Abstract
Mitochondrial impairment is hypothesized to contribute to the pathogenesis of insulin resistance. Mitofusin (Mfn) proteins regulate the biogenesis and maintenance of the mitochondrial network, and when inactivated, cause a failure in the mitochondrial architecture and decreases in oxidative capacity and glucose oxidation. Exercise increases muscle mitochondrial content, size, oxidative capacity and aerobic glucose oxidation. To address if Mfn proteins are implicated in these exercise-induced responses, we measured Mfn1 and Mfn2 mRNA levels, pre-, post-, 2 and 24 h post-exercise. Additionally, we measured the expression levels of transcriptional regulators that control mitochondrial biogenesis and functions, including PGC-1α, NRF-1, NRF-2 and the recently implicated ERRα. We show that Mfn1, Mfn2, NRF-2 and COX IV mRNA were increased 24 h post-exercise, while PGC-1α and ERRα mRNA increased 2 h post-exercise. Finally, using in vitro cellular assays, we demonstrate that Mfn2 gene expression is driven by a PGC-1α programme dependent on ERRα. The PGC-1α/ERRα-mediated induction of Mfn2 suggests a role of these two factors in mitochondrial fusion. Our results provide evidence that PGC-1α not only mediates the increased expression of oxidative phosphorylation genes but also mediates alterations in mitochondrial architecture in response to aerobic exercise in humans.
Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) is a nuclear transcriptional coactivator that regulates several important metabolic processes, including mitochondrial biogenesis, adaptive thermogenesis, respiration, insulin secretion and gluconeogenesis (Knutti & Kralli, 2001; Puigserver & Spiegelman, 2003; Kelly & Scarpulla, 2004). PGC-1α induces mitochondrial biogenesis by coactivating specific transcription factors, such as the nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) (Wu et al. 1999) and the nuclear receptor oestrogen receptor alpha (ERRα) (Mootha et al. 2004; Schreiber et al. 2004). The expression of PGC-1α mRNA is prominent in tissues with high energy demands, such as heart and skeletal muscle (Knutti & Kralli, 2001; Puigserver & Spiegelman, 2003), and is induced in response to signals of metabolic stress, such as exercise (Goto et al. 2000; Baar et al. 2002; Pilegaard et al. 2003; Russell et al. 2003). The activation and/or overexpression of PGC-1α in cells and rodents regulates the expression of genes involved in mitochondrial biogenesis and oxidative phosphorylation (Wu et al. 1999; Lin et al. 2002; Mootha et al. 2004; Schreiber et al. 2004). Although an increase in human skeletal muscle PGC-1α mRNA is associated with exercise it has yet to be established whether this is a causal relationship for the regulation of human muscle biogenesis.
The impairment of mitochondrial function has been suggested to be a crucial factor in the pathogenesis of insulin resistance (Kelley et al. 2002). PGC-1α expression, as well as several other oxidative phosphorylation genes, have been claimed to be reduced in subjects with diabetes and insulin resistance, when compared with healthy subjects (Mootha et al. 2003; Patti et al. 2003). However, neither of these studies adequately controlled for physical activity levels making the interpretation of the data complex. Although reductions in PGC-1α and PGC-1α-dependent pathways seem plausible to contribute to the impaired bioenergetic capacity of skeletal muscle mitochondria observed in type II diabetes (Kelley et al. 2002; Mootha et al. 2003; Patti et al. 2003), further investigations are required.
Endurance exercise is used as an intervention for the treatment of type II diabetes because it increases mitochondrial content, size and skeletal muscle oxidative capacity (Holloszy & Coyle, 1984), as well as improves insulin sensitivity (Dela et al. 1992). The exercise-induced adaptations are likely to be due, in part, to an increase in PGC-1α expression (Goto et al. 2000; Baar et al. 2002; Pilegaard et al. 2003; Russell et al. 2003; Short et al. 2003) and the effect of PGC-1α on the aforementioned metabolic functions. Exercise is also reported to lead to the increased expression of some of the transcription factors that cooperate with PGC-1α, such as NRF-1 and NRF-2 (Murakami et al. 1998; Baar et al. 2002; Short et al. 2003). The effects of exercise on the expression of ERRα, which was recently implicated in the process of mitochondrial biogenesis, have not yet been studied.
Skeletal muscle mitochondria from insulin resistant obese compared with healthy subjects also demonstrates a reduced gene and protein expression of mitofusin-2 (Mfn2), a key player involved in the formation and maintenance of the mitochondrial network (Bach et al. 2003). Inactivation of Mfn2 causes a dramatic failure in mitochondrial architecture, due to the lack of mitochondrial fusion, and leads to a significant decrease in oxidative capacity and glucose oxidation (Bach et al. 2003). Because exercise increases not only mitochondrial biogenesis but also oxidative capacity, we tested the hypothesis that exercise may also increase the expression of Mfn2 and/or of the related isoform, Mfn1.
We report here for the first time that acute exercise increases the expression of Mfn1, Mfn2 and ERRα mRNA in human skeletal muscle. We also demonstrate in vitro that Mfn2 can be induced directly by the cooperative actions of PGC-1α and ERRα. Our findings suggest that the metabolic signals that are initiated by contracting skeletal muscle are capable of stimulating the transcription of genes involved not only in mitochondrial biogenesis, but also in mitochondrial fusion, possibly via a PGC-1α/ERRα-dependent pathway.
