Abstract
The survival and growth of tumor cells in a foreign environment is considered a rate-limiting step during metastasis. To identify genes that may be essential for this process, we isolated highly metastatic variants from a poorly metastatic human melanoma cell line and performed expression analyses of metastases and primary tumors from these cells. GPR56 is among the genes markedly down-regulated in the metastatic variants. We show that overexpression of GPR56 suppresses tumor growth and metastasis, whereas reduced expression of GPR56 enhances tumor progression. Levels of GPR56 do not correlate with growth rate in vitro, suggesting that GPR56 may mediate growth suppression by interaction with a component in the tumor microenvironment in vivo. We show that GPR56 binds specifically to tissue transglutaminase, TG2, a widespread component of tissue and tumor stroma previously implicated as an inhibitor of tumor progression. We discuss the mechanisms whereby GPR56-TG2 interactions may suppress tumor growth and metastasis.
Keywords: tumor microenvironment, extracellular matrix
Metastasis is the main cause of death in cancer patients and is generally considered to be a multistep process (1): (i) cells detach from their neighboring cells in the primary tumor; (ii) some of the detached cells enter the circulation via blood vessels or lymphatics (intravasation); (iii) a fraction of the cells in the circulation arrest and transmigrate through blood vessels or lymphatics and invade into a distant tissue or organ (extravasation); (iv) some of the invading cells survive and proliferate in the new environment as metastases. To produce any clinically relevant metastases, a tumor cell must complete all these steps.
Abundant clinical and experimental data suggest that the survival and growth step (step iv) is a rate-limiting step during metastasis (1, 2). Frequently, tumor cells are able to enter the circulation and settle in many organs but are not able to proliferate or are only able to proliferate in certain organs. Experimental metastasis assays have been developed to study these steps of metastasis. In these assays, a pool of poorly metastatic tumor cells is injected into the circulation of immunodeficient mice and gives rise to metastases at low frequency. Cells in these rare metastases can be selected from the original pool as variants which, through genetic or epigenetic changes, have gained the ability to invade, survive, and grow in a foreign environment. When these cells are isolated and amplified in vitro, they largely maintain their enhanced metastatic potential. Genes involved in metastasis can then be identified by comparing the gene expression profiles between the highly metastatic variants and the poorly metastatic pool through microarray analyses. With this method, RhoC has previously been discovered to play important roles during melanoma metastasis to lung (3), and a five-gene signature has been discovered to be essential for breast cancer metastasis to bone (4).
In this article, we report that a member of a newly described family of G protein-coupled receptors (GPCRs), GPR56, contributes to suppression of melanoma metastasis and tumor growth. This suppression is not cell-autonomous, because cells with altered levels of GPR56 grow at similar rates in vitro. Therefore, GPR56 may interact with a factor in the tissue or tumor microenvironment to suppress metastases and tumor growth. Indeed, using biochemical methods, we identified a ubiquitously expressed extracellular matrix protein, transglutaminase (TG)2, as a ligand of GPR56, which may cooperate in the suppressive role of GPR56.
Results
GPR56 Is Down-Regulated in Tumors Derived from Highly Metastatic Melanoma Cell Lines.
Several highly metastatic melanoma cell lines were derived from pools of poorly metastatic cells (A375eco) by using the experimental metastasis assay (for details see Supporting Materials and Methods and Fig. 5A, which are published as supporting information on the PNAS web site). When 2 × 105 cells from each cell line were injected intravenously into immunodeficient mice, the derived MA and MC cell lines produced more lung metastases than did the parental line at 1 month after injection (Fig. 5B), indicating that the selected variants were progressively more metastatic. Expression levels of genes in tumors derived from the highly metastatic variants and the poorly metastatic parental line were compared by using Affymetrix arrays and dchip software (5). In parallel, we also compared the expression profiles between a previously described highly metastatic melanoma variant (SM cells) and its parental line (6) (Fig. 5A). GPR56 was among the genes that were significantly down-regulated in samples from all of the highly metastatic cells (Fig. 5C). Consistent with this finding, GPR56 mRNA has been reported to be reduced in several highly metastatic melanoma cell lines compared with poorly metastatic cells in vitro (7).
We confirmed by real-time PCR that GPR56 mRNA was down-regulated in the tumors from highly metastatic cells (ranging from −1.9- to −55.1-fold among different tumor samples). To examine whether GPR56 is also down-regulated at the protein level in tumor samples from highly metastatic cells, we generated peptide antibodies against the C terminus of GPR56 (denoted anti-GPRC). This antibody specifically recognized a band of ≈25 kDa in total lysates from cells expressing GPR56 (Fig. 6A, which is published as supporting information on the PNAS web site). This ≈25-kDa band is much smaller than the predicted molecular mass of unglycosylated protein (≈76 kDa), suggesting that the protein might be processed upon maturation. In support of this suggestion, the ≈25-kDa protein also appeared as the predominant band when the total lysates from cells expressing HA-tagged GPR56 were probed with anti-HA antibody (Fig. 6B). We also generated antibodies against a peptide at the N terminus of GPR56 (denoted anti-GPRN). This antibody specifically recognized a group of bands between 60 and 65 kDa in the total lysate of GPR56-expressing cells that were not detected by the anti-GPRC antibody (Fig. 6A), suggesting that the N terminus and C terminus of GPR56 are separated.
