the Present Study
| Percentage of SNPs Captured |
||||
| Study | tSNPs | Testsa | r2⩾.5 | r2⩾.8 |
| Van der Walt et al.47 | 9b | 9 HGs + 9 SNPs | 35 | 30 |
| Huerta et al.49 | 12b | 9 HGs + 12 SNPs | 33 | 26 |
| Niemi et al.42 | 8 | 9 HGs | 34 | 26 |
| Mohlke et al.27 | 32c | 9 HGs + 32 SNPs | 54 | 42 |
| Present study | 64 | All variants with frequency >1% | 100 | 100 |
HGs = haplogroups.
One D-loop–region SNP was used to specify haplogroup I; this SNP was excluded from our evaluation of the coverage of coding-region mtDNA SNPs.
Ten SNPs chosen by Mohlke et al.27 (in Finns) were observed at a frequency <1% in the 928 European sequences of the reference panel.
We selected a set of 64 SNPs that capture each of the 144 sites as well as the nine haplogroups with r2⩾0.8, using Tagger54; tSNPs and predictive tests are listed in table 4. Details on the assay conditions are available online (Broad Institute Tagger: SNPs in human mtDNA Web site).
Table 4. .
tSNPs and Tests Used to Predict All Coding-Region Variants with Frequency >1% in European mtDNA
| SNP or Haplogroup | Position (rCRSa) | Alleles (Major/Minor) | Rarer Allele Frequency (Reference Panel) | Captured by Test (tSNP or Haplotype) | Test Allele(s) | Test Allele Frequency (Reference Panel) | r2 for Test vs. Predicted Allele | SNP Type |
| SNP: | ||||||||
| mt709 | 709 | G/A | .20 | mt709 | tSNP | |||
| mt750 | 750 | G/A | .01 | mt750 | tSNP | |||
| mt930 | 930 | G/A | .05 | mt930 | tSNP | |||
| mt1189 | 1189 | T/C | .07 | mt1189 | tSNP | |||
| mt1243 | 1243 | T/C | .05 | mt11674 | T | .05 | 1 | |
| mt1438 | 1438 | G/A | .03 | mt4769 | A | .02 | .872 | |
| mt1719 | 1719 | G/A | .06 | mt709, mt12007, mt12705 | G, G, T | .05 | .836 | |
| mt1811 | 1811 | A/G | .13 | mt3348, mt9477, mt12372 | A, G, A | .13 | .944 | |
| mt1888 | 1888 | G/A | .11 | mt11812, mt12633 | A, C | .89 | .978 | |
| mt2158 | 2158 | T/C | .01 | mt11914, mt13879 | G, C | .01 | .838 | |
| mt2706 | 2706 | G/A | .38 | mt7028 | C | .38 | .982 | |
| mt3010 | 3010 | G/A | .23 | mt3010 | tSNP | |||
| mt3197 | 3197 | T/C | .06 | mt9477 | A | .05 | .959 | |
| mt3348 | 3348 | A/G | .02 | mt3348 | tSNP | |||
| mt3394 | 3394 | T/C | .01 | mt3394 | tSNP | |||
| mt3480 | 3480 | A/G | .09 | mt1189, mt9716 | T, T | .90 | .988 | |
| mt3505 | 3505 | A/G | .05 | mt11674 | T | .05 | .978 | |
| mt3720 | 3720 | A/G | .01 | mt5426, mt12372 | C, A | .01 | 1 | |
| mt3915 | 3915 | G/A | .01 | mt3915 | tSNP | |||
| mt3990 | 3990 | C/T | .01 | mt10915, mt15924 | C, G | .01 | 1 | |
| mt3992 | 3992 | C/T | .02 | mt7028, mt9123 | C, A | .02 | .9 | |
| mt4024 | 4024 | A/G | .02 | mt7028, mt9123 | C,A | .02 | 1 | |
| mt4216 | 4216 | T/C | .22 | mt12705, mt9123, mt11719, mt12372 | C, G, A, G | .21 | .969 | |
| mt4336 | 4336 | T/C | .02 | mt4336 | tSNP | |||
| mt4529 | 4529 | A/T | .03 | mt9123, mt10034 | G, C | .03 | 1 | |
| mt4561 | 4561 | T/C | .02 | mt709, mt9716 | A, C | .02 | .847 | |
| mt4580 | 4580 | G/A | .07 | mt7028, mt11719, mt15218 | T, G, A | .08 | .844 | |
| mt4639 | 4639 | T/C | .02 | mt8869 | G | .02 | .88 | |
| mt4769 | 4769 | G/A | .02 | mt4769 | tSNP | |||
| mt4793 | 4793 | A/G | .01 | mt4793 | tSNP | |||
| mt4917 | 4917 | A/G | .11 | mt11812, mt12633 | A, C | .89 | .978 | |
| mt4928 | 4928 | T/C | .02 | mt4928 | tSNP | |||
| mt5004 | 5004 | T/C | .02 | mt7028, mt9123 | C, A | .02 | .907 | |
| mt5046 | 5046 | G/A | .05 | mt709, mt12705 | A, T | .05 | 1 | |
| mt5147 | 5147 | G/A | .05 | mt709, mt930 | A, A | .05 | .953 | |
| mt5263 | 5263 | C/T | .01 | mt8869 | G | .02 | .865 | |
| mt5390 | 5390 | A/G | .01 | mt12372, mt13020 | A, C | .01 | .922 | |
| mt5426 | 5426 | T/C | .02 | mt5426 | tSNP | |||
| mt5460 | 5460 | G/A | .07 | mt11674, mt13879 | C, T | .94 | .857 | |
| mt5465 | 5465 | T/C | .02 | mt5465 | tSNP | |||
| mt5495 | 5495 | T/C | .03 | mt5495 | tSNP | |||
| mt5656 | 5656 | A/G | .02 | mt5656 | tSNP | |||
| mt6045 | 6045 | C/T | .01 | mt5426, mt13020 | C, C | .01 | 1 | |
| mt6152 | 6152 | T/C | .01 | mt5426, mt11812 | C, A | .01 | .922 | |
| mt6221 | 6221 | T/C | .02 | mt12705, mt14470, mt15884 | T, C, G | .02 | .898 | |
| mt6260 | 6260 | G/A | .01 | mt6260 | tSNP | |||
| mt6365 | 6365 | T/C | .02 | mt6365 | tSNP | |||
| mt6371 | 6371 | C/T | .02 | mt11674, mt12705, mt14470 | C, T, C | .02 | 1 | |
| mt6719 | 6719 | T/C | .01 | mt6719 | tSNP | |||
| mt6734 | 6734 | G/A | .01 | mt10034, mt10915 | C, C | .01 | .844 | |
| mt6776 | 6776 | T/C | .04 | mt6776 | tSNP | |||
| mt7028 | 7028 | T/C | .38 | mt7028 | tSNP | |||
| mt7476 | 7476 | C/T | .01 | mt13708, mt15257 | A, A | .01 | 1 | |
| mt7768 | 7768 | A/G | .03 | mt10034, mt14182 | T, C | .03 | 1 | |
| mt7864 | 7864 | C/T | .04 | mt709, mt12705, mt15784 | A, T, T | .05 | .95 | |
| mt8251 | 8251 | G/A | .08 | mt12633, mt12705, mt13020, mt13105, mt13708, mt13734, mt13879, mt13934, mt13965, mt13966, mt14182 | C, T, T, A, G, T, T, C, T, A, T | .09 | .959 | |
| mt8269 | 8269 | G/A | .03 | mt8269 | tSNP | |||
| mt8557 | 8557 | G/A | .01 | mt8269, mt13708 | A, A | .01 | .916 | |
| mt8616 | 8616 | G/T | .01 | mt10915, mt12705 | C, T | .01 | .844 | |
| mt8697 | 8697 | G/A | .11 | mt11812, mt12633 | A, C | .89 | .946 | |
| mt8705 | 8705 | T/C | .01 | mt8705 | tSNP | |||
| mt8869 | 8869 | A/G | .02 | mt8869 | tSNP | |||
| mt8994 | 8994 | G/A | .05 | mt709, mt12705 | A, T | .05 | .978 | |
| mt9055 | 9055 | G/A | .10 | mt1189, mt9716 | T, T | .90 | .988 | |
| mt9093 | 9093 | A/G | .01 | mt11377, mt12372 | A, A | .01 | .922 | |
| mt9123 | 9123 | G/A | .04 | mt9123 | tSNP | |||
| mt9150 | 9150 | A/G | .01 | mt9150 | tSNP | |||
| mt9477 | 9477 | G/A | .05 | mt9477 | tSNP | |||
| mt9612 | 9612 | G/A | .01 | mt4928, mt11719 | C, A | .01 | 1 | |
| mt9667 | 9667 | A/G | .01 | mt9667 | tSNP | |||
| mt9698 | 9698 | T/C | .10 | mt1189, mt9716 | T, T | .90 | .964 | |
| mt9716 | 9716 | T/C | .02 | mt9716 | tSNP | |||
| mt9899 | 9899 | T/C | .02 | mt9899 | tSNP | |||
| mt9947 | 9947 | G/A | .01 | mt10034, mt10915 | C, C | .01 | .916 | |
| mt10034 | 10034 | T/C | .03 | mt10034 | tSNP | |||
| mt10044 | 10044 | A/G | .02 | mt8269, mt9123, mt15884 | A, A, G | .02 | .932 | |
| mt10084 | 10084 | T/C | .01 | mt10084 | tSNP | |||
| mt10238 | 10238 | T/C | .05 | mt5465, mt10034 | T, T | .95 | .884 | |
| mt10398 | 10398 | A/G | .20 | mt1189, mt13708, mt10034 | T, G, T | .78 | .855 | |
| mt10463 | 10463 | T/C | .11 | mt11812, mt12633 | A, C | .89 | .978 | |
| mt10550 | 10550 | A/G | .08 | mt1189, mt9716 | T, T | .90 | .853 | |
| mt10876 | 10876 | A/G | .01 | mt12372, mt13020 | A, C | .01 | 1 | |
| mt10915 | 10915 | T/C | .02 | mt10915 | tSNP | |||
| mt11251 | 11251 | A/G | .22 | mt11812, mt12633, mt13708 | A, C, G | .77 | .905 | |
| mt11299 | 11299 | T/C | .10 | mt1189, mt9716 | T, T | .90 | 1 | |
| mt11377 | 11377 | G/A | .02 | mt11377 | tSNP | |||
| mt11467 | 11467 | A/G | .20 | mt12372, mt12705 | A, C | .20 | .993 | |
| mt11470 | 11470 | A/G | .01 | mt1189, mt15924 | C, G | .01 | 1 | |
| mt11485 | 11485 | T/C | .02 | mt11485 | tSNP | |||
| mt11674 | 11674 | C/T | .05 | mt11674 | tSNP | |||
| mt11719 | 11719 | A/G | .47 | mt11719 | tSNP | |||
| mt11812 | 11812 | A/G | .08 | mt11812 | tSNP | |||
| mt11840 | 11840 | C/T | .01 | mt1189, mt6260 | C, A | .01 | 1 | |
| mt11914 | 11914 | G/A | .02 | mt11914 | tSNP | |||
| mt11947 | 11947 | A/G | .05 | mt709, mt12705 | A, T | .05 | 1 | |
| mt12007 | 12007 | G/A | .01 | mt12007 | tSNP | |||
| mt12239 | 12239 | C/C | .01 | mt5465, mt6719 | C, C | .01 | .908 | |
| mt12308 | 12308 | A/G | .20 | mt12372, mt12705 | A, C | .20 | 1 | |
| mt12372 | 12372 | G/A | .20 | mt12372 | tSNP | |||
| mt12414 | 12414 | T/C | .02 | mt12414 | tSNP | |||
| mt12501 | 12501 | G/A | .03 | mt709, mt12705, mt14470 | G, T, T | .03 | .936 | |
| mt12612 | 12612 | A/G | .11 | mt12372, mt12705, mt11719, mt13708, mt1189 | G, C, A, A, T | .11 | 1 | |
| mt12618 | 12618 | G/A | .02 | mt5656, mt14182, mt14470 | G, C, T | .02 | .943 | |
| mt12633 | 12633 | C/A | .03 | mt12633 | tSNP | |||
| mt12669 | 12669 | C/T | .02 | mt5495, mt14793 | C, A | .02 | 1 | |
| mt12705 | 12705 | C/T | .11 | mt12705 | tSNP | |||
| mt13020 | 13020 | T/C | .02 | mt13020 | tSNP | |||
| mt13105 | 13105 | A/G | .01 | mt13105 | tSNP | |||
| mt13368 | 13368 | G/A | .11 | mt11812, mt12633 | A, C | .89 | .989 | |
| mt13617 | 13617 | T/C | .05 | mt9477 | A | .05 | 1 | |
| mt13708 | 13708 | G/A | .12 | mt13708 | tSNP | |||
| mt13734 | 13734 | T/C | .02 | mt13734 | tSNP | |||
| mt13740 | 13740 | T/C | .01 | mt1189, mt6260 | C, A | .01 | 1 | |
| mt13780 | 13780 | A/G | .03 | mt12705, mt15043 | T, A | .03 | 1 | |
| mt13879 | 13879 | T/C | .01 | mt13879 | tSNP | |||
| mt13934 | 13934 | C/T | .02 | mt13934 | tSNP | |||
| mt13965 | 13965 | T/C | .02 | mt13965 | tSNP | |||
| mt13966 | 13966 | A/G | .02 | mt13966 | tSNP | |||
| mt14022 | 14022 | A/G | .01 | mt6719, mt9123 | C, A | .01 | 1 | |
| mt14167 | 14167 | C/T | .10 | mt1189, mt9716 | T, T | .90 | 1 | |
| mt14182 | 14182 | T/C | .03 | mt14182 | tSNP | |||
| mt14233 | 14233 | A/G | .08 | mt11812 | G | .08 | .942 | |
| mt14365 | 14365 | C/T | .02 | mt7028, mt9123 | C, A | .02 | 1 | |
| mt14470 | 14470 | T/C | .03 | mt14470 | tSNP | |||
| mt14582 | 14582 | A/G | .02 | mt7028, mt9123 | C, A | .02 | 1 | |
| mt14687 | 14687 | A/G | .02 | mt13965 | C | .02 | .932 | |
| mt14766 | 14766 | T/C | .47 | mt11719, mt11812 | G, A | .47 | .966 | |
| mt14793 | 14793 | A/G | .03 | mt14793 | tSNP | |||
| mt14798 | 14798 | T/C | .18 | mt14798 | tSNP | |||
| mt14905 | 14905 | G/A | .11 | mt11812, mt12633 | A, C | .89 | .989 | |
| mt15043 | 15043 | G/A | .04 | mt15043 | tSNP | |||
| mt15218 | 15218 | A/G | .02 | mt15218 | tSNP | |||
| mt15257 | 15257 | G/A | .02 | mt15257 | tSNP | |||
| mt15452 | 15452 | C/A | .22 | mt12372, mt12705, mt11719 | G, C, A | .23 | .904 | |
| mt15607 | 15607 | A/G | .11 | mt11812, mt12633 | A, C | .89 | .989 | |
| mt15746 | 15746 | A/A | .01 | mt5465, mt6719 | C, C | .01 | 1 | |
| mt15758 | 15758 | A/G | .02 | mt15758 | tSNP | |||
| mt15784 | 15784 | T/C | .01 | mt15784 | tSNP | |||
| mt15833 | 15833 | C/C | .02 | mt15833 | tSNP | |||
| mt15884 | 15884 | G/C | .06 | mt15884 | tSNP | |||
| mt15904 | 15904 | C/T | .07 | mt11719, mt15218, mt7028 | G, A, T | .08 | .844 | |
| mt15907 | 15907 | A/G | .01 | mt12372, mt13020 | A, C | .01 | 1 | |
| mt15924 | 15924 | A/G | .06 | mt15924 | tSNP | |||
| mt15928 | 15928 | G/A | .11 | mt11812, mt12633 | A, C | .89 | .978 | |
| Haplogroup: | ||||||||
| mt1719, mt7028, mt10398 (X) | A,T,A | .02 | mt709, mt12705, mt15043 | G, T, G | .02 | .868 | ||
| mt1719, mt7028, mt8251, mt10398 (I) | A,T,A,G | .03 | mt8269, mt10034, mt14182 | G, C, T | .03 | 1 | ||
| mt4580, mt7028, mt10398 (V) | A,T,A | .07 | mt7028, mt11719, mt15218 | T, G, A | .08 | .844 | ||
| mt7028, mt10398, mt12308 (U) | T,A,G | .13 | mt1189, mt12372, mt12705 | T, A, C | .13 | .981 | ||
| mt7028, mt10398, mt13708 (J) | T,G,A | .10 | mt7028, mt12705, mt13708 | T, C, A | .11 | .934 | ||
| mt7028, mt8251, mt10398 (W) | T,A,A | .06 | mt12705, mt13966, mt15043 | T, A, G | .06 | .94 | ||
| mt7028, mt10398, mt13368 (T) | T,A,A | .11 | mt11812, mt12633 | A,C | .89 | .989 | ||
| mt7028, mt9055, mt10398, mt12308 (K) | T,A,G,G | .07 | mt1189 | C | .07 | .984 | ||
| mt7028, mt10398 (H) | C,A | .38 | mt7028 | C | .38 | .995 |
rCRS = revised Cambridge Reference Sequence (see Human Mitochondrial DNA Revised Cambridge Reference Sequence Web site).