Methods
Subject details
The subjects consisted of 11 trained male cyclists, 36 ± 4.9 years of age (mean ±s.d.), maximal oxygen consumption 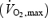 57.6 ± 7 ml kg−1 min−1, mass 75.3 ± 9 kg, body fat 16.4 ± 4.5%. An additional five subjects, matched for age (34 ± 7.2 years) and mass (78.1 ± 4 kg), were included as a control group for the effect of time. The control group did not complete the exercise trials but did undergo the muscle biopsies at the same time intervals as the exercising subjects. The entire study was approved by the local medical society ethical committee and all participants gave their informed consent and agreed to muscle biopsies and, where required, the physiological testing. The study conformed to the Declaration of Helsinki.
57.6 ± 7 ml kg−1 min−1, mass 75.3 ± 9 kg, body fat 16.4 ± 4.5%. An additional five subjects, matched for age (34 ± 7.2 years) and mass (78.1 ± 4 kg), were included as a control group for the effect of time. The control group did not complete the exercise trials but did undergo the muscle biopsies at the same time intervals as the exercising subjects. The entire study was approved by the local medical society ethical committee and all participants gave their informed consent and agreed to muscle biopsies and, where required, the physiological testing. The study conformed to the Declaration of Helsinki.
Measurement of oxygen consumption
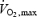 was measured using a Quark B2 metabolic cart (Cosmed, Rome, Italy) while subjects were cycling on an ergometer (Ergoline 900, Sensor Medic, Bitz, Germany). The subjects began cycling at a power of 90 W. The power was increased by 30 W every 3 min until the subject could not maintain a minimal revolution of 75 r.p.m. At the end of each step, lactate concentration was obtained (Lactate Pro, Axon Lab, Baden, Switzerland). The duration of the test lasted between 20 and 30 min. Heart rate (Polar, LMT Leuenberger Medizintechnik, Wallisellen, Switzerland) and oxygen consumption were measured continually throughout the test.
was measured using a Quark B2 metabolic cart (Cosmed, Rome, Italy) while subjects were cycling on an ergometer (Ergoline 900, Sensor Medic, Bitz, Germany). The subjects began cycling at a power of 90 W. The power was increased by 30 W every 3 min until the subject could not maintain a minimal revolution of 75 r.p.m. At the end of each step, lactate concentration was obtained (Lactate Pro, Axon Lab, Baden, Switzerland). The duration of the test lasted between 20 and 30 min. Heart rate (Polar, LMT Leuenberger Medizintechnik, Wallisellen, Switzerland) and oxygen consumption were measured continually throughout the test. 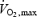 was calculated as the highest value averaged over a 30 second period (Russell et al. 2003).
was calculated as the highest value averaged over a 30 second period (Russell et al. 2003).
Fat free mass
Fat free mass was determined by plethysmographic measurement of body volume using the BOD POD Body Composition System (Life Measurement Instruments, Concord, USA) (Dempster & Aitkens, 1995). This device uses the relationship between pressure and volume to derive the body volume of a subject seated inside a fiberglass chamber.
Experimental procedure
The subjects performed a 10 km cycling time trial that included an increase in altitude from 500 to 1250 m. This 10 km route was chosen because it was familiar to all of the subjects participating in the study. The time trial for all subjects was performed on the same day. The diet of all the participants was matched 24 h before and after the test. The liquid consumption during and after the test was restricted to water. Muscle biopsies and blood samples were obtained at four time points: a week before the exercise trial, immediately after exercise, and 2 and 24 h post-exercise.
Muscle biopsy technique
Skeletal muscle samples were obtained under local anaesthesia (Rapidocaine, 1% plain) from the belly of the vastus lateralis muscle using a percutaneous needle biopsy technique (Pro-Mag, Medical Device Technologies Inc., Gainsville, FL, USA). An incision was made in the skin and three individual muscle samples were taken at each time point. The four muscle biopsies were taken from different incisions approximately 1.5–2.0 cm apart. The muscle samples were immediately frozen in liquid nitrogen and used for RNA (for samples for all four time points) and mitochondrial protein extraction (for samples pre- and 24 h post-exercise).
Cell culture and infections
SAOS2 cells cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 9% fetal calf serum were infected on day 1 with adenoviruses expressing siRNA for ERRα or the control AdSUPER at a multiplicity of infection of 100, and on day 4 with adenoviral vectors expressing either green fluorescent protein (GFP) or PGC-1α at a multiplicity of infection of 40. RNA and protein were isolated 24 h after the infection with GFP/PGC-1α viruses. All vectors have been previously described (Schreiber et al. 2003).