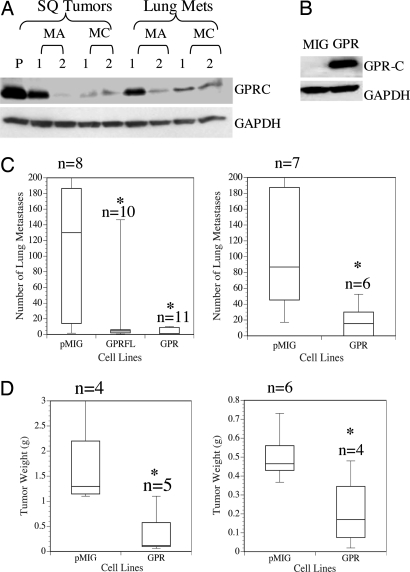
GPR56 belongs to a newly described family of GPCRs. Members of this family all contain a conserved proteolytic cleavage domain [GPS (GPCR proteolytic site)] (8). Many of them have been shown to be cleaved at this site upon maturation, and the cleavage was suggested to be essential for the cell-surface localization of the mature receptors (9). Our data strongly suggest that GPR56 is also cleaved and functions as a two-subunit receptor upon maturation. The two cleaved fragments were found to associate with each other, because the antibody against either one could specifically immunoprecipitate the other from the radioimmunoprecipitation assay (RIPA) lysate of GPR56-overexpressing cells (Fig. 6C). This association does not appear to be via covalent linkage by disulfide bonds, because the two fragments can be separated on an SDS/polyacrylamide gel in the absence of DTT (Fig. 6B). By using the anti-GPRC antibody we generated, GPR56 was confirmed to be down-regulated at the protein level in tumors derived from highly metastatic cell lines (Fig. 1A).
Fig. 1.
GPR56 is down-regulated in tumors from metastatic variants, and its reexpression suppresses tumor growth and metastasis in MC-1 cells. (A) Expression of GPR56 was examined by immunoblotting lysates of tumors from all of the cell lines by using anti-GPRC antibody. GAPDH was used as a loading control. GPR56 protein levels were reduced in both s.c. tumors and lung metastases derived from highly metastatic melanoma cells as compared with s.c. tumors of the poorly metastatic parental line (P). (B) GPR56 was expressed in MC-1 cells as determined by anti-GPR56 antibody on a Western blot. (C) Experimental metastasis assays of MC-1 cells with empty vector (pMIG) or expressing GPR56 (GPR) or FLAG-tagged GPR56 (GPRFL). Cells (5 × 105) were injected intravenously, and lung metastases were counted 2 months later. MC-1(pMIG-GPR) or MC-1(pMIG-GPRFL) cells produced significantly fewer metastases (two independent experiments). ∗, P < 0.05. (D) MC-1 cells expressing high levels of GPR56 also show significantly reduced primary tumor growth. Weights of s.c. tumors from MC-1(pMIG-GPR) or MC-1(pMIG) cells are shown. ∗, P < 0.05.
Expression of GPR56 Is Sufficient to Suppress Metastasis and Tumor Growth in MC-1 Cells.
To investigate whether expression of GPR56 in metastatic cells suppresses metastasis, we expressed GPR56 in MC-1 cells (Fig. 1B). Expression of GPR56 does not affect MC-1 cell proliferation in vitro (Fig. 7 which is published as supporting information on the PNAS web site), however, results from two independent experiments showed that the cells with ectopically expressed GPR56 [MC-1(pMIG-GPR)] resulted in significantly fewer lung metastases when tested by tail-vein injection assays (Fig. 1C). This reduction may be due, at least in part, to reduction of metastatic tumor growth, because MC-1(pMIG-GPR) cells grew significantly more slowly when injected s.c. (Fig. 1D). In similar experiments, overexpression of GPR56 in highly metastatic SM cells also reduced their metastasis (data not shown). These results suggest that overexpression of GPR56 suppresses both tumor growth and metastasis. This suppression must involve a factor in the microenvironment in vivo, because high levels of GPR56 do not affect cell proliferation in vitro.
Reduction of GPR56 Enhances Tumor Growth and Metastasis in A375eco Cells.
