Abstract
We previously defined a cholesterol recognition/interaction amino acid consensus (CRAC; ATVLNYYVWRDNS) in the carboxyl terminus of the peripheral-type benzodiazepine receptor (PBR), an outer mitochondrial membrane protein involved in the regulation of cholesterol transport into the mitochondria, the rate-determining step in steroid biosynthesis. We examined (i) the PBR–cholesterol interaction by UV crosslinking of the C17 side-chain containing progestin, promegestone, and (ii) the role of the CRAC domain of PBR in Leydig cell steroidogenesis by using a transducible peptide composed of the TAT domain of HIV and the CRAC domain of PBR. [3H]Promegestone photoincorporated into recombinant PBR, and this labeling was displaced by cholesterol. [3H]Promegestone also photoincorporated into the TAT-CRAC peptide. [3H]Promegestone crosslinking to TAT-CRAC could be displaced by cholesterol and promegestone, with IC50 values of 1 and 200 μM, respectively. TAT-CRAC efficiently transduced into MA-10 Leydig cells and inhibited the hCG- and cAMP-stimulated steroid production in a dose-dependent manner. TAT-CRAC did not affect the hCG-induced cAMP synthesis and the 22R-hydroxycholesterol-supported steroidogenesis. Mutated TAT-CRAC lost its ability to bind [3H]promegestone and to inhibit the hCG-stimulated steroidogenesis. These results show that TAT-CRAC binds cholesterol and competes for cholesterol interaction with endogenous PBR, suggesting that the cytosolic carboxyl-terminal domain of PBR is responsible for taking up and bringing steroidogenic cholesterol into the mitochondria.
The peripheral-type benzodiazepine receptor (PBR) is an 18-kDa protein found in many tissues of the body, but present at high levels in steroid-synthesizing tissues, such as gonads, adrenal, placenta, and brain (1–3), and in cells proliferating in high rate, such as in various aggressive tumor cells of the breast (4), brain (5, 6), and liver (7). In steroid-synthesizing cells, PBR is primarily localized in the outer mitochondrial membrane, where it is involved in the uptake of cholesterol, the precursor of the steroids formed into the mitochondria (2, 8). Cholesterol uptake into the mitochondria and transport into the inner mitochondrial membrane, where the first steroidogenic enzyme cytochrome P450 side chain cleavage (P450scc) is located, is the rate-determining step in steroid biosynthesis and the step where hormones act to accelerate steroid formation (9). Mitochondrial PBR has been also involved in other functions such as respiration (10, 11) and apoptosis (12, 13) that might also relate to its ability to bring cholesterol into the mitochondria. In proliferating at high rate tumor cells, PBR was found to be primarily localized in and/or around the nucleus, where it is involved in the uptake of cholesterol into the nucleus and regulation of cell proliferation (4, 6, 14). These studies suggested that PBR might serve a more general function in cholesterol compartmentalization (15).
In an effort to determine the function of PBR, we used an in vitro bacterial paradigm devoid of endogenous PBR and cholesterol (15). We demonstrated that deletions in the cytoplasmic carboxyl terminus of PBR dramatically reduced the cholesterol uptake function of PBR, although the full capacity to bind drug ligands, such as PK 11195, was retained. In these studies, we identified amino acids Tyr-153 and Arg-156 to be critical for the interaction of the receptor with cholesterol (15), and we proposed the presence of a cholesterol recognition/interaction amino acid consensus (CRAC; VLNYYVWR) sequence at the carboxyl terminus of PBR (15). However, the evidence for a direct interaction/binding of cholesterol to PBR and for the consequences of this interaction/binding on a PBR-dependent function, such as steroidogenesis, was missing.
To address these issues, we first examined the direct interaction/binding of cholesterol to purified recombinant 18-kDa PBR protein and to its carboxyl-terminal domain, CRAC; second, we investigated the functional role of CRAC in hormone-stimulated steroidogenesis by transducing the peptide in the MA-10 mouse Leydig tumor cells.