RNA extraction and real time quantitative PCR
RNA from skeletal muscle (approximately 15 mg of muscle) was extracted using a commercially available preparation, peqGOLD Tri-Fast (Peqlab, Germany). Three micrograms of RNA was reverse transcribed to cDNA using Random Hexomer primers and a Stratascript enzyme (Stratagene, the Netherlands). RNA from the SAOS2 cells was extracted using Trizol and reverse transcribed using a Superscript enzyme (Invitrogen, USA). Real-time PCR was performed using an MX3000p thermal cycler system and Brilliant SYBER Green QPCR Master Mix (Stratagene, the Netherlands). The PCR conditions for all genes consisted of one denaturing cycle at 90°C for 10 min, followed by 40 cycles consisting of denaturing at 90°C for 30 s, annealing at 60°C for 60 s and elongation at 72°C for 60 s. At the end of the PCR the samples were subjected to a melting curve analysis. To control for any variations due to efficiencies of the reverse transcription and PCR, 18S and acidic ribosomal phosphoprotein PO (36B4) were used as internal controls for the skeletal muscle and SAOS2 RNA, respectively. mRNA expression was calculated as follows. The number of cycles at which the best-fit line through the log-linear portion of each amplified curve intersects the noise band is inversely proportional to the log copy number (Higuchi et al. 1993). This value is refereed to as the critical threshold (CT) value. The ΔCT was calculated by subtracting the CT for 18S, or 36B4, from the CT for the gene of interest. The relative expression of the gene of interest is calculated using the expression 2−ΔCT and reported as arbitrary units. All PCR runs were performed in triplicate. PCR primer sequences are provided in Table 1.
Table 1.
Primer sequences and annealing temperatures used for the PCR
| Gene | Sequence 5′–3′ | Temp |
|---|---|---|
| Mfn1 | sense TGT TTT GGT CGC AAA CTC TG | 60 |
| Anti CTG TCT GCG TAC GTC TTC CA | ||
| Mfn2 | sense ATG CAT CCC CAC TTA AGC AC | 60 |
| Anti CCA GAG GGC AGA ACT TTG TC | ||
| PGC-1α | sense TCA GTC CTC ACT GGT GGA CA | 62 |
| Anti TGC TTC GTC GTC AAA AAC AG | ||
| ERRα | Sense TTCTCATCGCTGTCGCTGTCT | 64 |
| Anti CAGCCGCCGCACTAGTTG | ||
| NRF-1 | Sense GGT GCA GCA CCT TTG GAG AA | 60 |
| Anti CCA GAG CAG ACT CCA GGT CTT C | ||
| NRF-2 | Sense CAA GAA CGC CTT GGG ATA CC | 60 |
| Anti AAA CCA CCC AAT GCA GGA CTT | ||
| COXIV | Sense CAT GTG GCA GAA GCA CTA TGT GT | 60 |
| Anti GCC ACC CAC TCT TTG TCA AAG | ||
| 18S | Sense GAG GAT GAG GTG GAA CGT GT | 60 |
| Anti GGA CCT GGC TGT ATT TTC CA | ||
| 36B4 | Sense GTG ATG TGC AGC TGA TCA AGA CT | 60 |
| Anti GAT GAC CAG CCC AAA GGA GA |
Mfn1, mitofusin-1; Mfn2, mitofusin-2; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; ERRα, oestrogen related receptor-α; NRF-1, nuclear respiratory factor-1; NRF-2, nuclear respiratory factor-2; COXIV, cytochrome c oxidase subunit IV; 18S, ribosomal 18S; 36B4, acidic ribosomal phosphoprotein PO.
Mitochondrial extraction
Mitochondria were extracted from approximately 25 mg of skeletal muscle. The muscle was immersed in ice-cold medium buffer A (0.1 m KCl, 5 mm MgCl2, 5 mm EGTA, 5 mm sodium pyrophosphate, pH adjusted to 7.4, 2 μm leupeptin, 2 μm pepstatin, 0.5 mm phenylmethylsulphonylfluoride (PMSF) for 2 × 10 min. Following this, the muscle was finely minced in 1/10 (w/v) buffer B (0.25 m sacharose, 50 mm KCl, 5 mm EDTA, 1 mm sodium pyrophosphate, 5 mm MgCl2, pH adjusted to 6.8, 2 μm leupeptin, 2 μm pepstatin, 0.5 mm PMSF) and then disrupted with a motor driven Teflon/glass homogenizer. The entire procedure was performed at 0–4°C. The homogenate was centrifuged at 1300 g for 10 min at 4°C, and the supernatant was removed and kept on ice (SN1). The pellet was resuspended with buffer B and centrifuged again at 1300 g for 10 min at 4°C. The second supernatant obtained (SN2) was mixed with the SN1 and centrifuged at 9000 g for 15 min at 4°C. The mitochondrial pellet was re-suspended in buffer B and protein concentration determined using a Micro BCA protein assay (Pierce, Rockford, IL, USA).
Western blotting
Forty micrograms of muscle mitochondrial protein fractions were subjected to Western analysis using an antibody raised against Mfn2 (Bach et al. 2003). Electrophoresis was performed using a 10% SDS PAGE gel in cold (4°C) buffer containing 25 mm Tris pH 8.8, 192 mm glycine and 20% methanol. After protein transfer, PVDF membranes were blocked with 5% non-fat dry milk in phosphate-buffered saline (PBS) containing 0.05% Tween-20, and thereafter antibody incubation was performed with gentle shaking overnight at 4°C at a dilution of 1: 400 in the 5% non-fat dry milk in 0.5% Tween-20/PBS. After the primary antibody incubation, the membranes were incubated at room temperature for 60 min with a horseradish peroxidase-conjugated goat antirabbit IgG (Cell Signalling, Beverly, MA, USA) at a dilution of 1: 2000. The membranes were then washed for 4 × 5 min in 0.05% Tween-20/PBS and treated for 1 min with chemiluminescence substrate (Pierce, Rockford, IL, USA). Finally, X-ray film was exposed to the PVDF membranes for 1 min. Porin was used to control for the amount of protein loaded into each well (Sigma, Basel). The reaction product of each blot was analysed by densitometry using Scion imageware. SAOS2 protein extracts were prepared by lysing cells in 100 mm Tris pH 7.5, 1% NP40, 250 mm NaCl, 1 mm EDTA buffer. Extracts were then subjected to western analysis using antibodies against PGC-1α (Schreiber et al. 2003) and ERRα (Johnston et al. 1997).