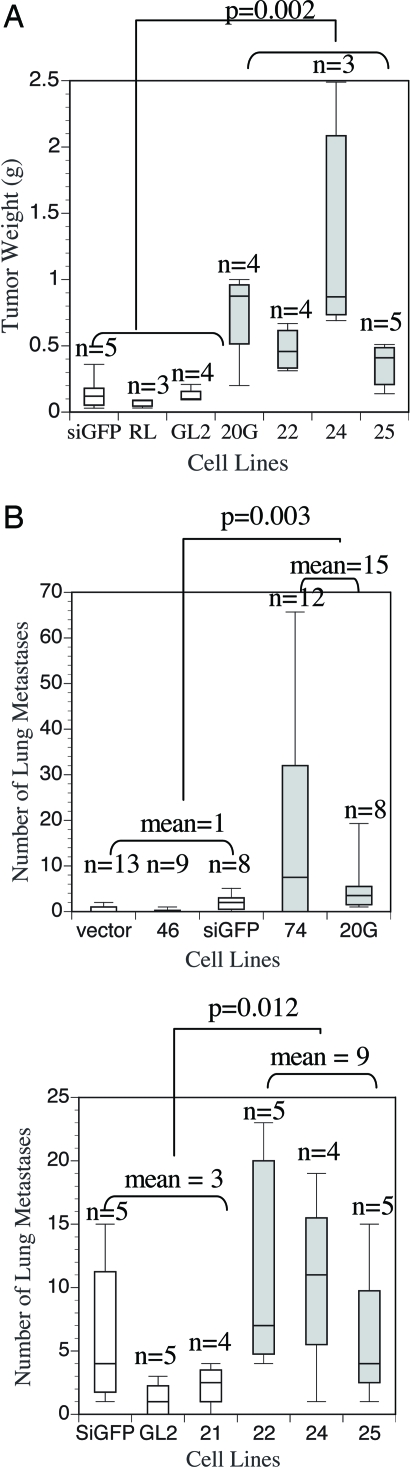
We next examined whether reduction of GPR56 is sufficient to enhance metastasis in poorly metastatic cells. We selected several sequences in human GPR56 cDNA and expressed them from a retroviral vector as short hairpin RNAs (shRNAs). Several of these shRNAs (74, 20G, 22, 24, 25), when expressed in A375eco cells [A375-RNAi (RNA interference)], suppressed the expression of GPR56 significantly (Fig. 8A, which is published as supporting information on the PNAS web site). shRNAs targeting GFP (siGFP), firefly luciferase (GL2), and renilla luciferase (RL) were used as controls. We also included as controls vector only and two shRNAs (46 and 21) that contain GPR56 sequences but have no suppressing effects. A375eco cells with reduced levels of GPR56 grow slightly faster than the controls in vitro, but there is also some variation in the growth rate among the control RNAi lines (Fig. 8B). When these cells were injected s.c. into immunodeficient mice, the A375-RNAi cells grew faster than the controls (Fig. 2A). This increase of growth cannot be attributed entirely to the increase of growth in vitro, because some A375-RNAi lines (22 and 24) grew at similar rates as the controls (siGFP and GL2) in vitro, but they grew faster as tumors in vivo. When these cells were injected intravenously into immunodeficient mice, there was a statistically significant increase in the number of metastases from the A375-RNAi cells as compared with controls (Fig. 2B). These data suggest that down-regulation of GPR56 leads to enhanced tumor growth and metastasis. Consistent with the overexpression data, this enhancement of tumor growth and metastasis may also reflect an influence of the microenvironment in vivo.
Fig. 2.
Reduction of GPR56 by RNAi promotes tumor growth and metastasis. (A) Reduction of GPR56 resulted in increased s.c. tumor growth in vivo. Weights of s.c. tumors from A375-RNAi cells are shown. Box plots of A375-RNAi cells with significant reduction of GPR56 protein (see Fig. 8) are shaded. Statistical significance is shown for all of these A375-RNAi lines versus controls. (B) Reduction of GPR56 resulted in an enhancement of metastasis in vivo as shown by experimental metastasis assays. A375eco-RNAi cells (5 × 105) or controls were injected intravenously into immunodeficient nude mice (Upper) and nonobese diabetic severe combined immunodeficient (NOD-SCID) mice (Lower). Lung metastases were counted 2 months later. Box plots of A375eco-RNAi cells with significant reduction of GPR56 protein (see Fig. 8) are shaded. Statistical significance is shown for all of these A375-RNAi lines versus controls.
N Terminus of GPR56 Binds to TG2.
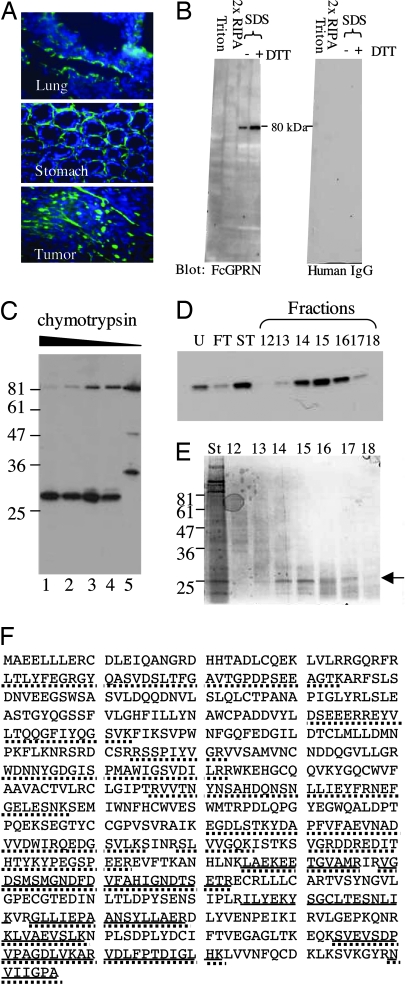
The above results suggest that a factor in the tumor microenvironment may cooperate with GPR56 to suppress tumor growth and metastasis. We speculated that this factor might be an extracellular binding partner or a ligand of GPR56. To search for such a ligand, we fused the portion of GPR56 N terminus that is predicted to be cleaved at the GPS motif to human IgG Fc fragment and expressed it in MC-1 cells as FcGPRN. FcGPRN was purified from the supernatant of expressing cells and used as a probe for the potential ligand of GPR56. On frozen sections from several tissues, including lung, FcGPRN bound in a pattern characteristic of extracellular matrix (Fig. 3A), suggesting that the ligand of GPR56 may be an extracellular matrix protein. Because extracellular matrix proteins tend to be insoluble in mild detergents, we fractionated mouse lungs using detergents of increasing strength (Fig. 9, which is published as supporting information on the PNAS web site), and found that FcGPRN used in an overlay assay recognized a protein of ≈80 kDa in a Triton- and radioimmunoprecipitation assay (RIPA)-insoluble fraction (Fig. 3B). These data strongly suggested to us that GPR56 binds to an extracellular matrix protein.