Materials and Methods
Materials.
[1,2,6,7-N-3H]Progesterone [specific activity, 94.1 Ci/mmol (1 Ci = 37 GBq)], 17,21-dimethyl-19-nor-pregn-4,9-diene-3,20-dione (promegestone), and [17α-methyl-3H]promegestone (specific activity, 94.1 Ci/mmol) were obtained from NEN. Cholesterol (water-soluble), progesterone, pregnenolone, 17β-estradiol, and testosterone were obtained from Sigma. Cell culture supplies were purchased from Life Technologies (Grand Island, NY). Tissue culture plasticware was from Corning. Electrophoresis reagents and materials were supplied from NOVEX (San Diego). All other chemicals used were of analytical grade and were obtained from various commercial sources.
Expression and Purification of Recombinant PBR.
pET15PBR vector (15) was used to transform the BL21(DE3) Escherichia coli strain (Novagen) where the expression of recombinant mouse PBR protein was induced by 1 mM isopropyl-1-thiol-β-D-galactopyranoside. PBR protein expression was monitored by SDS/PAGE followed by Coomassie Blue staining or immunoblot analysis by using an anti-PBR antibody (described below). Cells were harvested and resuspended in binding buffer (20 mM Tris-HCl, pH 7.9/0.5 M NaCl/5 mM imidazole) and sonicated thoroughly. The pellet was collected at 20,000 × g centrifugation and dissolved in binding buffer containing 1% SDS. The recombinant PBR was purified by the His⋅Bind metal chelation resin (Novagen) and stored in binding buffer with 1% SDS.
PBR Antibody Generation and Immunoblot Analysis.
Rabbit anti-mouse PBR antibody was prepared by sequential immunization with a peptide (amino acids 7–19) VGLTLVPSLGGFMGAYFVR of mouse PBR protein, which was coupled to KLH. PBR antibodies were purified by an affinity resin containing the same peptide immobilized onto agarose (Bethyl Laboratories, Montgomery, TX). Protein samples were solubilized in sample buffer (25 mM Tris-HCl, pH 6.8/1% SDS/5% β-mercaptoethanol/1 mM EDTA/4% glycerol/0.01% bromphenol blue), boiled for 5 min, and loaded onto a 15% SDS/PAGE minigel (MiniProtein II System, Bio-Rad). The PBR-rich mitochondria of MA-10 mouse tumor Leydig cells were isolated and used as a control of the native receptor (16). Electrophoresis was performed at 25 mA/gel by using a standard SDS/PAGE running buffer (25 mM Tris/192 mM glycine/0.1% SDS). The proteins were electrophoretically transferred to nitrocellulose membrane (Schleicher & Schuell). The membrane was incubated in blocking TTBS (20 mM Tris-HCl, pH 7.5/0.5 M NaCl/0.05% Tween 20) buffer containing 10% nonfat milk at room temperature for 1 h, followed by incubation with a primary antibody against PBR (1:2,000) for 2 h. The membrane was then washed with TTBS three times for 10 min each time. After a 1-h incubation with the secondary antibody, goat anti-rabbit IgG conjugated with horseradish peroxidase (1:5,000; Transduction Laboratories, Lexington, KY), the membrane was washed with TTBS three times for 10 min each time. Specific protein bands were detected by chemiluminescence by using the Renaissance kit (DuPont/NEN). The specificity of the bands recognized by the antibody was demonstrated by using preabsorbed antibody prepared by incubating the antibody with the peptide used for the immunization.
Peptide Synthesis, Modeling, and Labeling.