Chromatin immunoprecipitation (ChIP)
SAOS2 cells (∼4.0 × 106) cultured in 15 cm plates were infected with GFP or PGC-1α adenoviruses at a multiplicity of infection of 50. Twenty-four hours post-infection, cells were treated with 1% formaldehyde (cross-linker) for 7.5 min, harvested, and sonicated to ∼500 bp fragments, as described in (Chen et al. 1999). Sonicated chromatin was precleared for 2 h at 4°C with a mixture of protein A-/protein G-sepharose (Amersham Biosciences, Uppsala Sweden. One-tenth of each sample was used to estimate input or ‘total’ DNA. The remaining 9/10 was split three ways and incubated overnight at 4°C with control (anti-GFP), anti-ERRα (generous gift of Dr V. Giguere; Sladek et al. 1997), or anti-PGC-1α antibodies (Schreiber et al. 2003). After an additional incubation with protein A-/protein G-sepharose for 2 h at 4°C, immunocomplexes were processed as previously described (Chen et al. 1999). DNA from each immunoprecipitation was then quantified with real-time PCR, using the Chromo4 system (Bio-Rad, Waltham, USA) and the following primers: 5′-AAATACAGCGGTGGATGTTAGAGA-3′ and 5′-CCAGGCCTAGGGTGAAGTGA-3′ for the Mfn2 gene; 5′-TTTCCAGCCCCCAATCTCA-3′ and 5′-TCAGCGCCACCTGGTTCTT-3′ for the Hsp70 gene. Genomic copy number was determined based on a dilution series of sonicated SAOS2 genomic DNA. Values were normalized against ‘total’ DNA for each sample.
Statistics
A one-way ANOVA with repeated measures, followed by linear contrasts, was used to compare the effects of exercise or time on the expression of Mfn1, Mfn2, PGC-1α, ERRα, NRF-1, NRF-2 and COX IV. A one-way ANOVA, followed by a Tukey-Kramer post hoc test, was used to compare the expression of Mfn1 and Mfn2 in the different infected cells. The α level of significance for the ANOVA was set at 0.05.
Results
Characteristics of the subjects performing the exercise trial are presented in Table 2. The performance time of the 10 km cycling time trial was correlated with 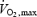 (r=−0.85; P = 0.001), lactate threshold (LT) (r=−0.85; P = 0.001) and maximal power output (Pmax) (r=−0.88; P = 0.0001), which were measured in the laboratory.
(r=−0.85; P = 0.001), lactate threshold (LT) (r=−0.85; P = 0.001) and maximal power output (Pmax) (r=−0.88; P = 0.0001), which were measured in the laboratory. 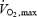 and LT are indicators of physical fitness levels and strong predictors of performance (Grant et al. 1997).
and LT are indicators of physical fitness levels and strong predictors of performance (Grant et al. 1997).
Table 2.
Subjects characteristics (exercising group)
| Variable | Mean ±s.d. |
|---|---|
| Age (years) | 36 ± 4.9 |
| Weight (kg) | 75.3 ± 9.0 |
| Body fat (%) | 16.4 ± 4.5 |
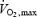 (ml kg−1 min−1) (ml kg−1 min−1) |
57.6 ± 7.0 |
| Pmax (W kg−1) | 4.4 ± 0.5 |
| LT (ml kg−1 min−1) | 46.7 ± 7.5 |
| 10 km time (min) | 46.2 ± 7.4 |
Pmax, maximal power output; LT, lactate threshold.
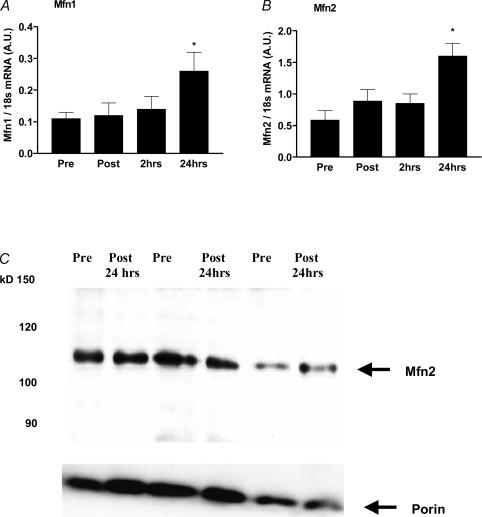
Although it is well known that exercise improves energy metabolism via the stimulation of mitochondrial biogenesis (Irrcher et al. 2003), it is unknown if exercise also regulates the genes involved in mitochondrial fusion. Using quantitative PCR, we show here that Mfn1 and Mfn2 mRNA levels were increased in human skeletal muscle by 2.4-fold and 2.7-fold, respectively (P < 0.05), 24 h after an acute exercise bout (Fig. 1). We could not detect a change in the levels of Mfn2 protein (Fig. 1) at 24 h post-exercise. It is possible that the increases in the Mfn2 protein level are transient and not detectable after 24 h, or that the protein increases at a later time point.