Fig. 3.
Identification of TG2 as a candidate GPR56-binding protein. (A) The N terminus of GPR56 that was fused to human Fc fragment (FcGPRN) recognizes extracellular matrix on tissue and tumor sections by immunohistochemistry. Blue, DAPI; green, FcGPRN. (B) FcGPRN recognized a band of ≈80 kDa in the radioimmunoprecipitation assay (RIPA)-insoluble fraction of lung lysate. The extraction scheme is shown in Fig. 9. Human IgG was used as a negative control. (C) FcGPRN recognizes an ≈28-kDa fragment after chymotryptic digestion of mouse lungs. Lanes 1–4 are 1:100, 1:250, 1:500, and 1:1,000 dilutions of 2.5% chymotrypsin. Lane 5 is a negative control with no chymotrypsin treatment. (D) The fractions after purification (indicated in Fig. 10) were probed by FcGPRN on a Western blot. (E) Some of the samples shown in D were stained with SimplyBlue stain and the ≈28-kDa bands were excised for mass spectrometric analysis (arrow). (F) Identification of mouse TG2 as a candidate protein from mass spectrometric analysis. The sequence of TG2 protein is shown. Bold lines indicate the sequences found in the ≈28-kDa fragment, and the dotted lines indicate the sequences found in the uncleaved ≈80-kDa protein. The ≈28-kDa fragment is located at the C terminus of the protein.
We designed a purification scheme to isolate this binding partner of GPR56 (Fig. 10, which is published as supporting information on the PNAS web site). It could be partially digested by chymotrypsin to an ≈28-kDa fragment and still maintain its binding capability for FcGPRN (Fig. 3C). The digested fragment could then be further purified by ion exchange and gel-filtration chromatography. Half of the fractions after purification were run on a polyacrylamide gel and probed with FcGPRN (Fig. 3D). The other half of those fractions that contained the target protein were run again on a polyacrylamide gel and stained by SimplyBlue stain (Fig. 3E). The protein bands at the correct molecular mass (Fig. 3E, arrow) were excised and subjected to mass spectrometric analysis. We also ran the RIPA-insoluble fraction of mouse lungs on polyacrylamide gels before chymotryptic digestion and excised the ≈80-kDa band for mass spectrometric analysis. TG2 appeared in both analyses as the top candidate protein (Fig. 3F; and see Table 1, which is published as supporting information on the PNAS web site). Therefore, we investigated further whether TG2 is a true binding partner of GPR56.
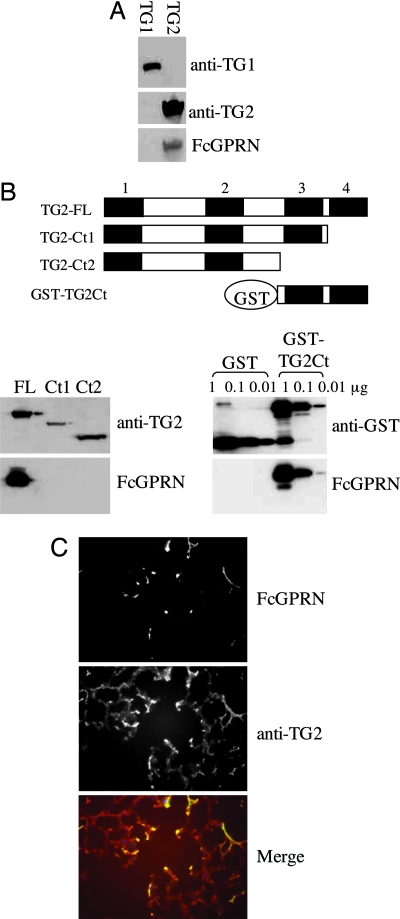
We first transiently expressed murine TG1 and -2 in 293T cells by cDNA transfection. The lysates from transfected cells were probed for FcGPRN binding in an overlay assay. We found that GPR56-binding protein was present only in TG2-transfected cells and not in TG1-transfected cells (Fig. 4A), strongly suggesting that TG2 is a binding partner of GPR56. TG2 consists of four protein domains: an N-terminal β-sandwich domain, a catalytic core, and two C-terminal β-barrel domains. Mass spectrometric analyses showed that the ≈28-kDa fragment recognized by FcGPRN is located at the C terminus of TG2 (Fig. 3F), so we tested whether the C-terminal domains of TG2 are necessary or sufficient for its binding to FcGPRN. When the C-terminal β-barrel domains of TG2 were deleted, ability to bind FcGPRN was lost, indicating that the C terminus of TG2 is necessary for its binding to FcGPRN (Fig. 4B). To investigate whether the C-terminal β-barrel domains are also sufficient to mediate the binding of TG2 to FcGPRN, we fused them to GST and expressed and purified the fusion protein from Escherichia coli. This fusion protein could still bind to FcGPRN in the overlay assay (Fig. 4B), suggesting that the C-terminal β-barrel domains are sufficient for the binding of TG2 to FcGPRN. Furthermore, on tissue sections, FcGPRN showed overlapping expression patterns with the extracellular form of TG2 (Fig. 4C), supporting again the conclusion that TG2 is a true binding partner of GPR56 in the extracellular matrix.