Twenty-five-mer TAT-CRAC and TAT-mCRAC peptides were chemically synthesized by Bethyl Laboratories. Each peptide contained an amino-terminal 11-mer TAT protein transduction domain (YGRKKRRQRRR; ref. 17) followed by a glycine residue and either a 13-mer mouse PBR CRAC sequence (ATVLNYYVWRDNS) or a mutated CRAC sequence (ATGLNSSVWLDNS). The three-dimensional structure estimate of the peptides was made starting from an extended conformation (φ = −135 deg, ψ = −45 deg, and ω = 180 deg), and energy was minimized by using the optirot program from tinker with an AMBER atom model (18). The docked location of the cholesterol (C29H50O) ligand to the TAT-CRAC target shown was predicted by using Monte Carlo simulated annealing as implemented in autodock (19). Peptides were labeled with Oregon Green 488 by using the protein labeling kit (Molecular Probes) and conditions as described by the manufacturer.
[3H]Promegestone Photolabeling.
Various concentrations of recombinant PBR, TAT-CRAC, or TAT-mCRAC in PBS were incubated with [3H]promegestone at a final concentration of 120 nM in the absence or presence of cholesterol or various steroids in a 100-μl final volume. After a 1-h incubation at 4°C, samples were photoirradiated for 30 min at a distance of <0.5 cm by using a UV light with maximum emission at 366 nm (Ultraviolet Products, Gabriel, CA). Sample loading buffer was applied to the samples, and they were submitted to SDS/PAGE by using the Tricine buffer system from NOVEX. Proteins were transferred to nitrocellulose membrane. The membrane was exposed to tritium-sensitive screen and analyzed by a Cyclone Storage phosphor system from Packard. Image analysis of the phosphoimages was performed by using the optiquant software from Packard. In separate experiments, samples were incubated with increasing concentrations of [3H]promegestone in the absence or presence of unlabeled promegestone (2 mM). Samples were photoirradiated and incubated as described above; at the end of the incubation, the samples were filtered through GF/B filters (Brandel, Gaithersburg, MD) equilibrated in 0.5% polyethyleneimine and washed with 40 ml of ice-cold PBS. Radioactivity trapped on the filters was determined by liquid scintillation counting. The dissociation constant (Kd) was defined following Scatchard analysis of the specific binding as previously described (15).
Cell Culture and Treatments.
MA-10 cells were grown in modified Waymouth's MB752/1 medium containing 15% horse serum, as previously described (16). To determine the efficiency of TAT-CRAC incorporation into the cells, MA-10 cells were cultured overnight on 8-chambered SuperCell culture slides (Fisher Scientific) at a concentration of approximately 25,000 cells/chamber. Media were replaced 24 h later with fresh media, and cells were treated with various concentrations of Oregon Green 488-labeled peptides for various time periods. After the incubation period, cells were washed with PBS and examined by fluorescent microscopy by using an Olympus BX40 fluorescence microscope.
For steroid synthesis experiments, MA-10 cells were plated into 96-well plate at the density of 2.5 × 104 cells/well; 24 h later, media were replaced with fresh media, and cells were treated with the indicated concentrations of peptides for 30 min. Cells were then stimulated with 50 ng/ml hCG in serum-free medium for 2 h. At the end of the incubation, the culture media were collected and tested for progesterone production by RIA. The RIA assay was carried out by using anti-progesterone antisera (ICN) and following the conditions recommended by the manufacturer. Progesterone production was normalized against the amount of protein in each well. RIA data were analyzed by using the MultiCalc software (EG & G Wallac, Gaithersburg, MD).
cAMP Assay.
MA-10 cells were cultured and treated as described above. cAMP was measured by using the cAMP[125I] RIA system from Amersham Pharmacia.
Protein Measurement.
Proteins were quantified by using the dye-binding assay of Bradford (20) with BSA as the standard.
Statistics.
Statistical analysis was performed by ANOVA followed by the Student–Newman–Keuls test or the Dunnett multiple comparisons test by using the instat (v.3.0) package from GraphPad (San Diego).