Figure 1. The effect of acute exercise on Mfn1 mRNA (A), Mfn2 mRNA (B) and Mfn2 protein (C) levels measured pre-, post- and 2 and 24 h post-exercise.
Mfn2 protein content was normalized against porin protein content. The blot shown is from 3 subjects representative of the 11 subjects used in the analysis. *P < 0.05, significantly different from pre-exercising levels.
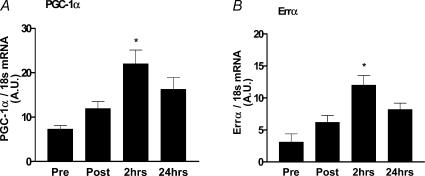
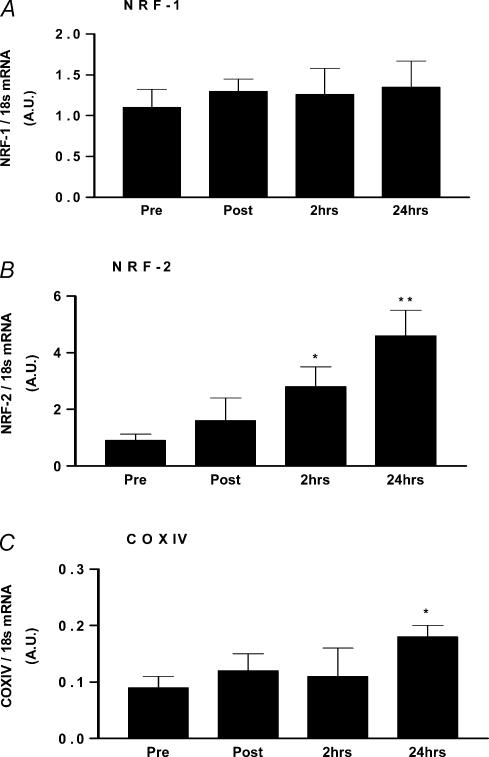
PGC-1α is an important player in mitochondrial biogenesis (Wu et al. 1999) and its expression has been shown to be increased in human muscle after exercise (Pilegaard et al. 2003; Russell et al. 2003; Short et al. 2003). In cell culture systems, PGC-1α is reported to also lead to the induction of some of the transcription factors with which it cooperates in mitochondrial biogenesis, such as NRF-1, NRF-2, and ERRα (Wu et al. 1999; Schreiber et al. 2003). To evaluate which of these transcriptional regulators are induced after a single bout of acute exercise in human muscle, we measured the mRNA levels of PGC-1α, ERRα, NRF-1 and NRF-2. PGC-1α and ERRα mRNA levels increased immediately after exercise, with a peak at 2 h post-exercise (3-fold increase at 2 h, P < 0.05) (Fig. 2). NRF-1 levels did not change significantly at any of the times tested, while NRF-2 levels increased 2 h (3-fold; P < 0.05) and 24 h post-exercise (5-fold; P < 0.01) (Fig. 3). As PGC-1α is also known to up-regulate COX IV (Wu et al. 1999), a nuclear encoded protein of the mitochondrial respiratory chain and a marker of mitochondrial biogenesis (Hood, 2001), we also measured its mRNA expression. We observed a 2-fold increase in COX IV mRNA at 24 h post-exercise (P < 0.05) (Fig. 3). To establish that the changes in gene expression were due to the exercise intervention and not due to a variation of time, a control group completed the muscle biopsy time coarse without exercise. As shown in Table 3 the expression of all genes measured was stable across the four biopsy sampling times demonstrating that the changes in gene expression observed in the present study was due to the exercise intervention employed.
Figure 2. The effect of acute exercise on PGC-1α (A) and ERRα (B) mRNA measured pre-, post-, and 2 and 24 h post-exercise.
*P < 0.05, significantly different from pre exercising levels.
Figure 3. The effect of acute exercise on NRF-1 (A), NRF-2 (B) and COX IV (C) mRNA levels measured pre-, post-, and 2 and 24 h post-exercise.
*P < 0.05, significantly different from pre exercising levels;
**P < 0.01, significantly different from all other time points.
Table 3.
Gene expression values from the non-exercising control group
| Gene | Pre | Post | 2 h | 24 h |
|---|---|---|---|---|
| Mfn1 | 0.09 ± 0.03 | 0.11 ± 0.06 | 0.10 ± 0.09 | 0.11 ± 0.06 |
| Mfn2 | 0.70 ± 0.12 | 0.62 ± 0.21 | 0.83 ± 0.17 | 0.84 ± 0.223 |
| PGC-1α | 7.67 ± 0.9 | 8.21 ± 1.23 | 8.66 ± 0.6 | 7.00 ± 0.92 |
| ERRα | 3.76 ± 0.22 | 3.59 ± 0.34 | 3.99 ± 0.51 | 3.55 ± 0.22 |
| NRF-1 | 1.40 ± 0.38 | 1.00 ± 0.22 | 1.20 ± 0.32 | 1.86 ± 0.25 |
| NRF-2 | 1.23 ± 0.62 | 1.77 ± 0.66 | 2.06 ± 0.52 | 1.99 ± 0.49 |
| COXIV | 0.11 ± 0.06 | 0.09 ± 0.07 | 0.13 ± 0.04 | 0.12 ± 0.01 |
The headings Pre, Post, 2 h and 24 h are with respect to the time intervals between biopsies and correspond to the time intervals for the exercising group. Mfn1, mitofusin-1; Mfn2, mitofusin-2; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; ERRα, oestrogen related receptor-α; NRF-1, nuclear respiratory factor-1; NRF-2, nuclear respiratory factor-2; COXIV, cytochrome c oxidase subunit IV.