Fig. 4.
TG2 is a binding partner of GPR56. (A) FcGPRN recognizes murine TG2, but not TG1, from transfected 293T cells. The 293T cells were transiently transfected with murine TG1 or TG2 in pCMV-SPORT6 vector for 48 h, and total cell lysates were run on polyacrylamide gels and probed with anti-TG1 antibody, anti-TG2 antibody, or FcGPRN. (B) The C-terminal TG2 domains are both necessary and sufficient for the binding of GPR56. The TG2 protein and its fragments tested for FcGPRN binding are schematically shown. The four conserved domains are presented as four black boxes and numbered as 1–4. Domain 1, N-terminal β-sandwich domain; domain 2, catalytic core; domains 3 and 4, the two C-terminal β-barrel domains. The sequences between domains are shown as white boxes. TG2-FL, full-length TG2; TG2-Ct1, TG2 protein lacking the last β-barrel domain; TG2-Ct2, TG2 protein lacking the last two β-barrel domains; and GST-TG2Ct, the last two β-barrel domains were expressed in E. coli as a GST fusion protein. FcGPRN binds to the C terminus of TG2 as well as the full-length TG2 but not to TG1 or truncated TG2 lacking C-terminal domain(s). The C terminus of TG2 is also sufficient to mediate the binding between TG2 and FcGPRN. (C) FcGPRN and anti-TG2 antibody show overlapping staining patterns on lung sections. In the merged picture, green is from FcGPRN staining, and red is from anti-TG2 staining.
Discussion
GPR56, an Orphan GPCR, Suppresses Metastasis.
We report that a GPCR, GPR56, is down-regulated in tumors from several highly metastatic melanoma cell lines compared with the poorly metastatic parental cells. We also show that GPR56 plays important roles in suppressing tumor growth and metastasis in our experimental model: overexpression of GPR56 leads to suppression of tumor growth and metastasis of melanoma, and reduction of GPR56 leads to an enhancement of tumor growth and metastasis. Because cells with altered levels of GPR56 grow at similar rates in vitro, the suppression by GPR56 must be mediated by factors in the tissue or tumor microenvironment. Indeed, through a series of biochemical purifications, we discovered that the extracellular portion of GPR56 binds to TG2, an extracellular matrix protein ubiquitously expressed in tissues and organs. These results provide a starting point for investigating the crosstalk between tumor cells and stroma and its effects on tumor progression.
GPCRs play important roles in a wide range of biological processes, which render them the main therapeutic targets in various diseases (10). Their roles during tumor progression have also been reported. For example, chemokine receptors, such as CXCR4 and CXCR7 (11), and protease-activated receptors (12–14), were both reported to enhance metastasis. In contrast, we found that GPR56 plays an inhibitory role during tumor progression. Consistent with our results, GPR56 was discovered recently to be up-regulated by the tumor suppressor gene, VHL, in renal carcinoma cell lines (15). We are currently testing whether GPR56 suppresses metastasis in naturally occurring melanoma by using spontaneous melanoma models. It should also be noted that the function of GPR56 might be different in different cancer types or stages. In two recent publications, GPR56 has been reported to be up-regulated in gliomas and esophageal squamous cell carcinoma (16, 17), although the effects of malignant progression were not examined.
GPR56 Is a GPCR Implicated in Multiple Biological Processes.
GPR56 belongs to a recently described family of ≈30 GPCRs called long N-terminal class B, 7-transmembrane proteins (LNB-7TM) (8, 18, 19). The LNB-7TM proteins are hypothesized to function in cell–cell or cell–matrix adhesion and in G protein-coupled signaling, because their long N termini often contain motifs or domains involved in cell adhesion. LNB-7TM proteins are thought to function in a range of biological processes, such as leukocyte trafficking (20), angiogenesis (21), and establishment of cell polarity (22). Their functions and mechanisms of action, however, remain largely unknown.