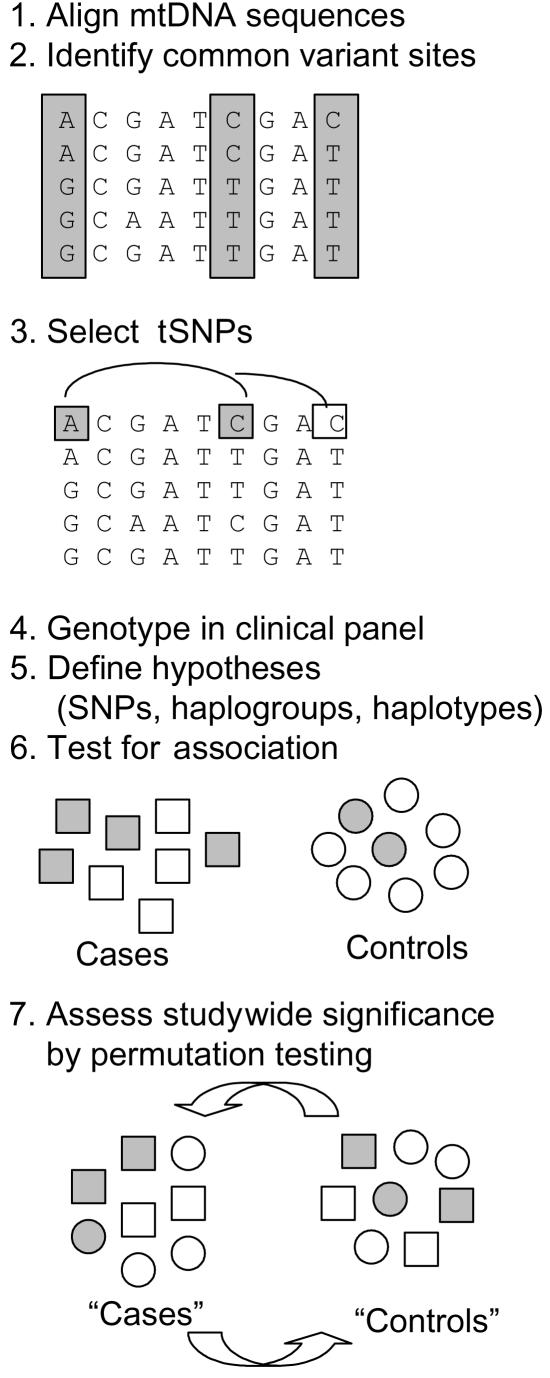
As a framework to test common mtDNA variants for association with clinical traits (fig. 1), we first enumerated all hypotheses to be tested for association. Specifically, we sought to test for association with each phenotype for each of the individual 144 variant sites with frequency >1% as well as for the nine previously codified European haplogroups. Association tests for type 2 diabetes (the dichotomous trait) were performed by 2×2 χ2 comparisons, and, for quantitative phenotypes, by linear regression. Studywide significance (Pstudy) was empirically evaluated by permutation testing (association studies were repeated 50–1,000 times by using randomized phenotype labels within each case-control pair for diabetes and by using randomized trait values within each of five populations for quantitative traits). We also tested all pairwise combinations of single variants with a nominal P value (Pnom) <.1 (another ∼600 correlated hypotheses) for association with diabetes and BMI.55
Figure 1. .
Procedure for identification and comprehensive disease-association testing of all common variants in the mtDNA coding region. 1 and 2, A total of 928 mtDNA sequences of European origin were aligned to identify 144 variants with frequency >1%. 3, tSNPs and multimarker haplotypes of tSNPs were selected to capture all 144 variant sites and haplogroups with r2⩾0.8 (a haplotype of tSNPs [shaded box] captures an untyped SNP [unshaded box]). 4, tSNPs were genotyped in a diabetic case-control panel with available metabolic phenotypes. 5, All hypotheses to be tested were enumerated. 6, Association tests were performed. 7, Studywide significance of results was assessed by permutation (multiple rounds of association testing with randomization of case-control labels or, for quantitative measures, shuffling within a population).
As a test of the hypothesis that common mtDNA variation plays a causal role in disease, we considered association with type 2 diabetes and metabolic traits. We genotyped each of the 64 tSNPs in a diabetes case-control sample of 6,608 subjects, with quantitative measurements available for a subset of subjects (table 1). Under the assumption of ∼100 independent tests performed in this study, this sample has >77% power to reject the null hypothesis of no association between mtDNA variants and type 2 diabetes at P<.05 for a 5% risk allele with a 1.5 genotype relative risk and has >98% power for risk alleles with frequency >10%.56 Allele frequencies for SNPs differed across the populations tested (table 5).
Table 5. .
Allele Frequencies of SNPs and Haplogroups across Five Study Populations
| Allele Frequency in Population | ||||||
| SNP or Haplogroup | Allele | Scandinavia(n = 918) | Sweden(n = 1,010) | Canada(n = 246) | United States(n = 2,428) | Poland(n = 2,006) |
| SNP: | ||||||
| mt709 | A | .130 | .146 | .199 | .136 | .154 |
| mt750 | A | .029 | .028 | .016 | .026 | .013 |
| mt930 | A | .026 | .031 | .050 | .039 | .058 |
| mt1189 | C | .053 | .057 | .073 | .075 | .038 |
| mt1243 | C | .029 | .011 | .012 | .020 | .019 |
| mt1438 | A | .067 | .045 | .017 | .039 | .027 |
| mt1719 | A | .031 | .053 | .126 | .061 | .049 |
| mt1811 | G | .105 | .220 | .140 | .160 | .135 |
| mt1888 | A | .074 | .112 | .159 | .088 | .123 |
| mt2158 | C | .015 | .010 | .033 | .010 | .014 |
| mt2706 | A | .460 | .431 | .322 | .419 | .404 |
| mt3010 | A | .262 | .227 | .149 | .229 | .222 |
| mt3197 | C | .161 | .107 | .041 | .076 | .113 |
| mt3348 | G | .002 | .003 | .000 | .002 | .001 |
| mt3394 | C | .018 | .007 | .004 | .010 | .005 |
| mt3480 | G | .066 | .069 | .094 | .099 | .044 |
| mt3505 | G | .029 | .011 | .012 | .020 | .019 |
| mt3720 | G | .007 | .014 | .000 | .015 | .022 |
| mt3915 | A | .012 | .035 | .025 | .025 | .020 |
| mt3990 | T | .003 | .010 | .004 | .007 | .009 |
| mt3992 | T | .010 | .018 | .000 | .013 | .013 |
| mt4024 | G | .010 | .018 | .000 | .013 | .013 |
| mt4216 | C | .149 | .217 | .238 | .194 | .221 |
| mt4336 | C | .021 | .017 | .025 | .023 | .033 |
| mt4529 | T | .011 | .025 | .114 | .031 | .018 |
| mt4561 | C | .010 | .007 | .017 | .017 | .004 |
| mt4580 | A | .061 | .050 | .045 | .061 | .056 |
| mt4639 | C | .019 | .007 | .004 | .004 | .010 |
| mt4769 | A | .067 | .045 | .017 | .039 | .027 |
| mt4793 | G | .014 | .014 | .008 | .021 | .014 |
| mt4917 | G | .074 | .112 | .159 | .088 | .123 |
| mt4928 | C | .004 | .001 | .000 | .000 | .003 |
| mt5004 | C | .010 | .018 | .000 | .013 | .013 |
| mt5046 | A | .029 | .013 | .017 | .019 | .020 |
| mt5147 | A | .025 | .030 | .008 | .036 | .054 |
| mt5263 | T | .019 | .007 | .004 | .004 | .010 |
| mt5390 | G | .007 | .014 | .000 | .016 | .023 |
| mt5426 | C | .011 | .022 | .000 | .020 | .026 |
| mt5460 | A | .044 | .021 | .045 | .030 | .033 |
| mt5465 | C | .000 | .000 | .000 | .001 | .000 |
| mt5495 | C | .020 | .023 | .004 | .010 | .015 |
| mt5656 | G | .081 | .013 | .012 | .014 | .038 |
| mt6045 | T | .007 | .015 | .000 | .016 | .022 |
| mt6152 | C | .007 | .014 | .000 | .016 | .022 |
| mt6221 | C | .001 | .020 | .004 | .010 | .006 |
| mt6260 | A | .007 | .019 | .020 | .012 | .006 |
| mt6365 | C | .003 | .001 | .000 | .008 | .006 |
| mt6371 | T | .001 | .022 | .004 | .012 | .007 |
| mt6719 | C | .000 | .002 | .000 | .001 | .004 |
| mt6734 | A | .003 | .011 | .008 | .007 | .009 |
| mt6776 | C | .043 | .028 | .069 | .038 | .016 |
| mt7028 | C | .460 | .431 | .322 | .419 | .404 |
| mt7476 | T | .010 | .033 | .021 | .019 | .008 |
| mt7768 | G | .073 | .025 | .020 | .038 | .049 |
| mt7864 | T | .026 | .011 | .012 | .015 | .015 |
| mt8251 | A | .040 | .036 | .149 | .060 | .049 |
| mt8269 | A | .023 | .022 | .033 | .021 | .026 |
| mt8557 | A | .016 | .008 | .033 | .010 | .009 |
| mt8616 | T | .003 | .010 | .008 | .007 | .009 |
| mt8697 | A | .074 | .112 | .159 | .088 | .123 |
| mt8705 | C | .027 | .016 | .000 | .004 | .003 |
| mt8869 | G | .019 | .007 | .004 | .004 | .010 |
| mt8994 | A | .029 | .013 | .017 | .019 | .020 |
| mt9055 | A | .066 | .069 | .094 | .099 | .044 |
| mt9093 | G | .013 | .008 | .000 | .010 | .012 |
| mt9123 | A | .010 | .019 | .000 | .014 | .014 |
| mt9150 | G | .000 | .009 | .000 | .006 | .002 |
| mt9477 | A | .161 | .107 | .041 | .076 | .113 |
| mt9612 | A | .004 | .001 | .000 | .000 | .001 |
| mt9667 | G | .023 | .012 | .000 | .010 | .015 |
| mt9698 | C | .066 | .069 | .094 | .099 | .044 |
| mt9716 | C | .012 | .012 | .021 | .023 | .007 |
| mt9899 | C | .016 | .023 | .008 | .018 | .030 |
| mt9947 | A | .003 | .011 | .008 | .007 | .009 |
| mt10034 | C | .011 | .028 | .114 | .031 | .018 |
| mt10044 | G | .001 | .013 | .000 | .007 | .009 |
| mt10084 | C | .001 | .005 | .000 | .009 | .005 |
| mt10238 | C | .011 | .027 | .114 | .032 | .018 |
| mt10398 | G | .146 | .204 | .278 | .231 | .168 |
| mt10463 | C | .074 | .112 | .159 | .088 | .123 |
| mt10550 | G | .066 | .069 | .094 | .099 | .044 |
| mt10876 | G | .007 | .014 | .000 | .016 | .023 |
| mt10915 | C | .006 | .012 | .008 | .009 | .010 |
| mt11251 | G | .156 | .226 | .250 | .212 | .231 |
| mt11299 | C | .066 | .069 | .094 | .099 | .044 |
| mt11377 | A | .024 | .036 | .004 | .023 | .020 |
| mt11467 | G | .267 | .248 | .183 | .239 | .254 |
| mt11470 | G | .005 | .002 | .000 | .014 | .004 |
| mt11485 | C | .009 | .021 | .025 | .017 | .012 |
| mt11674 | T | .029 | .011 | .012 | .020 | .019 |
| mt11719 | G | .477 | .470 | .416 | .481 | .457 |
| mt11812 | G | .055 | .081 | .114 | .064 | .083 |
| mt11840 | T | .007 | .017 | .020 | .011 | .004 |
| mt11914 | A | .048 | .014 | .004 | .041 | .022 |
| mt11947 | G | .029 | .013 | .017 | .019 | .020 |
| mt12007 | A | .014 | .007 | .049 | .012 | .016 |
| mt12308 | G | .267 | .248 | .183 | .239 | .254 |
| mt12372 | A | .267 | .248 | .183 | .239 | .255 |
| mt12414 | C | .034 | .012 | .012 | .019 | .019 |
| mt12501 | A | .018 | .028 | .135 | .050 | .042 |
| mt12612 | G | .073 | .099 | .071 | .102 | .067 |
| mt12618 | A | .058 | .012 | .012 | .013 | .022 |
| mt12633 | A | .018 | .030 | .042 | .023 | .038 |
| mt12669 | T | .018 | .001 | .004 | .000 | .000 |
| mt12705 | T | .059 | .065 | .161 | .083 | .070 |
| mt13020 | C | .012 | .016 | .004 | .017 | .028 |
| mt13105 | G | .012 | .009 | .029 | .010 | .008 |
| mt13368 | A | .074 | .112 | .159 | .088 | .123 |
| mt13617 | C | .161 | .107 | .041 | .076 | .113 |
| mt13708 | A | .081 | .119 | .087 | .123 | .114 |
| mt13734 | C | .007 | .017 | .000 | .016 | .023 |
| mt13740 | C | .007 | .017 | .020 | .011 | .004 |
| mt13780 | G | .010 | .028 | .082 | .039 | .019 |
| mt13879 | C | .015 | .010 | .033 | .010 | .014 |
| mt13934 | T | .004 | .015 | .037 | .028 | .031 |
| mt13965 | C | .007 | .006 | .000 | .005 | .007 |
| mt13966 | G | .015 | .028 | .000 | .019 | .014 |
| mt14022 | G | .000 | .002 | .000 | .000 | .000 |
| mt14167 | T | .066 | .069 | .094 | .099 | .044 |
| mt14182 | C | .073 | .025 | .029 | .040 | .049 |
| mt14233 | G | .055 | .081 | .114 | .064 | .083 |
| mt14365 | T | .010 | .018 | .000 | .013 | .013 |
| mt14470 | C | .001 | .030 | .004 | .025 | .019 |
| mt14582 | G | .010 | .018 | .000 | .013 | .013 |
| mt14687 | G | .007 | .006 | .000 | .005 | .007 |
| mt14766 | C | .477 | .470 | .416 | .480 | .456 |
| mt14793 | G | .067 | .077 | .023 | .041 | .072 |
| mt14798 | C | .093 | .127 | .100 | .176 | .119 |
| mt14905 | A | .074 | .112 | .159 | .088 | .123 |
| mt15043 | A | .010 | .040 | .082 | .047 | .035 |
| mt15218 | G | .033 | .053 | .057 | .034 | .049 |
| mt15257 | A | .010 | .034 | .021 | .022 | .014 |
| mt15452 | A | .149 | .219 | .236 | .196 | .222 |
| mt15607 | G | .074 | .112 | .159 | .088 | .123 |
| mt15758 | G | .012 | .014 | .111 | .016 | .005 |
| mt15784 | C | .003 | .003 | .008 | .012 | .016 |
| mt15833 | C | .004 | .015 | .025 | .016 | .024 |
| mt15884 | C | .036 | .034 | .012 | .032 | .022 |
| mt15904 | T | .061 | .050 | .045 | .062 | .056 |
| mt15907 | G | .007 | .014 | .000 | .016 | .023 |
| mt15924 | G | .017 | .047 | .107 | .062 | .036 |
| mt15928 | A | .074 | .112 | .159 | .088 | .123 |
| Haplogroup: | ||||||
| X | .015 | .025 | .018 | .024 | .030 | |
| I | .011 | .025 | .107 | .028 | .018 | |
| V | .061 | .050 | .045 | .061 | .056 | |
| U | .212 | .185 | .106 | .163 | .216 | |
| J | .075 | .100 | .083 | .106 | .083 | |
| W | .031 | .013 | .039 | .030 | .037 | |
| T | .074 | .112 | .159 | .088 | .123 | |
| K | .053 | .057 | .073 | .075 | .038 | |
| H | .460 | .431 | .322 | .419 | .404 | |
Results from association studies with type 2 diabetes and BMI are shown in tables 6, 7, and 8. For diabetes, only two hypotheses with Pnom<.05 were found, and these results were far from significant after permutation testing (50–1,000 iterations for each association test) (Pstudy=.55). For BMI, we found no significant association before (Pstudy=1.0) or after (Pstudy=.25) adjustment for sex and age. Tests for epistasis did not demonstrate any significant association with diabetes or BMI. Subanalysis of association with diabetes by population failed to show any strong effects in U.S., Scandinavian, and Canadian populations (tables 9 and 10). A significant association result was observed in the Polish subpopulation after permutation testing (PPolishstudy=.02 after 1,000 permutations); however, given that mtDNA variants from five populations were tested for association with diabetes, this result is consistent with noise and thus would need to be validated by replication in other large Polish samples.