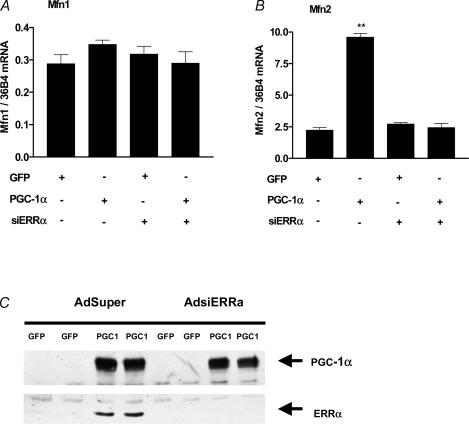
Our in vivo results showed that exercise leads to increases in the gene expression of the mitochondrial fusion proteins Mfn1 and Mfn2, as well as that of PGC-1α and its coactivators of mitochondrial biogenesis, ERRα and NRF-2. We next tested whether Mfn1 and Mfn2 could be regulated by PGC-1α and/or ERRα, using an in vitro cell system. Expression of PGC-1α in SAOS2 cells leads to mitochondrial biogenesis, in a manner that depends on endogenous ERRα, and which is similar to PGC-1α function in a muscle cell line (Mootha et al. 2004; Schreiber et al. 2004). Expression of PGC-1α in SAOS2 cells also led to the induction of Mfn2, but not Mfn1 expression (Fig. 4). The PGC-1α-driven induction of Mfn2 was mediated by the endogenous ERRα, since it was not seen in cells where ERRα expression was inhibited by small interfering (si) RNA (see Fig. 4C for ERRα protein levels). These results suggested that ERRα enables the induction of Mfn2 by PGC-1α, and that PGC-1α and ERRα are not sufficient for the induction of Mfn1. Mfn1 regulation may require a different PGC-1α controlled pathway that is not active in the SAOS2 cells, or a distinct exercise-induced stimulus that is independent of PGC-1α.
Figure 4. PGC-1α and ERRα induce Mfn2.
Mfn1 (A), and Mfn2 (B) mRNA levels were determined 24 h after infection of SAOS2 cells with GFP/PGC-1α viruses. **P < 0.01, significantly different from other infected cells. C, Western blot showing the effect of the siRNA for ERRα on the expression (or levels) of ERRα protein. Each example is shown in duplicate.
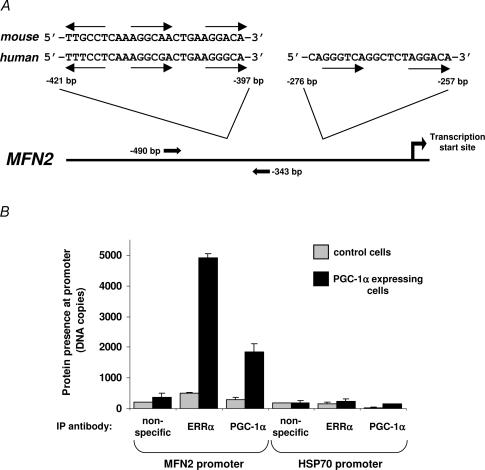
To test if ERRα and PGC-1α act directly at the Mfn2 gene to induce its mRNA expression we asked whether ERRα and PGC-1α interact physically with the Mfn2 promoter. Analysis of the Mfn2 promoter sequence indicated two regions with putative ERRα/nuclear receptor binding sites; the −421/−397 sites in particular are highly conserved in the mouse (top) and human (bottom) genes (Fig. 5A). Using chromatin immunoprecipitation assays, we detected specific binding of both PGC-1α and ERRα at the Mfn2 promoter in SAOS2 cells expression PGC-1α, but not in the control GFP-expressing cells.
Figure 5. ERRα and PGC-1α bind to sequences 5′ of the MFN2 promoter in SAOS2 cells.
A, diagram of the promoter and 5′ sequences of the human MFN2 gene. Two regions with putative ERRα/nuclear receptor binding sites are detailed above the MFN2 diagram; the −421/−397 sites are conserved in the mouse (top) and human (bottom) genes, as indicated. Arrows below the sequence depict putative recognition half-sites for nuclear receptors (consensus being AGGTCA). Numbering is relative to the MFN2 transcription start site. Also indicated are the positions of the oligonucleotides used to amplify the −343/−490 region. B, chromatin immunoprecipitation from SAOS2 cells after infection with GFP- (light grey) or PGC-1α- (dark grey) expressing adenoviruses. Quantitative PCR assays were used to detect the presence of the −343/−490 region of the Mfn2 promoter or, as control, the HSP70 promoter in DNA immunoprecipitated with the indicated antibodies (for ‘non-specific’, samples were immunoprecipitated with an antibody against GFP). Results shown are the mean ±s.e.m. from two of four representative independent experiments.