GPR56 has also been shown to be involved in brain development. Mutations at the N terminus of GPR56 cause a brain cortical malformation called bilateral frontoparietal polymicrogyria in human patients (23). The patients have abnormally numerous and small gyri in their cerebral cortex and are mentally retarded. GPR56 mRNA is preferentially expressed in the neuronal progenitor cells (23) as well as in hematopoietic stem cells (24, 25). Therefore, GPR56 may function to control the proliferation of pluripotent cells of different origins. Such a function could be similar to its role in melanoma progression.
These possibilities raise questions as to how GPR56 might affect cell proliferation and tumor progression. As a GPCR, GPR56 is likely to activate signal transduction pathways. GPR56 has been reported to interact with Gαq/11 and with tetraspanins CD9 and CD81 (26). However, little is known about the signal transduction properties of GPR56 and other LNB-7TM proteins (19), and significant work will be required to explore this issue. Key to any such future investigations will be identification of a ligand of GPR56.
GPR56 Interacts with TG2 in the Extracellular Matrix.
Our data show that the N terminus of GPR56 interacts with the C terminus of TG2 in the extracellular space. TG2, tissue TG, was the first TG recognized, based on its ability to catalyze the incorporation of primary amines into proteins in a Ca2+-dependent manner (27, 28). TG2 is localized both intracellularly and extracellularly. In the cytosol, it reportedly functions as a GTP-binding protein (29). Upon secretion, TG2 is activated by the high level of Ca2+ in the extracellular space and functions as a cross-linking enzyme in the matrix (28). There are numerous reports of down-regulation of TG2 in aggressive tumors and metastases (30–33). Recombinant TG2 applied to rat mammary adenocarcinomas implanted in dorsal skin window chambers produced significant growth delay in the tumors (34), and transfection of TG2 into a highly malignant hamster fibrosarcoma cell line led to significant reduction of tumor incidence (35). A recent report showed that exogenous TG2 inhibited angiogenesis and tumor growth, and tumor growth in TG2 knockout mice was enhanced (36). These results implicate TG2, like GPR56, as a suppressor of tumor growth.
TG2 has a number of properties that could contribute to its tumor-suppressive role. It acts to cross-link a wide variety of extracellular matrix proteins (28), it is up-regulated during wound healing (37–40), and has been implicated in several steps of wound healing (39). Levels of active TG2 are up-regulated at the tumor–stroma interface (34). TG2 promotes incorporation of TGFβ into matrix and its activation (41) and is, itself, up-regulated by TGFβ (42). TGFβ also up-regulates levels of extracellular matrix and integrins (43), and TGFβ is a well known suppressor of tumor growth and progression (44). TG2 is also known to bind to both matrix proteins such as fibronectin (45, 46) and integrins (47), independent of its enzymatic activities. These interactions promote cell adhesion and spreading as well as fibronectin fibril formation (42) and focal contact assembly (47). There is, therefore, a complex network of interactions among TG2, TGFβ, integrins, fibronectin, and other matrix proteins that has the net result of promoting normalization of cell behavior that could well contribute to suppression of tumor growth and metastasis.
Materials and Methods
For a detailed discussion, see Supporting Materials and Methods.
Cell Lines.
The A375 cell line (#CRL-1619; American Type Culture Collection) and derivatives and HEK293T cells were maintained in DMEM with 10% FBS and 2 mM glutamine. A375eco cells were generated by stable transfection of A375 cells with a plasmid containing the ecotropic receptor and selected for neomycin-resistance in the presence of 1.4 mg/ml G418.
Proliferation Assay.
MC-1, MC-1(pMIG), or MC-1(pMIG-GPR) cells were plated at 12,500 cells per well on 6-well plates, and the A375-RNAi series were plated at 20,000 cells per well on 12-well plates. The cells were trypsinized and counted every 24 h. Each time point was done in triplicate, and data were presented as the mean ± SD by using Microsoft excel software.
Transfection and Retroviral Infection.
Retrovirus was produced by transfecting the Phoenix packaging cell line with the corresponding plasmid DNA. The infected cells were selected 48 h later by either FACS for GFP-positive cells (for pMIG- or pLIPE-based virus) or by resistance to puromycin (2.5–7.5 μg/ml for pSIRISP-based virus) (see Supporting Materials and Methods for details of vectors). The selected cells were then pooled and amplified for subsequent analyses. To study the suppressive role of GPR56 in metastatic growth, two independent GPR56-overexpression lines (and the respective control cells) generated by separate viral infections were used. For transient transfections, HEK293T cells were transfected with 0.4 μg of plasmid DNA by using Effectene transfection reagent (Qiagen, Valencia, CA). Cells were directly lysed with Laemmli sample buffer 48 h later for Western blot analyses.
Anti-GPR56 Antibodies and FcGPRN Protein Production.
The C-terminal peptide of GPR56 (CGGPSPLKSNSDSARLPISSGSTSSSRI) and the N-terminal peptide of GPR56 (RSSLHYKPTPDLRISIENSEEALTVC) were synthesized, conjugated to keyhole limpet hemocyanin, and injected into rabbits for antiserum production (Covance Research Products, Denver, PA). The antisera were purified through columns with peptide cross-linked to Sulfolink coupling gel (Pierce) and eluted with 0.2 M glycine (pH 2.5), 0.5 M NaCl, and 0.01% BSA. FcGPRN was expressed in MC-1 cells from the pLIPE retroviral vector. Supernatant was collected 2 days after the cells became confluent and purified through a protein G column.