Results
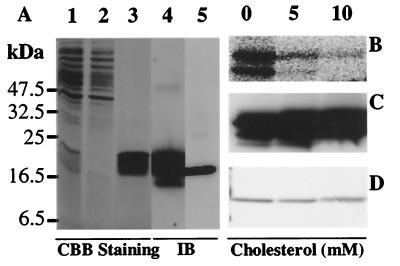
Recombinant mouse PBR with a HIS tag at the N terminus was expressed in bacteria. The recombinant PBR was purified by using the His⋅Bind metal chelation resin to nearly homogeneity. The purity of the purified protein was determined by Coomassie Brilliant Blue staining (Fig. 1A, lane 1, total extract; lane 2, flow-through; lane 3, purified recombinant PBR) and by immunoblot analysis by using an affinity-purified anti-PBR peptide antibody (Fig. 1A, lane 4, purified recombinant PBR). Because of the added HIS tag, the recombinant protein has a higher molecular mass of 20 kDa (Fig. 1A, lanes 1, 3 and 4) than the native PBR protein (18 kDa) present in isolated mouse Leydig cell mitochondria (Fig. 1A, lane 5).
Figure 1.
Purification of recombinant-PBR from E. coli and crosslinking of recombinant-PBR with [3H]promegestone. (A) SDS/PAGE followed by Coomassie Brilliant Blue (CBB) staining of the total extract (lane 1), flowthrough (lane 2), and recombinant PBR (lane 3) isolated as described in Materials and Methods. (B) Immunoblot analysis of 1 μg of purified recombinant PBR (lane 4) and of PBR-rich mitochondria of MA-10 mouse Leydig cells (lane 5) using an affinity-purified anti-PBR peptide antiserum. Isolated recombinant PBR (5 μM) was incubated with [3H]promegestone in the presence or absence of cholesterol. Samples were exposed to UV light; at the end of the incubation, the samples were separated by SDS/PAGE. Photoincorporation of [3H]promegestone to recombinant PBR was detected by phosphorimaging (B). Immunoblot analysis (C) and Coomassie Brilliant Blue staining of the gel (D) were used to monitor the equal protein loading.
To study the interaction of cholesterol with the isolated recombinant PBR, we used the progestin, [3H]promegestone. Promegestone was crosslinked by photoirradiation to the recombinant PBR (Fig. 1B). [3H]Promegestone crosslinking to the recombinant PBR was displaced by cholesterol (Fig. 1B) and promegestone (not shown). The presence of equal amounts of PBR protein in the crosslinked samples was monitored by immunoblot analysis (Fig. 1C) and Coomassie Brilliant Blue staining (Fig. 1D). The IC50 values for the inhibition of the photoincorporation of [3H]promegestone into recombinant PBR by cholesterol and promegestone were 300 and 3,000 μM, respectively (not shown). The low affinity of cholesterol and promegestone for recombinant PBR is probably because of the presence of detergent in the reaction buffer added to maintain the highly hydrophobic PBR protein in solution. Efforts to eliminate the detergent from the preparations used resulted in loosing the PBR [3H]promegestone and drug ligand binding capacity.
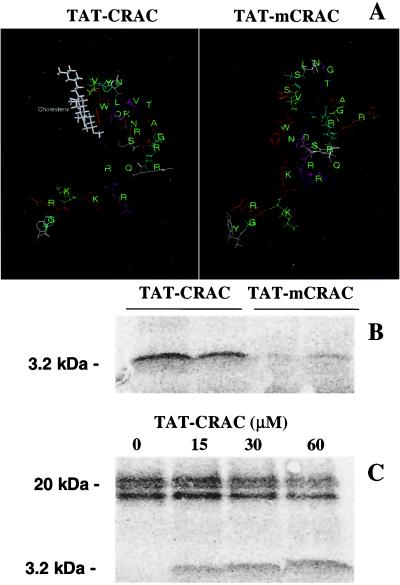
To resolve this issue better and characterize the [3H]promegestone–PBR interaction, we proceeded by investigating the ability of synthetic peptides designed according to the CRAC domain sequence of the 18-kDa PBR protein to photoincorporate [3H]promegestone. Moreover, to transduce these peptides into MA-10 Leydig cells for functional studies, we synthesized peptides containing the TAT domain of the HIV TAT protein coupled to the CRAC domain of PBR. To avoid synthesizing unusable peptides and hoping to predict their putative ability to interact with cholesterol, molecular modeling studies of the TAT-CRAC and TAT-mCRAC peptides were undertaken. The results show that the TAT-CRAC peptide forms a pocket where cholesterol may dock (Fig. 2A, left). This pocket is absent in the mutated TAT-mCRAC peptide, predicting the lack of interaction of this sequence with cholesterol (Fig. 2B, right). Computational docking simulations indicated that indeed cholesterol could dock in the pocket of the TAT-CRAC target and more specifically to amino acids YYWR (Fig. 2A, left).