Table 6. .
Results of Association Testing of All mtDNA Variants with Type 2 Diabetes and Metabolic Traits
| Phenotypea | n | Best Result | Effect Estimateb (95% CI) | Pnom | Pempiricalc |
| DM | 6,608 | mt12612 (syn ND5) | 1.25 (1.05–1.49) | .011 | >.55 (NS) |
| BMI | 6,523 | mt9123 (mis ND3) | −.04 (−.05 to −.03) | .0027 | 1.0 (NS) |
| Ins Index | 342 | mt3197 (16S rRNA); mt9477 (mis COIII); mt13617 (syn ND5) | −.14 (−.19 to −.09) | .012 | >.3 (NS) |
| HOMA-IR | 399 | mt4561 (mis ND2) | .102 (.052–.152) | .042 | >.26 (NS) |
| Cholesterol | 1,274 | mt15784 (syn cytB) | .10 (.07–.13) | .0003 | >.08 (NS) |
| Systolic BP | 2,047 | mt4561 (mis ND2) | −.045 (−.065 to −.025) | .042 | >.35 (NS) |
| Diastolic BP | 2,047 | mt3394 (tRNA S) | .022 (−.001 to .045) | .12 | >.5 (NS) |
BP = blood pressure; DM = diabetes mellitus, type 2; Ins = insulinogenic.
OR for DM and standardized regression coefficient (β) for all other phenotypes (the quantitative traits).
NS = not significant.
Table 7. .
Association Testing with Type 2 Diabetes, BMI, and Metabolic Traits[Note]
| Variable | Type 2 Diabetes | BMI | Insulinogenic Index |
HOMA-IR | Cholesterol | Systolic BP | Diastolic BP |
| n | 6,608 | 6,523 | 342 | 399 | 1,274 | 2,047 | 2,047 |
| Populations | All | All | Scandinavian/Swedish | Scandinavian/Swedish | All | All | All |
| Diabetics and controls | Yes | Yes | Controls only | Controls only | Yes | Yes | Yes |
| Log taken | … | Yes | Yes | Yes | No | No | No |
| Best result | mt12612 | mt9123 | mt3197, mt9477, mt13617 | mt4561 | mt15784 | mt4561 | mt3394 |
| Minimum P | .0106 | .0027 | .0439 | .0415 | .0003 | .04 | .1197 |
| Maximum test statistic (OR or β) | 1.25 | −.0250 | −.10 | .21 | 2.15 | −10.24 | −3.550 |
| No. of SNPs with Pnom < .05 | 3 | 26 | 3 | 2 | 13 | 1.00 | 0 |
| No. of SNPs with Pnom < .01 | 3 | 26 | 3 | 2 | 13 | 1.00 | 0 |
| Pstudy | >.55 | 1.00 | >.45 | >.26 | >.08 | >.35 | >.5 |
| Adjusted Pstudy | NA | >.25 | >.5 | >.42 | >.08 | >.1 | >.6 |
| Adjusted for | NA | Age, sex | Age, sex, BMI | Age, sex, BMI | Age, sex, BMI | Age, sex, BMI | Age, sex, BMI |
Note.— BP = blood pressure; NA = not applicable.
Table 8. .
Results of Association Testing of Each SNP and Haplogroup with Type 2 Diabetes, BMI, and Metabolic Traits[Note]
| Type 2 Diabetes | BMI | InsulinogenicIndex | HOMA-IR | Cholesterol | Systolic BP | Diastolic BP | |||||||||||
| SNP or Haplogroup | Allele | Allele Name | OR (95% CI) | Pnom | β | Pnom | β | Pnom | β | Pnom | β | Pnom | β | Pnom | β | Pnom | Percentage of Data |
| SNP: | |||||||||||||||||
| mt709 | A | 12SrRNA | .92 (.80–1.05) | .212 | −.001 | .621 | .02 | .688 | .01 | .824 | .09 | .3860 | −.91 | .509 | −.32 | .626 | 99.3 |
| mt750 | A | 12SrRNA | 1.08 (.78–1.50) | .627 | .000 | .997 | .01 | .924 | .07 | .407 | .15 | .4683 | .41 | .891 | −1.76 | .217 | 99.4 |
| mt930 | A | 12SrRNA | .88 (.69–1.12) | .288 | −.002 | .744 | −.06 | .754 | .04 | .689 | .01 | .9745 | .41 | .884 | .08 | .951 | 99.1 |
| mt1189 | C | 12SrRNA | .84 (.68–1.03) | .100 | −.002 | .561 | .07 | .412 | .02 | .734 | .09 | .4912 | .09 | .965 | .08 | .933 | 99.7 |
| mt1243 | C | 12SrRNA | 1.16 (.81–1.65) | .424 | .003 | .700 | .01 | .953 | −.05 | .481 | .12 | .6131 | .95 | .793 | 1.27 | .462 | 99.5 |
| mt1438 | A | 12SrRNA | 1.05 (.82–1.35) | .678 | −.003 | .619 | .09 | .293 | .08 | .120 | .20 | .1577 | −2.80 | .206 | −1.38 | .191 | 98.7 |
| mt1719 | A | 16SrRNA | 1.04 (.84–1.28) | .744 | −.001 | .752 | .01 | .918 | .02 | .790 | −.08 | .6530 | 1.00 | .648 | −.82 | .429 | 98.5 |
| mt1811 | G | 16SrRNA | .89 (.77–1.02) | .082 | .000 | .989 | .02 | .722 | .00 | .903 | −.09 | .3176 | −.57 | .673 | −.62 | .337 | 97.8 |
| mt1888 | A | 16SrRNA | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| mt2158 | C | 16SrRNA | 1.25 (.81–1.92) | .322 | −.004 | .633 | .23 | .093 | .06 | .506 | .26 | .4245 | −.22 | .956 | .52 | .782 | 98.7 |
| mt2706 | A | 16SrRNA | .99 (.90–1.09) | .884 | .002 | .446 | .06 | .135 | .01 | .572 | .10 | .1296 | −.26 | .789 | −.17 | .717 | 99.7 |
| mt3010 | A | 16SrRNA | .99 (.88–1.11) | .854 | −.004 | .089 | .04 | .354 | .01 | .829 | .02 | .8391 | −.54 | .639 | −.19 | .725 | 99.0 |
| mt3197 | C | 16SrRNA | 1.10 (.93–1.29) | .264 | −.004 | .252 | −.10 | .044 | −.03 | .314 | −.06 | .5465 | −.75 | .620 | −1.12 | .120 | 98.5 |
| mt3348 | G | synND1 | .37 (.10–1.34) | .131 | .039 | .097 | .04 | .915 | .16 | .522 | .05 | .9285 | 6.53 | .505 | −1.38 | .767 | 99.4 |
| mt3394 | C | misND1 | 1.29 (.77–2.18) | .336 | −.006 | .571 | .02 | .817 | .05 | .473 | −.74 | .0141 | −7.96 | .097 | −3.55 | .120 | 98.5 |
| mt3480 | G | synND1 | .84 (.69–1.01) | .059 | −.004 | .298 | .06 | .436 | .04 | .395 | .10 | .4205 | −.77 | .683 | −.17 | .845 | 98.7 |
| mt3505 | G | misND1 | 1.16 (.81–1.65) | .424 | .003 | .700 | .01 | .953 | −.05 | .481 | .12 | .6131 | .95 | .793 | 1.27 | .462 | 99.5 |
| mt3720 | G | synND1 | 1.22 (.82–1.80) | .327 | .016 | .041 | −.31 | .380 | −.20 | .422 | .09 | .8383 | −2.87 | .613 | −1.43 | .596 | 99.7 |
| mt3915 | A | synND1 | 1.05 (.76–1.45) | .763 | .016 | .013 | −.01 | .959 | .07 | .488 | .14 | .5599 | −3.00 | .333 | .58 | .694 | 98.2 |
| mt3990 | T | synND1 | 1.10 (.62–1.93) | .753 | −.001 | .941 | −.35 | .156 | −.06 | .742 | .47 | .3338 | 5.27 | .387 | −.89 | .758 | 99.1 |
| mt3992 | T | misND1 | .68 (.44–1.05) | .081 | −.025 | .004 | −.02 | .920 | −.16 | .141 | .16 | .5830 | 5.72 | .176 | 1.25 | .535 | 99.4 |
| mt4024 | G | misND1 | .68 (.44–1.05) | .081 | −.025 | .004 | −.02 | .920 | −.16 | .141 | .16 | .5830 | 5.72 | .176 | 1.25 | .535 | 99.4 |
| mt4216 | C | misND1 | .97 (.86–1.10) | .642 | .001 | .566 | −.07 | .188 | −.02 | .635 | .06 | .5276 | −.60 | .629 | .02 | .968 | 98.4 |
| mt4336 | C | tRNA Q | .87 (.64–1.19) | .384 | .001 | .873 | .02 | .847 | −.05 | .511 | −.17 | .4813 | −3.62 | .293 | −.80 | .625 | 99.5 |
| mt4529 | T | synND2 | 1.09 (.81–1.48) | .568 | .013 | .034 | −.35 | .157 | −.06 | .736 | .34 | .2124 | .35 | .902 | 1.37 | .310 | 98.7 |
| mt4561 | C | misND2 | .86 (.54–1.38) | .536 | −.007 | .484 | .03 | .837 | .21 | .042 | −.44 | .2737 | −10.24 | .042 | −2.89 | .230 | 99.1 |
| mt4580 | A | synND2 | 1.18 (.96–1.46) | .114 | .003 | .512 | .09 | .294 | −.04 | .474 | .40 | .0046 | −3.00 | .158 | −.37 | .715 | 99.5 |
| mt4639 | C | misND2 | 1.51 (.88–2.58) | .135 | −.010 | .344 | −.01 | .978 | .23 | .189 | −.70 | .0086 | .31 | .946 | .35 | .873 | 99.3 |
| mt4769 | A | synND2 | 1.05 (.82–1.35) | .678 | −.003 | .619 | .09 | .293 | .08 | .120 | .20 | .1577 | −2.80 | .206 | −1.38 | .191 | 98.7 |
| mt4793 | G | synND2 | .96 (.65–1.41) | .831 | .009 | .257 | .10 | .522 | −.13 | .207 | −.60 | .0326 | .87 | .833 | .63 | .752 | 99.2 |
| mt4917 | G | misND2 | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| mt4928 | C | synND2 | 1.51 (.43–5.30) | .522 | −.002 | .938 | −.34 | .341 | −.27 | .263 | −.59 | .3957 | 2.00 | .874 | 3.68 | .540 | 99.2 |
| mt5004 | C | synND2 | .68 (.44–1.05) | .081 | −.025 | .004 | −.02 | .920 | −.16 | .141 | .16 | .5830 | 5.72 | .176 | 1.25 | .535 | 99.4 |
| mt5046 | A | misND2 | 1.10 (.77–1.55) | .608 | .001 | .841 | .02 | .832 | −.08 | .245 | .18 | .4425 | −2.74 | .445 | .10 | .954 | 99.3 |
| mt5147 | A | synND2 | .88 (.68–1.13) | .314 | −.001 | .879 | −.06 | .763 | .04 | .684 | −.05 | .8222 | 1.76 | .568 | −.33 | .821 | 98.7 |
| mt5263 | T | misND2 | 1.51 (.88–2.58) | .135 | −.010 | .344 | −.01 | .978 | .23 | .189 | −.70 | .0086 | .31 | .946 | .35 | .873 | 99.3 |
| mt5390 | G | synND2 | 1.29 (.87–1.90) | .201 | .016 | .042 | −.31 | .379 | −.20 | .420 | .09 | .8404 | −2.86 | .613 | −1.41 | .601 | 99.4 |
| mt5426 | C | synND2 | 1.08 (.76–1.52) | .673 | .012 | .087 | −.15 | .453 | −.06 | .617 | .05 | .8639 | −4.80 | .257 | −1.34 | .508 | 99.7 |
| mt5460 | A | misND2 | 1.20 (.91–1.59) | .187 | .001 | .917 | .09 | .277 | −.01 | .842 | .17 | .3751 | .43 | .873 | .95 | .461 | 99.0 |
| mt5465 | C | synND2 | 1.01 (.06–16.09) | .997 | .111 | .045 | 99.3 | ||||||||||
| mt5495 | C | synND2 | 1.09 (.72–1.64) | .683 | −.009 | .274 | .05 | .736 | −.07 | .425 | −.24 | .3009 | 1.61 | .652 | .92 | .590 | 98.8 |
| mt5656 | G | ncnc | .89 (.67–1.18) | .426 | −.003 | .561 | −.13 | .062 | −.06 | .177 | −.09 | .5272 | .85 | .732 | .53 | .654 | 99.4 |
| mt6045 | T | synCO1 | 1.17 (.79–1.72) | .437 | .017 | .033 | −.31 | .380 | −.20 | .422 | .09 | .8383 | −2.04 | .710 | −.36 | .889 | 99.7 |
| mt6152 | C | synCO1 | 1.19 (.81–1.76) | .380 | .017 | .030 | −.31 | .380 | −.20 | .422 | .09 | .8383 | −2.87 | .613 | −1.43 | .596 | 99.7 |
| mt6221 | C | synCO1 | 1.22 (.74–2.04) | .437 | .003 | .740 | .39 | .260 | −.01 | .973 | −.05 | .9119 | −2.97 | .526 | 1.16 | .603 | 99.3 |
| mt6260 | A | synCO1 | .80 (.50–1.27) | .341 | −.001 | .908 | .11 | .542 | .04 | .758 | .00 | .9962 | 3.23 | .422 | −.02 | .991 | 98.7 |
| mt6365 | C | synCO1 | .94 (.49–1.83) | .863 | −.005 | .708 | .09 | .712 | −.09 | .589 | −.62 | .2985 | −14.10 | .196 | −2.14 | .681 | 99.3 |
| mt6371 | T | synCO1 | 1.16 (.72–1.88) | .547 | .002 | .832 | .39 | .263 | −.01 | .972 | −.05 | .9128 | −1.92 | .668 | 1.91 | .372 | 98.9 |
| mt6719 | C | synCO1 | 1.60 (.53–4.84) | .407 | −.025 | .251 | .76 | .5213 | −11.58 | .452 | −4.65 | .527 | 99.3 | ||||
| mt6734 | A | synCO1 | 1.17 (.67–2.05) | .575 | −.003 | .806 | −.35 | .155 | −.06 | .736 | .11 | .8173 | 2.63 | .643 | −1.13 | .676 | 99.7 |
| mt6776 | C | synCO1 | 1.09 (.82–1.43) | .557 | .004 | .438 | −.04 | .598 | .03 | .583 | .20 | .2676 | 1.76 | .487 | −1.25 | .302 | 99.5 |
| mt7028 | C | synCO1 | .99 (.90–1.09) | .884 | .002 | .446 | .06 | .135 | .01 | .572 | .10 | .1296 | −.26 | .789 | −.17 | .717 | 99.7 |
| mt7476 | T | tRNA S | .98 (.67–1.44) | .915 | .007 | .393 | .31 | .126 | .08 | .555 | −.39 | .0993 | .86 | .793 | −1.90 | .222 | 98.9 |