Supplementary Material
Acknowledgments
We thank Aaron Cook for i.v. injections; Denise Crowley for processing tissue and tumor sections; Jiqing Ye for technical advice and assistance with the biochemical purification of TG2; and Sophie Astrof, Marc Barry, Kaan Certel, Adam Lacy-Hulbert, and Arjan Van Der Flier for critical reading of the manuscript. This work was supported by National Institutes of Health Grant CA17007 (to R.O.H.), the Virginia and Daniel K. Ludwig Fund for Cancer Research (R.O.H.), the Howard Hughes Medical Institute (R.O.H.), and an Anna Fuller Postdoctoral Fellowship (to L.X.).
Abbreviations
- GPCR
G protein-coupled receptor
- RNAi
RNA interference
- TG
transglutaminase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Fidler I. J. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald I. C., Groom A. C., Chambers A. F. BioEssays. 2002;24:885–893. doi: 10.1002/bies.10156. [DOI] [PubMed] [Google Scholar]
- 3.Clark E. A., Golub T. R., Lander E. S., Hynes R. O. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 4.Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordon-Cardo C., Guise T. A., Massague J. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 5.Li C., Wong W. H. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozlowski J., Hart I. R., Fidler I. J., Hanna N. J. Natl. Cancer Inst. 1984;72:913–917. [PubMed] [Google Scholar]
- 7.Zendman A. J., Cornelissen I. M., Weidle U. H., Ruiter D. J., van Muijen G. N. FEBS Lett. 1999;446:292–298. doi: 10.1016/s0014-5793(99)00230-6. [DOI] [PubMed] [Google Scholar]
- 8.Fredriksson R., Gloriam D. E., Hoglund P. J., Lagerstrom M. C., Schioth H. B. Biochem. Biophys. Res. Commun. 2003;301:725–734. doi: 10.1016/s0006-291x(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 9.Volynski K. E., Silva J. P., Lelianova V. G., Atiqur Rahman M., Hopkins C., Ushkaryov Y. A. EMBO J. 2004;23:4423–4433. doi: 10.1038/sj.emboj.7600443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierce K. L., Premont R. T., Lefkowitz R. J. Nat. Rev. Mol. Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 11.Muller A., Homey B., Soto H., Ge N., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N., et al. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 12.Camerer E., Qazi A. A., Duong D. N., Cornelissen I., Advincula R., Coughlin S. R. Blood. 2004;104:397–401. doi: 10.1182/blood-2004-02-0434. [DOI] [PubMed] [Google Scholar]
- 13.Pei D. Cancer Cell. 2005;7:207–208. doi: 10.1016/j.ccr.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Boire A., Covic L., Agarwal A., Jacques S., Sherifi S., Kuliopulos A. Cell. 2005;120:303–313. doi: 10.1016/j.cell.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Maina E. N., Morris M. R., Zatyka M., Raval R. R., Banks R. E., Richards F. M., Johnson C. M., Maher E. R. Oncogene. 2005;24:4549–4558. doi: 10.1038/sj.onc.1208649. [DOI] [PubMed] [Google Scholar]
- 16.Shashidhar S., Lorente G., Nagavarapu U., Nelson A., Kuo J., Cummins J., Nikolich K., Urfer R., Foehr E. D. Oncogene. 2005;24:1673–1682. doi: 10.1038/sj.onc.1208395. [DOI] [PubMed] [Google Scholar]
- 17.Sud N., Sharma R., Ray R., Chattopadhyay T. K., Ralhan R. Cancer Lett. 2006;233:265–270. doi: 10.1016/j.canlet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Stacey M., Lin H. H., Gordon S., McKnight A. J. Trends Biochem. Sci. 2000;25:284–289. doi: 10.1016/s0968-0004(00)01583-8. [DOI] [PubMed] [Google Scholar]
- 19.Kwakkenbos M. J., Kop E. N., Stacey M., Matmati M., Gordon S., Lin H. H., Hamann J. Immunogenetics. 2004;55:655–666. doi: 10.1007/s00251-003-0625-2. [DOI] [PubMed] [Google Scholar]
- 20.Leemans J. C., te Velde A. A., Florquin S., Bennink R. J., de Bruin K., van Lier R. A., van der Poll T., Hamann J. J. Immunol. 2004;172:1125–1131. doi: 10.4049/jimmunol.172.2.1125. [DOI] [PubMed] [Google Scholar]
- 21.Yoon K. C., Ahn K. Y., Lee J. H., Chun B. J., Park S. W., Seo M. S., Park Y. G., Kim K. K. Gene Ther. 2005;12:617–624. doi: 10.1038/sj.gt.3302442. [DOI] [PubMed] [Google Scholar]