Discussion
A major adaptation to exercise is the improvement in skeletal muscle mitochondrial function and capacity, which are important determinants of insulin sensitivity. Improvements in mitochondrial function require an increase in the proteins that participate in the generation of a dynamic mitochondrial network. We show in this study that, in response to acute endurance exercise, there is an increase in the gene expression of the mitochondrial fusion proteins Mfn1 and Mfn2, in human skeletal muscle. Additionally, there is an increase in the mRNA levels of the transcriptional coactivator PGC-1α and the PGC-1α-interacting transcription factors ERRα and NRF-2. Notably, this is the first study to report changes in ERRα mRNA levels following exercise. Furthermore, there is an increase in COX IV mRNA, a PGC-1α target gene and marker of mitochondrial biogenesis. We have also show that PGC-1α and ERRα recognize the Mfn2 promoter and induce Mfn2 expression in cultured cells, suggesting that PGC-1α and ERRα mediate the Mfn2 induction by exercise in vivo.
Previous studies in HeLa cells have shown that Mfn1 and Mfn2 are essential for the maintenance of mitochondrial morphology, and that the two mitofusins function co-operatively in mitochondria morphogenesis (Eura et al. 2003). Knock down of Mfn2 in L6E9 myotubes results in a dramatic discontinuity of the mitochondrial network (Bach et al. 2003). Mfn proteins also appear to play a major role in mitochondrial metabolism; the repression of Mfn2 in L6E9 myotubes and 10T1/2 cells reduces glucose oxidation by 30 and 70%, respectively, and oxygen consumption by 30% (Bach et al. 2003). In line with these observations, obese humans, who demonstrate a lower insulin-stimulated glucose disposal and a reduced oxidative capacity, also have 36 and 43%, respectively, lower skeletal muscle Mfn2 mRNA and protein expression (Bach et al. 2003). Exercise improves mitochondrial function in both insulin resistant and healthy subjects by increasing mitochondrial oxidative metabolism. Our results suggest that this adaptation may be, in part, a response to increases in Mfn1 and Mfn2. Although we did not detect an increase in Mfn2 protein content at 24 h post-exercise it is still possible that Mfn2 protein levels either rise transiently or with considerably slower kinetics than the RNA.
PGC-1α has been shown to increase in response to exercise in human (Pilegaard et al. 2003; Russell et al. 2003; Short et al. 2003; Norrbom et al. 2004) and rodent skeletal muscle (Goto et al. 2000; Baar et al. 2002; Terada et al. 2002). PGC-1α is a major player in mitochondrial biogenesis, an adaptation stimulated by exercise, so it would seem likely that PGC-1α is involved in exercise-induced mitochondrial biogenesis. In addition to the role of PGC-1α as a regulator of mitochondrial biogenesis, we demonstrated that PGC-1α-activated programmes are also involved in mitochondrial fusion. PGC-1α can activate several pathways regulating mitochondrial biogenesis. Recently, it was demonstrated that PGC-1α interacts with and activates the transcriptional function of ERRα (Huss et al. 2002; Schreiber et al. 2003); this activation then leads to the induction of ERRα target genes (including ERRα itself), many of which are important for mitochondrial biogenesis (Mootha et al. 2004; Schreiber et al. 2004). Our in vivo results presented here show that, during recovery from acute exercise, there is an increase in both PGC-1α and ERRα mRNA, suggesting that this is an important pathway for the exercise-stimulated mitochondrial biogenesis and mitochondrial function observed in human skeletal muscle. The fast induction of ERRα, parallel to that of PGC-1α, is consistent with ERRα being an early direct target of PGC1, as shown in studies in cultured cells (Schreiber et al. 2003; Laganiere et al. 2004).
Another PGC-1α coactivated circuit that regulates mitochondrial biogenesis includes the nuclear respiratory factors NRF-1 and NRF-2 (Wu et al. 1999). These NRFs activate genes encoding proteins of the respiratory chain, such as β-ATP synthase, COX IV and CytC (Virbasius & Scarpulla, 1991; Kelly & Scarpulla, 2004), which are also targets of PGC-1α/ERRα coactivation (Schreiber et al. 2004). In rodents, NRF-1 and NRF-2 (Murakami et al. 1998; Baar et al. 2002) are up-regulated 6–18 h after a single exercise bout, parallel to increases in PGC-1α (Baar et al. 2002). In humans, NRF-1 and NRF-2 expression does not change after 3 h of single leg exercise nor after 4 weeks of single leg exercise training (Pilegaard et al. 2003). Additionally, NRF-1 did not change after 45 min (Norrbom et al. 2004) or 80 min of exhaustive single leg exercise nor after 4 h of whole body cycling exercise (Pilegaard et al. 2003). It has therefore been suggested that the basal level of NRF-1 in human skeletal muscle, if involved in acute exercise stimulated gene transcription, is sufficient to meet the needs of the cells (Pilegaard et al. 2003). In contrast, 16 weeks of whole body cycling exercise resulted in a small but significant 15% increase in NRF-1 mRNA in human skeletal muscle (Short et al. 2003). Our results show that acute whole body endurance exercise, results in an increase in NRF-2, but not NRF-1, at 2 and at 24 h post-exercise. With respect to exercise, the extent to which NRF-1 and NRF-2 are regulated at the level of expression may depend on the species, the specific muscle, and/or the type of exercise.