| mt7768 | G | synCOII | .87 (.69–1.11) | .267 | .007 | .141 | −.14 | .054 | −.02 | .691 | .25 | .0985 | 2.15 | .372 | −.93 | .418 | 98.0 |
| mt7864 | T | synCOII | 1.14 (.78–1.69) | .497 | −.003 | .663 | .02 | .832 | −.10 | .153 | .06 | .8159 | −3.99 | .299 | −1.18 | .517 | 99.3 |
| mt8251 | A | synCOII | 1.18 (.95–1.47) | .134 | −.003 | .437 | −.07 | .531 | .01 | .857 | −.29 | .1104 | 2.21 | .332 | −.51 | .641 | 96.6 |
| mt8269 | A | nc | 1.02 (.74–1.40) | .911 | −.016 | .012 | .04 | .716 | −.07 | .332 | .23 | .3098 | 1.69 | .589 | .49 | .745 | 99.2 |
| mt8557 | A | synATP8; misATP6 | 1.29 (.81–2.05) | .281 | −.016 | .094 | .22 | .095 | .06 | .518 | .23 | .4652 | −1.87 | .646 | −.80 | .680 | 99.1 |
| mt8616 | T | misATP6 | 1.28 (.73–2.25) | .381 | .003 | .797 | −.35 | .156 | −.06 | .742 | .47 | .3338 | 3.89 | .507 | −1.42 | .610 | 99.2 |
| mt8697 | A | synATP6 | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| mt8705 | C | misATP6 | 1.04 (.61–1.76) | .896 | −.016 | .122 | .01 | .896 | −.06 | .396 | −.18 | .4415 | 3.75 | .277 | −.65 | .694 | 99.5 |
| mt8869 | G | misATP6 | 1.51 (.88–2.58) | .135 | −.010 | .344 | −.01 | .978 | .23 | .189 | −.70 | .0086 | .31 | .946 | .35 | .873 | 99.3 |
| mt8994 | A | synATP6 | 1.10 (.77–1.55) | .608 | .001 | .841 | .02 | .832 | −.08 | .245 | .18 | .4425 | −2.74 | .445 | .10 | .954 | 99.3 |
| mt9055 | A | misATP6 | .84 (.69–1.01) | .059 | −.004 | .298 | .06 | .436 | .04 | .395 | .10 | .4205 | −.77 | .683 | −.17 | .845 | 98.7 |
| mt9093 | G | synATP6 | 1.52 (.93–2.47) | .091 | .001 | .892 | −.01 | .955 | −.01 | .933 | −.27 | .3518 | 1.86 | .706 | 2.51 | .285 | 99.3 |
| mt9123 | A | synATP6 | .76 (.50–1.15) | .194 | −.025 | .003 | −.02 | .928 | −.16 | .146 | .16 | .5823 | 5.72 | .178 | 1.25 | .537 | 99.0 |
| mt9150 | G | synATP6 | 2.38 (1.07–5.32) | .034 | .030 | .048 | .53 | .3708 | .15 | .985 | 1.49 | .687 | 99.5 | ||||
| mt9477 | A | misCOIII | 1.10 (.93–1.29) | .264 | −.004 | .252 | −.10 | .044 | −.03 | .314 | −.06 | .5465 | −.75 | .620 | −1.12 | .120 | 98.5 |
| mt9612 | A | misCOIII | 2.51 (.52–12.26) | .254 | −.041 | .246 | −.34 | .341 | −.27 | .263 | −.59 | .3957 | 2.00 | .874 | 3.68 | .540 | 99.2 |
| mt9667 | G | misCOIII | 1.27 (.83–1.94) | .280 | −.001 | .929 | −.09 | .474 | −.04 | .687 | .55 | .0268 | −1.50 | .709 | 1.29 | .500 | 99.6 |
| mt9698 | C | synCOIII | .84 (.69–1.01) | .059 | −.004 | .298 | .06 | .436 | .04 | .395 | .10 | .4205 | −.77 | .683 | −.17 | .845 | 98.7 |
| mt9716 | C | synCOIII | .80 (.54–1.20) | .290 | −.008 | .320 | .01 | .936 | .09 | .284 | .12 | .7185 | −4.65 | .290 | −1.34 | .524 | 98.8 |
| mt9899 | C | synCOIII | 1.04 (.74–1.45) | .826 | −.002 | .765 | .17 | .214 | .02 | .798 | .26 | .3337 | 1.25 | .726 | −.53 | .755 | 98.7 |
| mt9947 | A | misCOIII | 1.17 (.67–2.05) | .575 | −.003 | .806 | −.35 | .155 | −.06 | .736 | .11 | .8173 | 2.63 | .643 | −1.13 | .676 | 99.7 |
| mt10034 | C | tRNA G | 1.08 (.80–1.46) | .602 | .015 | .013 | −.35 | .155 | −.06 | .736 | .36 | .1523 | .30 | .915 | 1.15 | .389 | 99.7 |
| mt10044 | G | tRNA G | .75 (.43–1.32) | .322 | −.025 | .030 | .00 | .16 | .6998 | 9.15 | .118 | 2.10 | .453 | 99.0 | |||
| mt10084 | C | misND3 | .61 (.31–1.17) | .138 | −.005 | .705 | .15 | .669 | .02 | .938 | −.41 | .5478 | 7.00 | .433 | 3.62 | .396 | 99.5 |
| mt10238 | C | synND3 | 1.10 (.82–1.49) | .523 | .017 | .005 | −.35 | .156 | −.06 | .739 | .36 | .1620 | .35 | .901 | 1.18 | .375 | 99.0 |
| mt10398 | G | misND3 | 1.04 (.92–1.18) | .501 | .002 | .464 | .05 | .321 | .02 | .588 | −.05 | .5965 | .80 | .519 | .30 | .617 | 98.6 |
| mt10463 | C | tRNA R | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| mt10550 | G | synND4L | .84 (.69–1.01) | .059 | −.004 | .298 | .06 | .436 | .04 | .395 | .10 | .4205 | −.77 | .683 | −.17 | .845 | 98.7 |
| mt10876 | G | synND4 | 1.29 (.87–1.90) | .201 | .016 | .042 | −.31 | .379 | −.20 | .420 | .09 | .8404 | −2.86 | .613 | −1.41 | .601 | 99.4 |
| mt10915 | C | synND4 | 1.15 (.69–1.92) | .586 | −.004 | .708 | −.29 | .151 | .00 | .975 | .57 | .1545 | 6.83 | .187 | −1.02 | .679 | 99.2 |
| mt11251 | G | synND4 | 1.02 (.91–1.15) | .740 | −.001 | .668 | .05 | .308 | .00 | .928 | −.05 | .5786 | .13 | .915 | −.01 | .988 | 96.7 |
| mt11299 | C | synND4 | .84 (.69–1.01) | .059 | −.004 | .298 | .06 | .436 | .04 | .395 | .10 | .4205 | −.77 | .683 | −.17 | .845 | 98.7 |
| mt11377 | A | synND4 | 1.09 (.79–1.50) | .589 | −.003 | .660 | .13 | .272 | .04 | .638 | −.44 | .0188 | 1.52 | .603 | .04 | .978 | 99.3 |
| mt11467 | G | synND4 | .94 (.84–1.06) | .314 | .001 | .773 | .08 | .050 | .02 | .516 | .00 | .9659 | .50 | .659 | .18 | .739 | 99.6 |
| mt11470 | G | synND4 | .75 (.42–1.32) | .314 | .012 | .277 | −.07 | .785 | −.13 | .354 | .09 | .8644 | 2.33 | .779 | −.54 | .891 | 99.7 |
| mt11485 | C | synND4 | 1.03 (.69–1.53) | .893 | −.005 | .505 | .11 | .539 | .04 | .760 | −.07 | .7835 | 1.58 | .679 | −1.05 | .564 | 99.2 |
| mt11674 | T | synND4 | 1.16 (.81–1.65) | .424 | .003 | .700 | .01 | .953 | −.05 | .481 | .12 | .6131 | .95 | .793 | 1.27 | .462 | 99.5 |
| mt11719 | G | synND4 | 1.04 (.95–1.15) | .399 | .001 | .592 | .03 | .463 | .02 | .485 | .02 | .7837 | .55 | .574 | .00 | .996 | 99.6 |
| mt11812 | G | synND4 | .87 (.73–1.05) | .158 | −.003 | .417 | −.03 | .760 | −.04 | .497 | .09 | .4970 | −1.49 | .416 | −.53 | .541 | 99.5 |
| mt11840 | T | synND4 | .88 (.53–1.45) | .606 | .002 | .830 | .11 | .543 | .04 | .764 | −.20 | .4944 | 2.51 | .547 | −.70 | .725 | 99.7 |
| mt11914 | A | synND4 | .91 (.69–1.21) | .511 | .000 | .999 | −.03 | .737 | .03 | .603 | −.06 | .7572 | −.76 | .803 | 1.42 | .328 | 99.2 |
| mt11947 | G | synND4 | 1.10 (.77–1.55) | .608 | .001 | .841 | .02 | .832 | −.08 | .245 | .18 | .4425 | −2.74 | .445 | .10 | .954 | 99.3 |
| mt12007 | A | synND4 | 1.43 (.94–2.16) | .095 | −.005 | .522 | .21 | .152 | .04 | .678 | .35 | .2934 | −2.22 | .576 | −.12 | .947 | 99.2 |
| mt12308 | G | tRNA L2 | .94 (.84–1.06) | .314 | .001 | .773 | .08 | .050 | .02 | .516 | .00 | .9659 | .50 | .659 | .18 | .739 | 99.6 |
| mt12372 | A | synND5 | .94 (.84–1.05) | .300 | −.001 | .759 | −.08 | .050 | −.02 | .516 | .00 | .9659 | −.50 | .659 | −.18 | .739 | 99.6 |
| mt12414 | C | synND5 | 1.15 (.81–1.62) | .435 | .005 | .461 | −.04 | .696 | −.09 | .149 | .04 | .8653 | −.68 | .843 | 1.23 | .454 | 99.4 |
| mt12501 | A | synND5 | 1.01 (.80–1.28) | .928 | .004 | .408 | −.32 | .110 | −.03 | .854 | .37 | .1120 | −1.50 | .564 | .37 | .764 | 98.8 |
| mt12612 | G | synND5 | 1.25 (1.05–1.49) | .011 | −.003 | .441 | −.08 | .229 | −.04 | .337 | .21 | .0900 | −2.51 | .149 | −.19 | .819 | 98.9 |
| mt12618 | A | synND5 | .97 (.70–1.35) | .862 | .005 | .443 | −.13 | .122 | −.03 | .547 | −.06 | .7141 | 2.21 | .440 | .20 | .886 | 99.1 |
| mt12633 | A | synND5 | .94 (.71–1.27) | .703 | .001 | .815 | .16 | .228 | .02 | .846 | .24 | .3547 | 2.52 | .409 | 1.18 | .415 | 97.9 |
| mt12669 | T | synND5 | 1.57 (.61–4.02) | .347 | −.013 | .510 | .00 | .996 | −.02 | .847 | −.04 | .9000 | 1.62 | .767 | 2.51 | .337 | 98.8 |
| mt12705 | T | synND5 | 1.08 (.9–1.30) | .384 | .002 | .513 | .01 | .949 | −.06 | .272 | .11 | .4386 | −1.69 | .361 | .61 | .492 | 99.7 |
| mt13020 | C | synND5 | 1.18 (.83–1.67) | .369 | .009 | .224 | −.20 | .325 | −.13 | .276 | −.26 | .4577 | −1.10 | .811 | .23 | .916 | 99.5 |
| mt13105 | G | misND5 | 1.06 (.65–1.72) | .812 | −.005 | .610 | .04 | .841 | .29 | .042 | −.26 | .3799 | 1.36 | .748 | .96 | .636 | 98.8 |
| mt13368 | A | synND5 | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| mt13617 | C | synND5 | 1.10 (.93–1.29) | .264 | −.004 | .252 | −.10 | .044 | −.03 | .314 | −.06 | .5465 | −.75 | .620 | −1.12 | .120 | 98.5 |
| mt13708 | A | misND5 | 1.15 (.98–1.34) | .079 | .001 | .859 | .06 | .340 | .02 | .609 | −.21 | .0768 | 1.54 | .342 | .27 | .727 | 99.0 |
| mt13734 | C | synND5 | 1.18 (.80–1.73) | .399 | .016 | .047 | −.31 | .375 | −.20 | .418 | .08 | .8423 | −1.24 | .816 | −1.49 | .559 | 98.4 |
| mt13740 | C | synND5 | .88 (.53–1.45) | .606 | .002 | .830 | .11 | .543 | .04 | .764 | −.20 | .4944 | 2.51 | .547 | −.70 | .725 | 99.7 |
| mt13780 | G | misND5 | 1.22 (.91–1.62) | .187 | .013 | .025 | −.35 | .155 | −.06 | .736 | .36 | .1937 | .13 | .966 | 1.29 | .376 | 99.4 |
| mt13879 | C | misND5 | 1.27 (.83–1.96) | .272 | −.003 | .768 | .22 | .095 | .06 | .518 | .25 | .4270 | −.26 | .947 | .51 | .788 | 99.5 |
| mt13934 | T | misND5 | 1.00 (.73–1.37) | .988 | .010 | .132 | −.08 | .815 | .11 | .521 | .33 | .4052 | −1.80 | .670 | 1.92 | .341 | 99.0 |
| mt13965 | C | synND5 | .73 (.38–1.38) | .333 | −.006 | .659 | −.13 | .457 | −.11 | .323 | 1.21 | .0076 | −2.09 | .741 | −.60 | .843 | 97.3 |
| mt13966 | G | misND5 | .90 (.63–1.3) | .574 | −.007 | .365 | .17 | .283 | −.08 | .405 | −.24 | .3534 | .32 | .926 | 1.40 | .395 | 99.5 |
| mt14022 | G | synND5 | 99.3 | ||||||||||||||
| mt14167 | T | synND6 | .84 (.69–1.01) | .059 | −.004 | .298 | .06 | .436 | .04 | .395 | .10 | .4205 | −.77 | .683 | −.17 | .845 | 98.7 |
| mt14182 | C | synND6 | .88 (.69–1.11) | .279 | .007 | .144 | −.14 | .054 | −.02 | .691 | −.25 | .0985 | 1.77 | .456 | −1.05 | .354 | 98.2 |
| mt14233 | G | synND6 | .87 (.73–1.05) | .158 | −.003 | .417 | −.03 | .760 | −.04 | .497 | .09 | .4970 | −1.49 | .416 | −.53 | .541 | 99.5 |
| mt14365 | T | synND6 | .68 (.44–1.05) | .081 | −.025 | .004 | −.02 | .920 | −.16 | .141 | .16 | .5830 | 5.72 | .176 | 1.25 | .535 | 99.4 |
| mt14470 | C | synND6 | 1.28 (.90–1.81) | .170 | .005 | .488 | .40 | .252 | −.01 | .984 | −.33 | .3817 | 2.31 | .558 | 1.83 | .327 | 98.2 |
| mt14582 | G | misND6 | .68 (.44–1.05) | .081 | −.025 | .004 | −.02 | .920 | −.16 | .141 | .16 | .5830 | 5.72 | .176 | 1.25 | .535 | 99.4 |
| mt14687 | G | tRNA E | .73 (.38–1.38) | .333 | −.006 | .659 | −.13 | .457 | −.11 | .323 | 1.21 | .0076 | −2.09 | .741 | −.60 | .843 | 97.3 |