- 22.Das G., Reynolds-Kenneally J., Mlodzik M. Dev. Cell. 2002;2:655–666. doi: 10.1016/s1534-5807(02)00147-8. [DOI] [PubMed] [Google Scholar]
- 23.Piao X., Hill R. S., Bodell A., Chang B. S., Basel-Vanagaite L., Straussberg R., Dobyns W. B., Qasrawi B., Winter R. M., Innes A. M., et al. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 24.Terskikh A. V., Easterday M. C., Li L., Hood L., Kornblum H. I., Geschwind D. H., Weissman I. L. Proc. Natl. Acad. Sci. USA. 2001;98:7934–7939. doi: 10.1073/pnas.131200898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terskikh A. V., Miyamoto T., Chang C., Diatchenko L., Weissman I. L. Blood. 2003;102:94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- 26.Little K. D., Hemler M. E., Stipp C. S. Mol. Biol. Cell. 2004;15:2375–2387. doi: 10.1091/mbc.E03-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar N. K., Clarke D. D., Waelsch H. Biochim. Biophys. Acta. 1957;25:451–452. doi: 10.1016/0006-3002(57)90512-7. [DOI] [PubMed] [Google Scholar]
- 28.Lorand L., Graham R. M. Nat. Rev. Mol. Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 29.Nakaoka H., Perez D. M., Baek K. J., Das T., Husain A., Misono K., Im M. J., Graham R. M. Science. 1994;264:1593–1596. doi: 10.1126/science.7911253. [DOI] [PubMed] [Google Scholar]
- 30.Barnes R. N., Bungay P. J., Elliott B. M., Walton P. L., Griffin M. Carcinogenesis. 1985;6:459–463. doi: 10.1093/carcin/6.3.459. [DOI] [PubMed] [Google Scholar]
- 31.Birckbichler P. J., Bonner R. B., Hurst R. E., Bane B. L., Pitha J. V., Hemstreet G. P., III Cancer. 2000;89:412–423. doi: 10.1002/1097-0142(20000715)89:2<412::aid-cncr29>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.Knight C. R., Rees R. C., Elliott B. M., Griffin M. Biochim. Biophys. Acta. 1990;1053:13–20. doi: 10.1016/0167-4889(90)90019-a. [DOI] [PubMed] [Google Scholar]
- 33.Hand D., Elliott B. M., Griffin M. Biochim. Biophys. Acta. 1987;930:432–437. doi: 10.1016/0167-4889(87)90016-4. [DOI] [PubMed] [Google Scholar]
- 34.Haroon Z. A., Lai T. S., Hettasch J. M., Lindberg R. A., Dewhirst M. W., Greenberg C. S. Lab. Invest. 1999;79:1679–1686. [PubMed] [Google Scholar]
- 35.Johnson T. S., Knight C. R., el-Alaoui S., Mian S., Rees R. C., Gentile V., Davies P. J., Griffin M. Oncogene. 1994;9:2935–2942. [PubMed] [Google Scholar]
- 36.Jones R. A., Kotsakis P., Johnson T. S., Chau D. Y., Ali S., Melino G., Griffin M. Cell Death Differ. 2005 doi: 10.1038/sj.cdd.4401816. 10.1038/sj.cdd.4401816. [DOI] [PubMed] [Google Scholar]
- 37.Bowness J. M., Tarr A. H., Wong T. Biochim. Biophys. Acta. 1988;967:234–240. doi: 10.1016/0304-4165(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 38.Bowness J. M., Henteleff H., Dolynchuk K. N. Connect. Tissue Res. 1987;16:57–70. doi: 10.3109/03008208709001994. [DOI] [PubMed] [Google Scholar]
- 39.Verderio E. A., Johnson T., Griffin M. Amino Acids. 2004;26:387–404. doi: 10.1007/s00726-004-0094-4. [DOI] [PubMed] [Google Scholar]
- 40.Haroon Z. A., Hettasch J. M., Lai T. S., Dewhirst M. W., Greenberg C. S. FASEB J. 1999;13:1787–1795. doi: 10.1096/fasebj.13.13.1787. [DOI] [PubMed] [Google Scholar]
- 41.Verderio E., Gaudry C., Gross S., Smith C., Downes S., Griffin M. J. Histochem. Cytochem. 1999;47:1417–1432. doi: 10.1177/002215549904701108. [DOI] [PubMed] [Google Scholar]
- 42.Akimov S. S., Belkin A. M. J. Cell Sci. 2001;114:2989–3000. doi: 10.1242/jcs.114.16.2989. [DOI] [PubMed] [Google Scholar]
- 43.Ignotz R. A., Heino J., Massague J. J. Biol. Chem. 1989;264:389–392. [PubMed] [Google Scholar]
- 44.Derynck R., Akhurst R. J., Balmain A. Nat. Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 45.Lorand L., Dailey J. E., Turner P. M. Proc. Natl. Acad. Sci. USA. 1988;85:1057–1059. doi: 10.1073/pnas.85.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hang J., Zemskov E. A., Lorand L., Belkin A. M. J. Biol. Chem. 2005;280:23675–23683. doi: 10.1074/jbc.M503323200. [DOI] [PubMed] [Google Scholar]
- 47.Akimov S. S., Krylov D., Fleischman L. F., Belkin A. M. J. Cell Biol. 2000;148:825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.