Figure 2.
Molecular modeling of TAT-CRAC and TAT-mCRAC and photoincorporation of [3H]promegestone into TAT-CRAC and TAT-mCRAC. (A) For TAT-CRAC (left) and TAT-mCRAC (right), the three-dimensional structure estimates and the cholesterol docking computational simulations were made as described in Materials and Methods. TAT-CRAC and mutant TAT-mCRAC peptides (160 μM) were incubated with [3H]promegestone and exposed to UV light as described in Materials and Methods. (B) After crosslinking, the samples were separated by SDS/PAGE, and the radiolabeled peptides were detected by phosphorimaging. (C) The crosslinking of [3H]promegestone to 5 μM recombinant PBR can be displaced by increasing concentrations of the TAT-CRAC peptide.
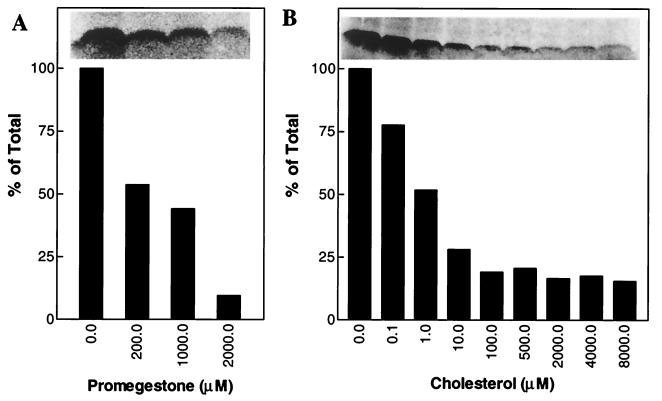
Fig. 2B shows that [3H]promegestone was UV crosslinked to TAT-CRAC. However, mutated TAT-mCRAC exhibited very low to no photoincorporation of [3H]promegestone (Fig. 2B). TAT-CRAC was also able to compete with the recombinant PBR for [3H]promegestone binding (Fig. 2C). Image analysis of the phosphoimages indicate that the effect of TAT-CRAC is dose-dependent as addition of increasing concentrations of TAT-CRAC results in increased labeling of the peptide and decreased labeling of the recombinant PBR protein (ratios of radiolabeled PBR/radiolabeled TAT-CRAC in the presence of 15, 30, and 60 μM TAT-CRAC were 1.4, 1.0, and 0.6, respectively). Cold promegestone displaced the interaction between [3H]promegestone and TAT-CRAC in a dose-dependent manner with an IC50 of 200 μM (Fig. 3A). However, cholesterol inhibited the crosslinking of [3H]promegestone to TAT-CRAC in a dose-dependent manner with an IC50 of 1 μM (Fig. 3B). Testosterone, 17β-estradiol, progesterone, and pregnenolone failed to inhibit the crosslinking of [3H]promegestone to TAT-CRAC (not shown).
Figure 3.