| mt14766 | C | misCyt B | 1.04 (.95–1.15) | .405 | −.001 | .624 | −.03 | .463 | −.02 | .485 | −.02 | .7837 | −.55 | .574 | .00 | .996 | 99.4 |
| mt14793 | G | misCyt B | 1.16 (.94–1.42) | .174 | −.003 | .489 | −.11 | .211 | .06 | .260 | −.06 | .6519 | 1.38 | .498 | .58 | .544 | 96.8 |
| mt14798 | C | misCyt B | .93 (.81–1.07) | .296 | −.002 | .591 | .05 | .371 | .04 | .274 | .03 | .8101 | 1.13 | .473 | .59 | .427 | 99.2 |
| mt14905 | A | synCyt B | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| mt15043 | A | synCyt B | 1.03 (.80–1.32) | .846 | .013 | .012 | −.35 | .155 | −.06 | .749 | .28 | .2715 | −1.49 | .591 | .67 | .613 | 98.5 |
| mt15218 | G | misCyt B | 1.16 (.91–1.48) | .232 | −.007 | .133 | −.15 | .137 | −.09 | .223 | .11 | .5128 | 2.70 | .251 | .99 | .380 | 98.8 |
| mt15257 | A | misCyt B | .87 (.61–1.23) | .428 | .006 | .393 | .31 | .126 | .09 | .549 | −.45 | .0515 | .39 | .903 | −1.94 | .207 | 99.5 |
| mt15452 | A | misCyt B | .96 (.85–1.09) | .538 | .002 | .497 | −.07 | .156 | −.02 | .569 | .06 | .4695 | −.60 | .625 | .00 | .995 | 99.2 |
| mt15607 | G | synCyt B | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| mt15758 | G | misCyt B | .90 (.61–1.34) | .603 | .017 | .030 | −.13 | .608 | .13 | .450 | −.02 | .9565 | −2.87 | .359 | 2.26 | .130 | 98.9 |
| mt15784 | C | synCyt B | .94 (.58–1.51) | .795 | −.001 | .937 | .18 | .459 | 2.15 | .0003 | 10.48 | .176 | 1.97 | .594 | 99.6 | ||
| mt15833 | C | synCyt B | 1.14 (.78–1.65) | .508 | .003 | .696 | .20 | .430 | −.30 | .086 | .23 | .5210 | .58 | .898 | 1.76 | .411 | 99.5 |
| mt15884 | C | misCyt B | 1.00 (.74–1.34) | .987 | .002 | .748 | −.01 | .902 | −.08 | .174 | .04 | .8212 | 1.96 | .473 | .88 | .498 | 94.3 |
| mt15904 | T | tRNA T | 1.18 (.96–1.45) | .122 | .003 | .513 | .09 | .295 | −.04 | .481 | .40 | .0047 | −3.02 | .155 | −.38 | .707 | 99.0 |
| mt15907 | G | tRNA T | 1.29 (.87–1.90) | .201 | .016 | .042 | −.31 | .379 | −.20 | .420 | .09 | .8404 | −2.86 | .613 | −1.41 | .601 | 99.4 |
| mt15924 | G | tRNA T | 1.00 (.79–1.26) | .989 | .011 | .017 | −.11 | .470 | −.09 | .351 | −.08 | .6910 | .88 | .717 | .77 | .507 | 98.6 |
| mt15928 | A | tRNA T | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 |
| Haplogroup: | |||||||||||||||||
| X | .88 (.64–1.20) | .410 | −.010 | .098 | .10 | .480 | −.01 | .921 | −.20 | .4564 | −1.14 | .736 | 1.43 | .373 | 98.8 | ||
| I | 1.07 (.78–1.46) | .677 | .015 | .016 | −.35 | .156 | −.06 | .736 | .34 | .2121 | .63 | .828 | 1.75 | .208 | 98.8 | ||
| V | 1.18 (.96–1.46) | .114 | .003 | .512 | .09 | .294 | −.04 | .474 | .40 | .0046 | −3.00 | .158 | −.37 | .715 | 99.5 | ||
| U | .99 (.87–1.12) | .857 | .000 | .989 | .12 | .010 | .03 | .369 | .05 | .5499 | .74 | .558 | .24 | .694 | 99.3 | ||
| J | 1.23 (1.04–1.45) | .017 | −.003 | .452 | −.06 | .379 | −.03 | .500 | .21 | .0822 | −2.02 | .241 | −.25 | .763 | 98.9 | ||
| W | .98 (.74–1.30) | .882 | −.003 | .589 | .00 | .973 | −.07 | .274 | .25 | .2813 | −3.03 | .353 | .06 | .969 | 99.4 | ||
| T | .89 (.76–1.05) | .160 | −.002 | .560 | .04 | .639 | −.03 | .585 | .13 | .2933 | −.49 | .761 | −.07 | .923 | 97.5 | ||
| K | .84 (.68–1.03) | .100 | −.002 | .564 | .07 | .412 | .02 | .734 | .09 | .4912 | .09 | .965 | .08 | .933 | 99.7 | ||
| H | .99 (.90–1`.09) | .884 | .002 | .446 | .06 | .135 | .01 | .572 | .10 | .1296 | −.26 | .789 | −.17 | .717 | 99.7 | ||
Note.— Some SNPs were monomorphic for subsets of individuals used for analysis; hence, values for those SNPs are missing. Standardized regression coefficients (β) for best results are listed in table 6. BP = blood pressure.
Table 9. .
Results of Association Testing of All mtDNA Variants with Type 2 Diabetes, by Population
| Sample | n | Best Result | OR (95% CI) | Pnom | Pempiricala |
| Scandinavian | 918 | mt8869 (misATP6); mt5263 (misND2); mt4639 (misND2) | 7.67 (2.18–27.04) | .0015 | >.5 (NS) |
| Swedish | 1,010 | mt8251 (synCOII) | 2.56 (1.25–5.26) | .0105 | >.5 (NS) |
| Canadian | 246 | mt15218 (misCytB) | 3.93 (1.16–13.28) | .0277 | 1 (NS) |
| U.S.b | 2,428 | mt5495 (synND2) | 3.60 (1.42–9.12) | .0070 | >.3 (NS) |
| Polandb | 2,006 | mt12612 (synND5) | 2.07 (1.44–2.98) | .000085 | .02 |
NS = not significant.
Sample from Genomics Collaborative Inc.
Table 10. .
Results of Association Testing of Each SNP and Haplogroup with Type 2 Diabetes, by Population[Note]
| Scandinavia | Sweden | Canada | United States | Poland | |||||||
| SNP or Haplogroup | Allele | OR (95% CI) | Pnom | OR (95% CI) | Pnom | OR (95% CI) | Pnom | OR (95% CI) | Pnom | OR (95% CI) | Pnom |
| SNP: | |||||||||||
| mt709 | A | .95 (.64–1.39) | .77788 | 1.04 (.73–1.48) | .8202 | .71 (.38–1.34) | .2872 | .92 (.73–1.17) | .5037 | .87 (.69–1.11) | .27722 |
| mt750 | A | 1.73 (.79–3.79) | .16969 | 1.00 (2.12–0.47) | .9957 | 1.02 (.14–7.34) | .9868 | .93 (.57–1.54) | .7896 | 1.08 (.49–2.38) | .85038 |
| mt930 | A | 1.69 (.74–3.87) | .21450 | .41 (.89–0.19) | .0242 | .70 (.22–2.24) | .5437 | 1.17 (.77–1.76) | .4637 | .74 (.51–1.09) | .12453 |
| mt1189 | C | .96 (.54–1.71) | .88946 | .76 (1.31–0.45) | .3280 | .61 (.23–1.61) | .3185 | .87 (.64–1.17) | .3550 | .83 (.52–1.30) | .41337 |
| mt1243 | C | 1.00 (.46–2.19) | .99543 | 4.53 (1.11–18.42) | .0349 | 2.00 (.19–21.34) | .5661 | .84 (.48–1.50) | .5617 | 1.31 (.68–2.53) | .41544 |
| mt1438 | A | 1.07 (.64–1.81) | .78927 | .96 (1.74–0.53) | .8896 | 1.01 (.14–7.28) | .9933 | 1.12 (.74–1.69) | .5998 | 1.01 (.59–1.74) | .96710 |
| mt1719 | A | 2.56 (1.15–5.72) | .02158 | 1.35 (.77–2.38) | .2903 | 1.59 (.73–3.45) | .2413 | .80 (.57–1.11) | .1845 | .92 (.61–1.38) | .68233 |
| mt1811 | G | .74 (.48–1.13) | .16548 | .76 (1.04–0.56) | .0864 | .69 (.33–1.43) | .3144 | .96 (.77–1.19) | .7146 | .97 (.75–1.25) | .80347 |
| mt1888 | A | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| mt2158 | C | .39 (.13–1.20) | .10167 | .67 (2.36–0.19) | .5313 | 1.72 (.41–7.26) | .4577 | 1.30 (.57–2.97) | .5349 | 2.53 (1.14–5.62) | .02222 |
| mt2706 | A | 1.00 (.77–1.30) | .97143 | 1.00 (.78–1.29) | .9710 | .78 (.46–1.33) | .3614 | .97 (.82–1.13) | .6678 | 1.04 (.87–1.25) | .64907 |
| mt3010 | A | .76 (.57–1.02) | .06835 | 1.33 (.99–1.80) | .0584 | .68 (.33–1.39) | .2904 | 1.03 (.85–1.24) | .7900 | .96 (.78–1.19) | .71873 |
| mt3197 | C | .94 (.66–1.33) | .70875 | 1.25 (.83–1.89) | .2942 | 1.55 (.43–5.59) | .5014 | 1.35 (1.00–1.83) | .0494 | .94 (.71–1.24) | .66619 |
| mt3348 | G | 1.00 (.06–16.11) | .99752 | .50 (5.28–0.05) | .5643 | .25 (.03–1.88) | .1775 | ||||
| mt3394 | C | .46 (.16–1.30) | .14431 | 6.01 (.93–38.79) | .0593 | 1.31 (.57–2.98) | .5257 | 2.38 (.64–8.85) | .19679 | ||
| mt3480 | G | .70 (.41–1.19) | .18747 | .85 (1.39–0.52) | .5269 | .61 (.26–1.47) | .2733 | .85 (.65–1.11) | .2230 | .95 (.62–1.45) | .80851 |
| mt3505 | G | 1.00 (.46–2.19) | .99543 | 4.53 (1.11–18.42) | .0349 | 2.00 (.19–21.34) | .5661 | .84 (.48–1.50) | .5617 | 1.31 (.68–2.53) | .41544 |
| mt3720 | G | .99 (.20–4.95) | .99357 | 2.50 (.81–7.72) | .1113 | 1.47 (.76–2.84) | .2472 | .87 (.48–1.58) | .64860 | ||
| mt3915 | A | .57 (.17–1.92) | .36280 | 1.13 (.57–2.24) | .7270 | .50 (.09–2.70) | .4200 | 1.40 (.83–2.34) | .2072 | .85 (.45–1.61) | .62083 |
| mt3990 | T | .51 (.05–5.43) | .57874 | 1.51 (.43–5.32) | .5250 | 1.13 (.43–2.92) | .8090 | .90 (.34–2.33) | .82373 | ||
| mt3992 | T | .80 (.21–2.98) | .73763 | 1.00 (2.53–0.39) | .9932 | .68 (.34–1.38) | .2824 | .49 (.22–1.09) | .08073 | ||
| mt4024 | G | .80 (.21–2.98) | .73763 | 1.00 (2.53–0.39) | .9932 | .68 (.34–1.38) | .2824 | .49 (.22–1.09) | .08073 | ||
| mt4216 | C | .74 (.51–1.07) | .11273 | .99 (1.33–0.73) | .9280 | .63 (.34–1.14) | .1273 | 1.14 (.93–1.40) | .1960 | .93 (.76–1.15) | .52925 |
| mt4336 | C | .58 (.23–1.46) | .24638 | 1.45 (.55–3.81) | .4554 | 2.03 (.38–10.95) | .4084 | .64 (.37–1.09) | .1008 | 1.03 (.63–1.68) | .89781 |
| mt4529 | T | 4.04 (.96–17.01) | .05732 | 1.07 (.49–2.38) | .8594 | 1.63 (.74–3.63) | .2284 | .76 (.48–1.21) | .2515 | 1.26 (.65–2.45) | .49148 |
| mt4561 | C | .12 (.02–0.71) | .01937 | 2.46 (.50–12.1) | .2664 | .98 (.14–7.10) | .9866 | .82 (.45–1.51) | .5282 | 2.51 (.51–12.27) | .25489 |
| mt4580 | A | 2.20 (1.25–3.87) | .00611 | 1.52 (.86–2.71) | .1529 | .84 (.25–2.83) | .7790 | 1.05 (.75–1.46) | .7945 | .96 (.66–1.41) | .83729 |
| mt4639 | C | 7.67 (2.18–27.04) | .00153 | .40 (1.96–0.08) | .2597 | .66 (.19–2.34) | .5228 | 1.23 (.51–2.98) | .64660 | ||
| mt4769 | A | 1.07 (.64–1.81) | .78927 | .96 (1.74–0.53) | .8896 | 1.01 (.14–7.28) | .9933 | 1.12 (.74–1.69) | .5998 | 1.01 (.59–1.74) | .96710 |
| mt4793 | G | 1.18 (.39–3.53) | .76772 | .55 (1.63–0.19) | .2800 | 1.03 (.06–16.58) | .9860 | .99 (.56–1.73) | .9596 | 1.08 (.50–2.30) | .84636 |
| mt4917 | G | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| mt4928 | C | 3.10 (.36–26.62) | .30349 | .67 (.11–3.97) | .65919 | ||||||
| mt5004 | C | .80 (.21–2.98) | .73763 | 1.00 (2.53–0.39) | .9932 | .68 (.34–1.38) | .2824 | .49 (.22–1.09) | .08073 | ||
| mt5046 | A | 1.00 (.46–2.19) | .99095 | 3.31 (.97–11.25) | .0556 | 2.97 (.34–26.13) | .3254 | .67 (.37–1.21) | .1830 | 1.37 (.73–2.57) | .33308 |
| mt5147 | A | 1.57 (.68–3.65) | .29093 | .36 (.80–0.16) | .0116 | 1.24 (.81–1.90) | .3296 | .76 (.52–1.12) | .16931 | ||
| mt5263 | T | 7.67 (2.18–27.04) | .00153 | .40 (1.96–0.08) | .2597 | .66 (.19–2.34) | .5228 | 1.23 (.51–2.98) | .64660 | ||
| mt5390 | G | 1.00 (.20–5.00) | .99571 | 2.53 (.82–7.79) | .1072 | 1.61 (.84–3.06) | .1492 | .92 (.51–1.65) | .77333 | ||
| mt5426 | C | .66 (.19–2.33) | .51804 | 1.75 (.74–4.17) | .2041 | 1.34 (.76–2.37) | .3124 | .79 (.45–1.37) | .39715 | ||
| mt5460 | A | .73 (.38–1.38) | .33182 | 1.64 (.68–3.95) | .2742 | 1.80 (.52–6.20) | .3546 | 1.00 (.63–1.60) | .9943 | 1.73 (1.04–2.86) | .03442 |
| mt5465 | C | 1.00 (.06–15.98) | .9991 | ||||||||