In the present study, the increase in PGC-1α, ERRα and NRF-2 was also associated with an increase in COXIV at 24 h post-exercise. From our in vivo results it is difficult to ascertain whether the exercise induced increase in COX IV is due to a PGC-1α coactivation of ERRα, NRF-1, NRF-2 or a combination of these factors. In vitro results from cellular assays have shown that COXIV induction by PGC-1α depends, at least partly, on ERRα function (Schreiber et al. 2004). Additionally, it has been demonstrated in mouse C2C12 myotubes that PGC-1α increases the transcriptional activity of both ERRα and the GA-repeat binding protein (GABP, the mouse homologue of NRF-2), forming a double-positive-feedback loop with PGC-1α and driving the early expression of mitochondrial genes (Mootha et al. 2004). Whether NRF-1 is downstream of ERRα and NRF-2 in human skeletal muscle, as seems to be the case in the C2C12 cells (Mootha, 2004), is unknown. Possibly, the effects of long-term exercise on human skeletal muscle COXIV mRNA regulation (Vogt et al. 2001; Russell et al. 2003; Short et al. 2003) are synergistically controlled by PGC-1α, ERRα, NRF-1 and NRF-2 pathways.
In summary, we have shown that acute endurance exercise induces the expression of Mfn1, Mfn2 and ERRα mRNA levels in human skeletal muscle, during the 24 h post-exercise. We have also demonstrated in vitro that Mfn2, but not Mfn1, is induced directly by the transcriptional couple of PGC-1α and ERRα. The PGC-1α/ERRα-mediated induction of Mfn2 implicates PGC-1α in mitochondrial fusion and adds yet another key metabolic function to its regulatory capabilities. Our results also suggest that exercise is not only responsible for providing signals that effect mitochondrial biogenesis, but also for promoting mitochondrial fusion, demonstrating another mechanism, targeted by exercise, which may assist in reducing insulin resistance.
Acknowledgments
We would like to thank Dr S. Leal for assistance with the muscle biopsies and J. Cardenas for technical assistance. This work was supported by the Loterie Suisse Romande. A.K. received funding from the NIH (grant DK064951).
References
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, Rabasa-Lhoret R, Wallberg-Henriksson H, Laville M, Palacin M, Vidal H, Rivera F, Brand M, Zorzano A. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem. 2003;278:17190–17197. doi: 10.1074/jbc.M212754200. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Dela F, Mikines KJ, von Linstow M, Secher NH, Galbo H. Effect of training on insulin-mediated glucose uptake in human muscle. Am J Physiol. 1992;263:E1134–E1143. doi: 10.1152/ajpendo.2006.263.6.E1134. [DOI] [PubMed] [Google Scholar]
- Dempster P, Aitkens S. A new air displacement method for the determination of human body composition. Med Sci Sports Exerc. 1995;27:1692–1697. [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Yokota S, Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem (Tokyo) 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. cDNA Cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem Biophys Res Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- Grant S, Craig I, Wilson J, Aitchison T. The relationship between 3 km running performance and selected physiological variables. J Sports Sci. 1997;15:403–410. doi: 10.1080/026404197367191. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol. 1984;56:831–838. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- Hood DA. Invited Review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90:1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- Irrcher I, Adhihetty PJ, Joseph AM, Ljubicic V, Hood DA. Regulation of mitochondrial biogenesis in muscle by endurance exercise. Sports Med. 2003;33:783–793. doi: 10.2165/00007256-200333110-00001. [DOI] [PubMed] [Google Scholar]
- Johnston SD, Liu X, Zuo F, Eisenbraun TL, Wiley SR, Kraus RJ, Mertz JE. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen response elements. Mol Endocrinol. 1997;11:342–352. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- Laganiere J, Tremblay GB, Dufour CR, Giroux S, Rousseau F, Giguere V. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor alpha (ERRalpha) promoter dictates peroxisome proliferator-activated receptor gamma coactivator-1alpha control of ERRalpha expression. J Biol Chem. 2004;279:18504–18510. doi: 10.1074/jbc.M313543200. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Murakami T, Shimomura Y, Yoshimura A, Sokabe M, Fujitsuka N. Induction of nuclear respiratory factor-1 expression by an acute bout of exercise in rat muscle. Biochim Biophys Acta. 1998;1381:113–122. doi: 10.1016/s0304-4165(98)00018-x. [DOI] [PubMed] [Google Scholar]
- Norrbom J, Sundberg CJ, Ameln H, Kraus WE, Jansson E, Gustafsson T. PGC-1alpha mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. J Appl Physiol. 2004;96:189–194. doi: 10.1152/japplphysiol.00765.2003. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J Physiol. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Deriaz O. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 2003;52:2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha) -induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- Short KR, Vittone JL, Bigelow ML, Proctor DN, Rizza RA, Coenen-Schimke JM, Nair KS. Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes. 2003;52:1888–1896. doi: 10.2337/diabetes.52.8.1888. [DOI] [PubMed] [Google Scholar]
- Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, Tabata I. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem Biophys Res Commun. 2002;296:350–354. doi: 10.1016/s0006-291x(02)00881-1. [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol Cell Biol. 1991;11:5631–5638. doi: 10.1128/mcb.11.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M, Puntschart A, Geiser J, Zuleger C, Billeter R, Hoppeler H. Molecular adaptations in human skeletal muscle to endurance training under simulated hypoxic conditions. J Appl Physiol. 2001;91:173–182. doi: 10.1152/jappl.2001.91.1.173. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]