| mt5495 | C | 1.26 (.49–3.21) | .63062 | .78 (1.78–0.34) | .5495 | 3.60 (1.42–9.12) | .0070 | .52 (.24–1.12) | .09332 | ||
| mt5656 | G | .89 (.55–1.43) | .62761 | .44 (1.40–0.14) | .1650 | 2.02 (.19–21.5) | .5613 | .94 (.47–1.86) | .8515 | .95 (.60–1.50) | .81851 |
| mt6045 | T | .99 (.20–4.95) | .99357 | 2.00 (.69–5.77) | .2001 | 1.38 (.72–2.64) | .3266 | .87 (.48–1.58) | .64860 | ||
| mt6152 | C | .99 (.20–4.95) | .99357 | 2.50 (.81–7.72) | .1113 | 1.38 (.72–2.64) | .3266 | .87 (.48–1.58) | .64860 | ||
| mt6221 | C | 1.51 (.61–3.69) | .3711 | .92 (.42–2.02) | .8309 | 1.60 (.53–4.86) | .40589 | ||||
| mt6260 | A | .50 (.09–2.64) | .41267 | .89 (2.20–0.36) | .7960 | .66 (.11–3.98) | .6514 | .87 (.42–1.79) | .7082 | .72 (.23–2.26) | .56962 |
| mt6365 | C | .50 (.05–5.27) | .56306 | 1.22 (.50–2.94) | .6641 | .84 (.25–2.75) | .76877 | ||||
| mt6371 | T | 1.19 (.51–2.78) | .6872 | 1.00 (.48–2.05) | .9909 | 1.60 (.53–4.84) | .40999 | ||||
| mt6719 | C | 2.00 (.19–21.04) | .5649 | ||||||||
| mt6734 | A | .50 (.05–5.27) | .56306 | 1.20 (.36–3.95) | .7643 | 1.28 (.47–3.43) | .6288 | 1.01 (.40–2.55) | .98979 | ||
| mt6776 | C | .61 (.32–1.17) | .13893 | .86 (1.82–0.40) | .6896 | 1.48 (.55–4.00) | .4403 | 1.21 (.80–1.85) | .3696 | 1.68 (.82–3.42) | .15645 |
| mt7028 | C | 1.00 (.77–1.30) | .97143 | 1.00 (.78–1.29) | .9710 | .78 (.46–1.33) | .3614 | .97 (.82–1.13) | .6678 | 1.04 (.87–1.25) | .64907 |
| mt7476 | T | 1.26 (.34–4.69) | .73514 | .94 (1.89–0.47) | .8687 | .24 (.03–1.84) | .1693 | .95 (.52–1.71) | .8560 | 1.51 (.54–4.23) | .43224 |
| mt7768 | G | .94 (.56–1.56) | .80245 | .46 (1.05–0.20) | .0638 | .67 (.11–4.01) | .6580 | 1.12 (.74–1.71) | .5869 | .79 (.52–1.19) | .25418 |
| mt7864 | T | 1.01 (.45–2.27) | .98696 | 4.46 (1.09–18.21) | .0370 | .63 (.32–1.23) | .1775 | 1.43 (.68–2.99) | .34545 | ||
| mt8251 | A | 1.99 (.99–4.00) | .05416 | 2.56 (1.25–5.26) | .0105 | 1.91 (.92–3.97) | .0834 | .82 (.58–1.15) | .2553 | 1.11 (.74–1.68) | .61329 |
| mt8269 | A | .50 (.20–1.24) | .13380 | 1.00 (.43–2.33) | .9962 | 1.70 (.40–7.15) | .4721 | 1.08 (.62–1.90) | .7795 | 1.18 (.68–2.05) | .55420 |
| mt8557 | A | .40 (.13–1.24) | .11350 | 1.68 (.4–6.95) | .4760 | 1.70 (.40–7.15) | .4721 | 1.50 (.68–3.34) | .3168 | 2.03 (.77–5.32) | .15049 |
| mt8616 | T | .51 (.05–5.43) | .57874 | 1.51 (.43–5.32) | .5250 | 1.43 (.55–3.75) | .4663 | 1.01 (.40–2.55) | .98461 | ||
| mt8697 | A | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| mt8705 | C | 1.28 (.58–2.84) | .54662 | 1.29 (.48–3.48) | .6142 | .50 (.13–1.95) | .3164 | .66 (.11–3.93) | .65108 | ||
| mt8869 | G | 7.67 (2.18–27.04) | .00153 | .40 (1.96–0.08) | .2597 | .66 (.19–2.34) | .5228 | 1.23 (.51–2.98) | .64660 | ||
| mt8994 | A | 1.00 (.46–2.19) | .99095 | 3.31 (.97–11.25) | .0556 | 2.97 (.34–26.13) | .3254 | .67 (.37–1.21) | .1830 | 1.37 (.73–2.57) | .33308 |
| mt9055 | A | .70 (.41–1.19) | .18747 | .85 (1.39–0.52) | .5269 | .61 (.26–1.47) | .2733 | .85 (.65–1.11) | .2230 | .95 (.62–1.45) | .80851 |
| mt9093 | G | 1.42 (.45–4.48) | .55082 | .59 (2.46–0.14) | .4725 | 1.77 (.79–3.97) | .1688 | 1.88 (.80–4.40) | .14472 | ||
| mt9123 | A | .79 (.21–2.97) | .73265 | 1.10 (.44–2.73) | .8340 | .78 (.40–1.54) | .4743 | .55 (.26–1.19) | .12787 | ||
| mt9150 | G | 2.02 (.52–7.89) | .3137 | 1.82 (.62–5.35) | .2795 | ||||||
| mt9477 | A | .94 (.66–1.33) | .70875 | 1.25 (.83–1.89) | .2942 | 1.55 (.43–5.59) | .5014 | 1.35 (1.00–1.83) | .0494 | .94 (.71–1.24) | .66619 |
| mt9612 | A | 3.10 (.36–26.62) | .30349 | 1.01 (.06–16.11) | .99660 | ||||||
| mt9667 | G | .91 (.38–2.17) | .83321 | .71 (2.24–0.23) | .5590 | 2.45 (1.04–5.77) | .0398 | 1.23 (.59–2.56) | .58585 | ||
| mt9698 | C | .70 (.41–1.19) | .18747 | .85 (1.39–0.52) | .5269 | .61 (.26–1.47) | .2733 | .85 (.65–1.11) | .2230 | .95 (.62–1.45) | .80851 |
| mt9716 | C | .10 (.02–0.52) | .00640 | 1.41 (.45–4.44) | .5613 | .66 (.11–3.94) | .6447 | .85 (.49–1.45) | .5463 | 1.33 (.46–3.84) | .59560 |
| mt9899 | C | .66 (.23–1.85) | .42724 | 1.57 (.68–3.64) | .2914 | 1.09 (.59–2.00) | .7912 | 1.04 (.62–1.74) | .89486 | ||
| mt9947 | A | .50 (.05–5.27) | .56306 | 1.20 (.36–3.95) | .7643 | 1.28 (.47–3.43) | .6288 | 1.01 (.40–2.55) | .98979 | ||
| mt10034 | C | 4.05 (.96–17.07) | .05642 | .86 (1.83–0.41) | .7015 | 1.63 (.74–3.63) | .2284 | .82 (.51–1.29) | .3891 | 1.26 (.65–2.45) | .49152 |
| mt10044 | G | .86 (2.57–0.29) | .7829 | .63 (.25–1.63) | .3422 | .70 (.27–1.84) | .47261 | ||||
| mt10084 | C | 1.51 (.25–8.96) | .6507 | .61 (.26–1.47) | .2748 | .42 (.11–1.58) | .20139 | ||||
| mt10238 | C | 4.17 (1.00–17.5) | .05071 | .93 (1.99–0.43) | .8453 | 1.65 (.74–3.66) | .2195 | .84 (.53–1.33) | .4550 | 1.21 (.62–2.36) | .58387 |
| mt10398 | G | .89 (.61–1.28) | .52021 | 1.10 (.81–1.50) | .5386 | 1.01 (.57–1.77) | .9810 | .93 (.77–1.13) | .4784 | 1.28 (1.01–1.62) | .04098 |
| mt10463 | C | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| mt10550 | G | .70 (.41–1.19) | .18747 | .85 (1.39–0.52) | .5269 | .61 (.26–1.47) | .2733 | .85 (.65–1.11) | .2230 | .95 (.62–1.45) | .80851 |
| mt10876 | G | 1.00 (.20–5.00) | .99571 | 2.53 (.82–7.79) | .1072 | 1.61 (.84–3.06) | .1492 | .92 (.51–1.65) | .77333 | ||
| mt10915 | C | .68 (.11–4.07) | .67551 | 1.41 (.45–4.44) | .5613 | 1.10 (.47–2.60) | .8280 | 1.01 (.42–2.44) | .98377 | ||
| mt11251 | G | .76 (.53–1.10) | .14209 | 1.06 (.79–1.43) | .6847 | .66 (.37–1.20) | .1761 | 1.09 (.90–1.33) | .3731 | 1.08 (.87–1.33) | .49302 |
| mt11299 | C | .70 (.41–1.19) | .18747 | .85 (1.39–0.52) | .5269 | .61 (.26–1.47) | .2733 | .85 (.65–1.11) | .2230 | .95 (.62–1.45) | .80851 |
| mt11377 | A | 1.22 (.52–2.84) | .65089 | .99 (1.93–0.51) | .9854 | .99 (.58–1.68) | .9624 | 1.36 (.72–2.55) | .34438 | ||
| mt11467 | G | .82 (.61–1.10) | .19156 | .76 (1.01–0.57) | .0625 | .85 (.44–1.62) | .6208 | 1.06 (.88–1.28) | .5415 | .99 (.81–1.21) | .91817 |
| mt11470 | G | .67 (.11–3.97) | .65737 | .99 (15.94–0.06) | .9966 | .89 (.45–1.75) | .7298 | .33 (.07–1.52) | .15606 | ||
| mt11485 | C | 1.03 (.26–4.13) | .96985 | 1.10 (.46–2.61) | .8291 | .50 (.09–2.72) | .4259 | 1.16 (.63–2.16) | .6346 | .92 (.41–2.10) | .84714 |
| mt11674 | T | 1.00 (.46–2.19) | .99543 | 4.53 (1.11–18.42) | .0349 | 2.00 (.19–21.34) | .5661 | .84 (.48–1.50) | .5617 | 1.31 (.68–2.53) | .41544 |
| mt11719 | G | .84 (.65–1.09) | .19783 | 1.12 (.88–1.44) | .3564 | .98 (.59–1.63) | .9394 | .98 (.83–1.15) | .7929 | 1.02 (.86–1.22) | .81946 |
| mt11812 | G | 1.00 (.56–1.76) | .99367 | .66 (1.04–0.42) | .0742 | .73 (.33–1.61) | .4352 | 1.10 (.79–1.52) | .5829 | .80 (.58–1.10) | .16912 |
| mt11840 | T | .50 (.09–2.65) | .41575 | 1.12 (.43–2.93) | .8164 | .66 (.11–3.94) | .6447 | .86 (.39–1.85) | .6918 | 1.00 (.25–4.01) | .99887 |
| mt11914 | A | .99 (.54–1.82) | .97616 | .75 (2.16–0.26) | .5877 | .88 (.58–1.31) | .5197 | .91 (.50–1.66) | .76814 | ||
| mt11947 | G | 1.00 (.46–2.19) | .99095 | 3.31 (.97–11.25) | .0556 | 2.97 (.34–26.13) | .3254 | .67 (.37–1.21) | .1830 | 1.37 (.73–2.57) | .33308 |
| mt12007 | A | .44 (.14–1.39) | .16011 | 1.34 (.30–5.98) | .7025 | 3.21 (.90–11.42) | .0710 | .93 (.45–1.93) | .8377 | 2.92 (1.35–6.33) | .00662 |
| mt12308 | G | .82 (.61–1.10) | .19156 | .76 (1.01–0.57) | .0625 | .85 (.44–1.62) | .6208 | 1.06 (.88–1.28) | .5415 | .99 (.81–1.21) | .91817 |
| mt12372 | A | .82 (.61–1.10) | .19156 | .76 (1.01–0.57) | .0625 | .85 (.44–1.62) | .6208 | 1.06 (.88–1.28) | .5415 | .98 (.80–1.20) | .87760 |
| mt12414 | C | .93 (.45–1.90) | .84046 | 3.03 (.87–10.57) | .0820 | 2.03 (.19–21.66) | .5564 | .87 (.49–1.56) | .6462 | 1.38 (.72–2.65) | .32421 |
| mt12501 | A | 4.47 (1.41–14.21) | .01107 | 1.31 (.62–2.80) | .4793 | 1.82 (.85–3.89) | .1220 | .78 (.54–1.12) | .1775 | .86 (.56–1.34) | .50957 |
| mt12612 | G | .68 (.41–1.13) | .13447 | 1.44 (.95–2.19) | .0861 | .90 (.33–2.41) | .8290 | 1.10 (.84–1.43) | .5011 | 2.07 (1.44–2.98) | .00009 |
| mt12618 | A | .92 (.52–1.61) | .76225 | .33 (1.15–0.09) | .0826 | 2.02 (.19–21.5) | .5613 | 1.06 (.52–2.16) | .8657 | 1.21 (.66–2.20) | .53536 |
| mt12633 | A | .60 (.22–1.65) | .32172 | 1.75 (.83–3.69) | .1396 | .42 (.11–1.59) | .2010 | 1.11 (.65–1.90) | .6962 | .80 (.50–1.27) | .33851 |
| mt12669 | T | 1.68 (.61–4.62) | .31301 | ||||||||
| mt12705 | T | 1.62 (.93–2.83) | .09073 | 1.64 (.98–2.73) | .0587 | 2.07 (1.03–4.17) | .0417 | .77 (.57–1.02) | .0725 | 1.08 (.77–1.52) | .66678 |
| mt13020 | C | .83 (.25–2.73) | .75609 | 2.23 (.79–6.29) | .1305 | 1.48 (.80–2.75) | .2130 | .93 (.55–1.58) | .78339 | ||
| mt13105 | G | 2.70 (.75–9.72) | .12933 | 1.26 (.34–4.70) | .7332 | .38 (.08–1.90) | .2410 | .71 (.32–1.6) | .4119 | 1.49 (.53–4.18) | .44614 |
| mt13368 | A | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| mt13617 | C | .94 (.66–1.33) | .70875 | 1.25 (.83–1.89) | .2942 | 1.55 (.43–5.59) | .5014 | 1.35 (1.00–1.83) | .0494 | .94 (.71–1.24) | .66619 |
| mt13708 | A | .70 (.43–1.13) | .14564 | 1.37 (.93–2.01) | .1113 | .88 (.36–2.17) | .7887 | 1.06 (.83–1.35) | .6467 | 1.41 (1.07–1.87) | .01525 |
| mt13734 | C | 1.00 (.20–4.96) | .99566 | 2.21 (.78–6.25) | .1336 | 1.28 (.68–2.42) | .4442 | .91 (.51–1.64) | .76543 | ||
| mt13740 | C | .50 (.09–2.65) | .41575 | 1.12 (.43–2.93) | .8164 | .66 (.11–3.94) | .6447 | .86 (.39–1.85) | .6918 | 1.00 (.25–4.01) | .99887 |
| mt13780 | G | 3.59 (.82–15.71) | .09011 | 1.55 (.72–3.33) | .2590 | 2.38 (.89–6.34) | .0826 | .78 (.52–1.18) | .2396 | 1.80 (.94–3.46) | .07579 |
| mt13879 | C | .39 (.13–1.22) | .10522 | .67 (2.36–0.19) | .5292 | 1.71 (.41–7.20) | .4650 | 1.41 (.62–3.17) | .4107 | 2.53 (1.14–5.60) | .02270 |
| mt13934 | T | .98 (.14–7.02) | .98766 | 1.14 (.41–3.16) | .8039 | .50 (.12–1.98) | .3202 | 1.32 (.81–2.15) | .2663 | .79 (.47–1.32) | .36476 |
| mt13965 | C | .20 (.03–1.43) | .10900 | .50 (2.67–0.09) | .4201 | 1.16 (.39–3.46) | .7903 | .85 (.29–2.55) | .77797 | ||
| mt13966 | G | 1.34 (.46–3.88) | .59012 | 1.56 (.73–3.35) | .2523 | .67 (.38–1.21) | .1840 | .70 (.33–1.47) | .34686 | ||
| mt14022 | G | ||||||||||
| mt14167 | T | .70 (.41–1.19) | .18747 | .85 (1.39–0.52) | .5269 | .61 (.26–1.47) | .2733 | .85 (.65–1.11) | .2230 | .95 (.62–1.45) | .80851 |
| mt14182 | C | .94 (.56–1.56) | .80245 | .46 (1.05–0.20) | .0646 | 1.36 (.30–6.16) | .6933 | 1.08 (.72–1.62) | .7233 | .78 (.52–1.18) | .24376 |
| mt14233 | G | 1.00 (.56–1.76) | .99367 | .66 (1.04–0.42) | .0742 | .73 (.33–1.61) | .4352 | 1.10 (.79–1.52) | .5829 | .80 (.58–1.10) | .16912 |
| mt14365 | T | .80 (.21–2.98) | .73763 | 1.00 (2.53–0.39) | .9932 | .68 (.34–1.38) | .2824 | .49 (.22–1.09) | .08073 | ||
| mt14470 | C | 1.30 (.62–2.69) | .4861 | 1.06 (.63–1.77) | .8292 | 1.72 (.89–3.32) | .10541 | ||||
| mt14582 | G | .80 (.21–2.98) | .73763 | 1.00 (2.53–0.39) | .9932 | .68 (.34–1.38) | .2824 | .49 (.22–1.09) | .08073 | ||
| mt14687 | G | .20 (.03–1.43) | .10900 | .50 (2.67–0.09) | .4201 | 1.16 (.39–3.46) | .7903 | .85 (.29–2.55) | .77797 | ||
| mt14766 | C | .84 (.65–1.09) | .19783 | 1.12 (.88–1.44) | .3564 | .98 (.59–1.63) | .9394 | .98 (.84–1.15) | .8257 | 1.01 (.85–1.21) | .87667 |
| mt14793 | G | .96 (.55–1.66) | .87265 | 1.02 (.64–1.62) | .9347 | 3.93 (.50–30.57) | .1916 | 1.56 (1.03–2.35) | .0356 | 1.05 (.75–1.48) | .78017 |
| mt14798 | C | .64 (.40–1.01) | .05530 | 1.09 (.75–1.59) | .6345 | .67 (.29–1.57) | .3546 | .94 (.77–1.16) | .5936 | .97 (.74–1.27) | .82976 |
| mt14905 | A | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| mt15043 | A | 3.52 (.80–15.47) | .09524 | 1.10 (.58–2.07) | .7716 | 2.31 (.87–6.17) | .0937 | .82 (.56–1.20) | .2983 | 1.00 (.62–1.61) | .98972 |
| mt15218 | G | .87 (.42–1.80) | .70133 | .89 (1.55–0.51) | .6774 | 3.93 (1.16–13.28) | .0277 | 1.63 (1.04–2.55) | .0349 | .96 (.64–1.43) | .82791 |
| mt15257 | A | 1.26 (.34–4.71) | .73015 | .88 (1.75–0.44) | .7182 | .25 (.03–1.90) | .1782 | .96 (.56–1.66) | .8895 | .75 (.35–1.59) | .44968 |
| mt15452 | A | .74 (.51–1.06) | .09985 | .97 (1.31–0.72) | .8596 | .63 (.34–1.14) | .1275 | 1.11 (.90–1.35) | .3229 | .95 (.77–1.17) | .61947 |
| mt15607 | G | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| mt15758 | G | 4.55 (1.12–18.51) | .03434 | .55 (1.64–0.19) | .2850 | 1.31 (.59–2.92) | .5109 | .49 (.25–0.94) | .0325 | 1.50 (.43–5.30) | .52502 |
| mt15784 | C | 1.96 (.19–20.80) | .57476 | 2.00 (.19–21.07) | .5654 | .75 (.35–1.58) | .4457 | 1.13 (.56–2.28) | .72367 | ||
| mt15833 | C | .33 (.04–2.86) | .31519 | 1.13 (.41–3.15) | .8099 | 2.09 (.39–11.20) | .3912 | 1.00 (.53–1.89) | .9939 | 1.29 (.73–2.30) | .37879 |
| mt15884 | C | .83 (.41–1.66) | .59473 | 1.13 (.57–2.24) | .7271 | 2.03 (.19–21.66) | .5564 | .99 (.61–1.59) | .9631 | 1.00 (.54–1.84) | .99182 |
| mt15904 | T | 2.21 (1.26–3.88) | .00595 | 1.53 (.86–2.71) | .1509 | .83 (.25–2.80) | .7683 | 1.04 (.74–1.45) | .8268 | .96 (.65–1.40) | .82022 |
| mt15907 | G | 1.00 (.20–5.00) | .99571 | 2.53 (.82–7.79) | .1072 | 1.61 (.84–3.06) | .1492 | .92 (.51–1.65) | .77333 | ||
| mt15924 | G | 1.51 (.54–4.26) | .43223 | .96 (1.73–0.53) | .8923 | 1.45 (.64–3.30) | .3698 | 1.04 (.74–1.44) | .8366 | .77 (.48–1.24) | .28244 |
| mt15928 | A | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 |
| Haplogroup: | |||||||||||
| X | 1.36 (.47–3.93) | .57301 | 1.25 (.56–2.77) | .5881 | .83 (.49–1.41) | .4886 | .61 (.36–1.04) | .06825 | |||
| I | 4.14 (.99–17.37) | .05238 | .92 (2.05–0.42) | .8474 | 1.41 (.62–3.21) | .4066 | .80 (.49–1.30) | .3599 | 1.21 (.62–2.36) | .58179 | |
| V | 2.20 (1.25–3.87) | .00611 | 1.52 (.86–2.71) | .1529 | .84 (.25–2.83) | .7790 | 1.05 (.75–1.46) | .7945 | .96 (.66–1.41) | .83729 | |
| U | .80 (.58–1.09) | .16011 | .80 (1.10–0.58) | .1671 | 1.18 (.52–2.66) | .6944 | 1.16 (.93–1.44) | .1824 | 1.02 (.82–1.26) | .87963 | |
| J | .68 (.41–1.12) | .13275 | 1.46 (.96–2.21) | .0787 | 1.02 (.41–2.54) | .9689 | 1.07 (.83–1.39) | .6023 | 1.79 (1.29–2.47) | .00047 | |
| W | 1.01 (.48–2.15) | .97179 | 2.25 (.71–7.14) | .1672 | 3.76 (.85–16.73) | .0819 | .73 (.45–1.16) | .1839 | .97 (.61–1.54) | .90282 | |
| T | .88 (.53–1.46) | .62894 | .86 (1.27–0.58) | .4465 | .61 (.30–1.23) | .1654 | 1.11 (.84–1.47) | .4713 | .79 (.60–1.03) | .08576 | |
| K | .96 (.54–1.71) | .88946 | .76 (1.31–0.45) | .3280 | .61 (.23–1.61) | .3185 | .87 (.64–1.17) | .3550 | .83 (.52–1.30) | .41337 | |
| H | 1.00 (.77–1.30) | .97143 | 1.00 (.78–1.29) | .9710 | .78 (.46–1.33) | .3614 | .97 (.82–1.13) | .6678 | 1.04 (.87–1.25) | .64907 | |
Note.— The best results are shaded. An absence of values indicates a monomorphic SNP.
The remaining quantitative metabolic traits were studied only in nondiabetic controls, and sample sizes for these additional studies were significantly smaller than for diabetes or BMI (tables 6, 7, and 8). No significant associations were identified using unadjusted models for insulin resistance (HOMA-IR), insulin secretion (insulinogenic index), cholesterol levels, and systolic and diastolic blood pressure (table 6) or after adjustment for sex, age, and BMI (tables 7 and 8).
We specifically examined the results for the three missense variants that were hypothesized to have been adaptive in cold climates and to be related to “energy deficiency” diseases today.18,19 No association with increased risk of diabetes, obesity, or other metabolic traits was observed (table 11). The putative association of mt16189C with type 2 diabetes reported elsewhere24–26 was not replicated here (n=6,608; odds ratio [OR] 1.03; 95% CI 0.88–1.21; Pnom=.36), which is consistent with other recent reports that did not support this hypothesis.27,28 With this sample size (under the assumption of an OR of 1.5 and an mt16189C allele frequency of 14%, consistent with previous reports), we have >99% power to reject the null hypothesis (of no association) at the P<.01 level.56
Table 11. .
Results of Association Testing of Three European mtDNA Variants Predicted to Be Selected for Adaptation to Cold Climates
|
mt4917G(ND2 N150D) |
mt14798C(cytB F18L) |
mt15257A(cytB D171N) |
|||||
| Phenotypea | n | Effect Estimateb | Pnom | Effect Estimateb | Pnom | Effect Estimateb | Pnom |
| DM | 6,608 | .89 (.76–1.05) | .16 | .93 (.81–1.07) | .30 | .87 (.61–1.23) | .47 |
| BMI | 6,523 | −.002 | .56 | −.002 | .59 | .006 | .39 |
| Ins index | 342 | .04 | .29 | .05 | .37 | .31 | .13 |
| HOMA-IR | 399 | −.03 | .59 | .04 | .27 | .09 | .55 |
| Cholesterol | 1,274 | .13 | .29 | .03 | .81 | −.45 | .05 |
| Systolic BP | 2,047 | −.49 | .76 | 1.13 | .47 | .39 | .90 |
| Diastolic BP | 2,047 | −.07 | .92 | .59 | .43 | −1.94 | .21 |
BP = blood pressure; DM = diabetes mellitus, type 2; Ins = insulinogenic.
OR (95% CI) for DM and β for all other phenotypes.
Discussion
We set out to develop a comprehensive approach to mtDNA association testing that (a) captures all common coding-region mtDNA variants and haplogroups efficiently by use of linkage disequilibrium, (b) tests all common variants for association, and (c) accounts for the multiple comparisons implicit in testing the many variants in mtDNA. Compared with the methods in the literature, this approach provides a more complete and statistically conservative assessment of the role of common mtDNA variants in disease; the current application to diabetes and BMI provides a sample size threefold larger than any previous study.
Despite this well-powered evaluation, our analyses do not support the hypothesis that common mtDNA variants play a role in type 2 diabetes and BMI, at least in the European samples studied. The results from our association tests of all common mtDNA variants and the risk of type 2 diabetes show that there is no single common coding-region mtDNA variant in European populations that strongly influences risk of type 2 diabetes or BMI, which is consistent with recent results obtained by Mohlke et al.27 Haplogroup association results for type 2 diabetes were also consistent with noise. There may be population-specific variants that confer altered risk of diabetes (as in the Polish sample tested here), and further studies, including replication and complete resequencing,57 will be needed to explore that hypothesis. Our results showed no significant association of mtDNA variants with other metabolic traits (blood pressure, cholesterol, insulin secretion, and insulin resistance traits); however, this study was not as well powered for these traits.
It is interesting and perhaps surprising that we could not detect any phenotypic consequence of missense variants that were predicted to have been selected for in colder climates.2,19,58 Although our study does not directly address the hypothesis that climatic influences have led to selective adaptation of mtDNA variants, if common mtDNA variation does influence such traits, it is not reflected in glucose-stimulated insulin secretion, insulin resistance, body weight, blood pressure, cholesterol level, or type 2 diabetes.
This study does not address the role of mtDNA heteroplasmy in diabetes. Acquired somatic mtDNA mutations in tissues of relevance to type 2 diabetes are undetected in the blood samples used here. Typically, blood DNA exhibits much less heteroplasmy than nondividing tissues.59 In the 6,608 DNA samples tested, a very small amount of heteroplasmy was observed (1.1% of samples had one or more “heterozygous” calls), although the mass spectroscopy–based genotyping platform is sensitive to low-frequency spectral peaks corresponding to the alternate allele. Rare mitochondrial diseases exhibit more pronounced heteroplasmy, perhaps because cells with mutant alleles are inviable without wild-type mtDNA molecules. Thus, we think it is unlikely that mtDNA heteroplasmy for inherited variants significantly influenced our results.
These results should not be taken as addressing the broader hypothesis that mitochondrial OXPHOS plays a causal role in type 2 diabetes; there are >70 nuclear-encoded OXPHOS subunits and an estimated 1,500 mitochondrial proteins that are as yet untested. Moreover, although the mtDNA variants tested here do not appear to influence metabolic disease, inherited variation in mtDNA may influence risk of other diseases, such as Alzheimer disease, Parkinson disease, cardiovascular disease, cardiomyopathy, HIV lipodystrophy, and prostate cancer. A systematic evaluation of mtDNA and nuclear mitochondrial genes for a causal role in these diseases and an investigation of epistatic interactions between the two genomes will reveal the extent to which mitochondrial defects play a causal role in human disease.
Acknowledgments
D.A. is a Charles E. Culpeper Scholar of the Rockefeller Brothers Fund and a Burroughs Wellcome Fund Clinical Scholar in Translational Research. R.S. is a recipient of a National Institutes of Health National Research Service Award fellowship. This work was funded by a grant from the National Institute of Diabetes & Digestive & Kidney Diseases (to D.A.; Diabetes Genome Anatomy Project). D.A., J.N.H., M.J.D., and V.M. are recipients of The Richard and Susan Smith Family Foundation/American Diabetes Association Pinnacle Program Project Award. We thank M. Sun, for assistance with genotyping; colleagues at Massachussetts General Hospital, the Broad Institute, and investigators from Diabetes Genome Anatomy Project, for useful discussions; Mitokor, for 536 publicly available mtDNA sequences; MITOMAP, for being a resource for information about mtDNA polymorphisms; and submitters of mtDNA sequences to GenBank.
Web Resources
The URLs for data presented herein are as follows:
- Broad Institute Tagger: SNPs in human mtDNA, http://www.broad.mit.edu/mpg/tagger/mito.html (for information on sequence alignments, tSNP assays, and assay conditions)
- Human Mitochondrial DNA Revised Cambridge Reference Sequence, http://www.mitomap.org/mitoseq.html
- MITOMAP, http://www.mitomap.org/ (for information on mtDNA)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Leigh syndrome; cardioencephalomyopathy; MELAS; KSS; MIDD; hypertension, hypercholesterolemia, and hypomagnesemia; type 2 diabetes; and Alzheimer, Parkinson, and Huntington diseases)
References
- 1.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115:629–640 10.1016/S0092-8674(03)00926-7 [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39:359–407 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimauro S, Davidzon G (2005) Mitochondrial DNA and disease. Ann Med 37:222–232 10.1080/07853890510007368 [DOI] [PubMed] [Google Scholar]
- 4.Smeitink JA, Zeviani M, Turnbull DM, Jacobs HT (2006) Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab 3:9–13 10.1016/j.cmet.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 5.Zeviani M, Di Donato S (2004) Mitochondrial disorders. Brain 127:2153–2172 10.1093/brain/awh259 [DOI] [PubMed] [Google Scholar]
- 6.Taylor RW, Turnbull DM (2005) Mitochondrial DNA mutations in human disease. Nat Rev Genet 6:389–402 10.1038/nrg1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett TG (2001) Mitochondrial diabetes, DIDMOAD and other inherited diabetes syndromes. Best Pract Res Clin Endocrinol Metab 15:325–343 10.1053/beem.2001.0149 [DOI] [PubMed] [Google Scholar]
- 8.van den Ouweland JM, Lemkes HH, Ruitenbeek W, Sandkuijl LA, de Vijlder MF, Struyvenberg PA, van de Kamp JJ, Maassen JA (1992) Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet 1:368–371 10.1038/ng0892-368 [DOI] [PubMed] [Google Scholar]
- 9.Wilson FH, Hariri A, Farhi A, Zhao H, Petersen KF, Toka HR, Nelson-Williams C, Raja KM, Kashgarian M, Shulman GI, Scheinman SJ, Lifton RP (2004) A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science 306:1190–1194 10.1126/science.1102521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simoneau JA, Kelley DE (1997) Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol 83:166–171 [DOI] [PubMed] [Google Scholar]
- 11.Kelley DE, He J, Menshikova EV, Ritov VB (2002) Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51:2944–2950 [DOI] [PubMed] [Google Scholar]
- 12.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE (2005) Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54:8–14 [DOI] [PubMed] [Google Scholar]
- 13.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 14.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 100:8466–8471 10.1073/pnas.1032913100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI (2003) Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300:1140–1142 10.1126/science.1082889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI (2004) Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350:664–671 10.1056/NEJMoa031314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen KF, Dufour S, Shulman GI (2005) Decreased insulin-stimulated ATP synthesis and phosphate transport in muscle of insulin-resistant offspring of type 2 diabetic parents. PLoS Med 2:e233 10.1371/journal.pmed.0020233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA 100:171–176 10.1073/pnas.0136972100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC, Coskun PE, Golik P, Macaulay V, Clark AG, Hosseini S, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A (2004) Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303:223–226 10.1126/science.1088434 [DOI] [PubMed] [Google Scholar]
- 20.Lowell BB, Shulman GI (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307:384–387 10.1126/science.1104343 [DOI] [PubMed] [Google Scholar]
- 21.Savage DB, Petersen KF, Shulman GI (2005) Mechanisms of insulin resistance in humans and possible links with inflammation. Hypertension 45:828–833 10.1161/01.HYP.0000163475.04421.e4 [DOI] [PubMed] [Google Scholar]
- 22.Schapira AH (1999) Mitochondrial involvement in Parkinson’s disease, Huntington’s disease, hereditary spastic paraplegia and Friedreich’s ataxia. Biochim Biophys Acta 1410:159–170 10.1016/S0005-2728(98)00164-9 [DOI] [PubMed] [Google Scholar]
- 23.Beal MF (2005) Mitochondria take center stage in aging and neurodegeneration. Ann Neurol 58:495–505 10.1002/ana.20624 [DOI] [PubMed] [Google Scholar]
- 24.Poulton J, Brown MS, Cooper A, Marchington DR, Phillips DI (1998) A common mitochondrial DNA variant is associated with insulin resistance in adult life. Diabetologia 41:54–58 10.1007/s001250050866 [DOI] [PubMed] [Google Scholar]
- 25.Poulton J, Bednarz AL, Scott-Brown M, Thompson C, Macaulay VA, Simmons D (2002) The presence of a common mitochondrial DNA variant is associated with fasting insulin levels in Europeans in Auckland. Diabet Med 19:969–971 10.1046/j.0742-3071.2002.00836.x [DOI] [PubMed] [Google Scholar]
- 26.Poulton J, Luan J, Macaulay V, Hennings S, Mitchell J, Wareham NJ (2002) Type 2 diabetes is associated with a common mitochondrial variant: evidence from a population-based case-control study. Hum Mol Genet 11:1581–1583 10.1093/hmg/11.13.1581 [DOI] [PubMed] [Google Scholar]
- 27.Mohlke KL, Jackson AU, Scott LJ, Peck EC, Suh YD, Chines PS, Watanabe RM, Buchanan TA, Conneely KN, Erdos MR, Narisu N, Enloe S, Valle TT, Tuomilehto J, Bergman RN, Boehnke M, Collins FS (2005) Mitochondrial polymorphisms and susceptibility to type 2 diabetes-related traits in Finns. Hum Genet 118:245–254 10.1007/s00439-005-0046-4 [DOI] [PubMed] [Google Scholar]
- 28.Chinnery PF, Elliott HR, Patel S, Lambert C, Keers SM, Durham SE, McCarthy MI, Hitman GA, Hattersley AT, Walker M (2005) Role of the mitochondrial DNA 16184–16193 poly-C tract in type 2 diabetes. Lancet 366:1650–1651 10.1016/S0140-6736(05)67492-2 [DOI] [PubMed] [Google Scholar]
- 29.Herrnstadt C, Howell N (2004) An evolutionary perspective on pathogenic mtDNA mutations: haplogroup associations of clinical disorders. Mitochondrion 4:791–798 10.1016/j.mito.2004.07.041 [DOI] [PubMed] [Google Scholar]
- 30.Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Florez JC, Sjogren M, Burtt N, Orho-Melander M, Schayer S, Sun M, Almgren P, Lindblad U, Tuomi T, Gaudet D, Hudson TJ, Daly MJ, Ardlie KG, Hirschhorn JN, Altshuler D, Groop L (2004) Association testing in 9,000 people fails to confirm the association of the insulin receptor substrate-1 G972R polymorphism with type 2 diabetes. Diabetes 53:3313–3318 [DOI] [PubMed] [Google Scholar]
- 32.Winckler W, Graham RR, de Bakker PI, Sun M, Almgren P, Tuomi T, Gaudet D, Hudson TJ, Ardlie KG, Daly MJ, Hirschhorn JN, Groop L, Altshuler D (2005) Association testing of variants in the hepatocyte nuclear factor 4α gene with risk of type 2 diabetes in 7,883 people. Diabetes 54:886–892 [DOI] [PubMed] [Google Scholar]
- 33.Tang K, Oeth P, Kammerer S, Denissenko MF, Ekblom J, Jurinke C, van den Boom D, Braun A, Cantor CR (2004) Mining disease susceptibility genes through SNP analyses and expression profiling using MALDI-TOF mass spectrometry. J Proteome Res 3:218–227 10.1021/pr034080s [DOI] [PubMed] [Google Scholar]
- 34.Autere J, Moilanen JS, Finnila S, Soininen H, Mannermaa A, Hartikainen P, Hallikainen M, Majamaa K (2004) Mitochondrial DNA polymorphisms as risk factors for Parkinson’s disease and Parkinson’s disease dementia. Hum Genet 115:29–35 10.1007/s00439-004-1123-9 [DOI] [PubMed] [Google Scholar]
- 35.Carrieri G, Bonafe M, De Luca M, Rose G, Varcasia O, Bruni A, Maletta R, Nacmias B, Sorbi S, Corsonello F, Feraco E, Andreev KF, Yashin AI, Franceschi C, De Benedictis G (2001) Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer’s disease. Hum Genet 108:194–198 10.1007/s004390100463 [DOI] [PubMed] [Google Scholar]
- 36.Dato S, Passarino G, Rose G, Altomare K, Bellizzi D, Mari V, Feraco E, Franceschi C, De Benedictis G (2004) Association of the mitochondrial DNA haplogroup J with longevity is population specific. Eur J Hum Genet 12:1080–1082 10.1038/sj.ejhg.5201278 [DOI] [PubMed] [Google Scholar]
- 37.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C (1999) Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J 13:1532–1536 [DOI] [PubMed] [Google Scholar]
- 38.De Benedictis G, Carrieri G, Varcasia O, Bonafe M, Franceschi C (2000) Inherited variability of the mitochondrial genome and successful aging in humans. Ann N Y Acad Sci 908:208–218 [DOI] [PubMed] [Google Scholar]
- 39.Giacchetti M, Monticelli A, De Biase I, Pianese L, Turano M, Filla A, De Michele G, Cocozza S (2004) Mitochondrial DNA haplogroups influence the Friedreich’s ataxia phenotype. J Med Genet 41:293–295 10.1136/jmg.2003.015289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khogali SS, Mayosi BM, Beattie JM, McKenna WJ, Watkins H, Poulton J (2001) A common mitochondrial DNA variant associated with susceptibility to dilated cardiomyopathy in two different populations. Lancet 357:1265–1267 10.1016/S0140-6736(00)04422-6 [DOI] [PubMed] [Google Scholar]
- 41.Momiyama Y, Furutani M, Suzuki Y, Ohmori R, Imamura S, Mokubo A, Asahina T, Murata C, Kato K, Anazawa S, Hosokawa K, Atsumi Y, Matsuoka K, Kimura M, Kasanuki H, Ohsuzu F, Matsuoka R (2003) A mitochondrial DNA variant associated with left ventricular hypertrophy in diabetes. Biochem Biophys Res Commun 312:858–864 10.1016/j.bbrc.2003.10.195 [DOI] [PubMed] [Google Scholar]
- 42.Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K (2003) Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet 112:29–33 10.1007/s00439-002-0843-y [DOI] [PubMed] [Google Scholar]
- 43.Niemi AK, Moilanen JS, Tanaka M, Hervonen A, Hurme M, Lehtimaki T, Arai Y, Hirose N, Majamaa K (2005) A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet 13:166–170 10.1038/sj.ejhg.5201308 [DOI] [PubMed] [Google Scholar]
- 44.Ross OA, McCormack R, Curran MD, Duguid RA, Barnett YA, Rea IM, Middleton D (2001) Mitochondrial DNA polymorphism: its role in longevity of the Irish population. Exp Gerontol 36:1161–1178 10.1016/S0531-5565(01)00094-8 [DOI] [PubMed] [Google Scholar]
- 45.Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K (1998) Mitochondrial genotype associated with longevity. Lancet 351:185–186 10.1016/S0140-6736(05)78211-8 [DOI] [PubMed] [Google Scholar]
- 46.Tanaka M (2002) Mitochondrial genotypes and cytochrome b variants associated with longevity or Parkinson’s disease. J Neurol Suppl 2 249:II/11–II/18 [DOI] [PubMed] [Google Scholar]
- 47.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, et al (2003) Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet 72:804–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Murali Doraiswamy P, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA (2004) Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett 365:28–32 10.1016/j.neulet.2004.04.051 [DOI] [PubMed] [Google Scholar]
- 49.Huerta C, Castro MG, Coto E, Blazquez M, Ribacoba R, Guisasola LM, Salvador C, Martinez C, Lahoz CH, Alvarez V (2005) Mitochondrial DNA polymorphisms and risk of Parkinson’s disease in Spanish population. J Neurol Sci 236:49–54 10.1016/j.jns.2005.04.016 [DOI] [PubMed] [Google Scholar]
- 50.Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kruglyak L, Nickerson DA (2001) Variation is the spice of life. Nat Genet 27:234–236 10.1038/85776 [DOI] [PubMed] [Google Scholar]
- 52.Reich DE, Gabriel SB, Altshuler D (2003) Quality and completeness of SNP databases. Nat Genet 33:457–458 10.1038/ng1133 [DOI] [PubMed] [Google Scholar]
- 53.The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D (2005) Efficiency and power in genetic association studies. Nat Genet 37:1217–1223 10.1038/ng1669 [DOI] [PubMed] [Google Scholar]
- 55.Marchini J, Donnelly P, Cardon LR (2005) Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet 37:413–417 10.1038/ng1537 [DOI] [PubMed] [Google Scholar]
- 56.Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19:149–150 10.1093/bioinformatics/19.1.149 [DOI] [PubMed] [Google Scholar]
- 57.Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, Chakravarti A (2004) The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res 14:812–819 10.1101/gr.2228504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis KK, Passarino G, Underhill PA, Scharfe C, Torroni A, Scozzari R, Modiano D, Coppa A, de Knijff P, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2006) The role of selection in the evolution of human mitochondrial genomes. Genetics 172:373–387 10.1534/genetics.105.043901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jazin EE, Cavelier L, Eriksson I, Oreland L, Gyllensten U (1996) Human brain contains high levels of heteroplasmy in the noncoding regions of mitochondrial DNA. Proc Natl Acad Sci USA 93:12382–12387 10.1073/pnas.93.22.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]