Inhibition of the photoincorporation of [3H]promegestone into TAT-CRAC by promegestone and cholesterol. The TAT-CRAC peptide (160 μM) was incubated with [3H]promegestone in the presence or absence of increasing concentrations of promegestone (A) or cholesterol (B) and exposed to UV light as described in Materials and Methods. After crosslinking, the samples were separated by SDS/PAGE, and the radiolabeled peptides were detected by phosphorimaging. The crosslinking of [3H]promegestone to TAT-CRAC peptide is displaced by promegestone (A) and cholesterol (B). Relative intensity of the radiolabeled peptide was determined as indicated in Materials and Methods.
To monitor the transduction into cells of the TAT-CRAC peptide, the peptide was labeled with Oregon Green 488 and added to the media at concentrations ranging from 0.03 to 30 μM. Fluorescent microscopy of treated cells showed that the TAT-CRAC peptide transduced rapidly into the majority of the cells in a dose-dependent manner (not shown). In 30 min, most of the cells were labeled (Fig. 4 A, free dye, and B, TAT-CRAC labeled with Oregon Green 488). Pretreatment of MA-10 cells with 10 and 30 μM TAT-CRAC for 30 min inhibited in a dose-dependent manner the hCG-stimulated progesterone formation by the cells (P < 0.0001; Fig. 4C). The same concentrations of TAT-CRAC also inhibited the dibutyryl-cAMP-stimulated progesterone synthesis by the cells (918 ± 101 ng/mg protein produced in control cells vs. 417 ± 21 ng/mg protein in cells treated with 30 μM TAT-CRAC). The mutant TAT-mCRAC was ineffective at all concentrations used (Fig. 4C). Neither TAT-CRAC nor TAT-mCRAC affected the 22R-hydroxycholesterol-supported steroid formation by the cells (Fig. 4D) and the hCG-stimulated cAMP formation by the cells (data not shown).
Figure 4.
Transduction and effect of TAT-CRAC on MA-10 Leydig cells. Fluorescent microscopy pictures of MA-10 cells treated for 30 min with either Oregon Green 488 (A) or 10 μM Oregon Green 488-labeled TAT-CRAC (B). MA-10 Leydig cells were treated with the indicated concentration of TAT-CRAC or TAT-mCRAC for 30 min. Cells were then exposed to 50 ng/ml hCG (C) or 20 μM 22R-hydroxycholesterol (D) for 2 h. At the end of the incubation, media were collected, and progesterone levels were determined by RIA. Results shown are means ±SD from four independent experiments (n = 12).
Discussion
To examine the interaction/binding of cholesterol to PBR, we used the photoreactive progestin promegestone and cholesterol in a series of ligand-binding studies by using purified recombinant PBR and a synthetic peptide containing the cholesterol recognition domain of PBR (CRAC). Affinity-labeling techniques have proved indispensable for the study of reversible biological recognition systems, as they conserve ligand–receptor interaction by covalent linkage. Because of the structure and the intrinsic properties of cholesterol, it was difficult to show specific binding and affinity labeling by using either radiolabeled cholesterol or radiolabeled azido-cholesterol (not shown). Thus, we turned our attention to the photoreactive progestin, promegestone, a steroid with a side chain at C17 similar to that present in cholesterol. Photoincorporation of [3H]promegestone has been used to identify several different progesterone receptors (21, 22) and nicotinic acetylcholine receptor (AChR) (23). There is strong evidence that cholesterol interacts directly with AChRs and that neutral lipids are required to maintain the AChR in a state responsive to agonist (24). Promegestone is a noncompetitive antagonist of the Torpedo AChR and reversibly inhibits acetylcholine-elicited currents for Torpedo AChRs expressed in Xenopus oocytes with the same potency as progesterone. Detailed studies demonstrated that the inhibition of acetylcholine-elicited currents by promegestone occurs because promegestone displaces cholesterol from a functionally important site (23). In this study, we took advantage of these properties of promegestone to detect the interaction/binding between PBR and cholesterol.
In our previous studies, we showed that the carboxyl terminus of PBR is crucial for its cholesterol uptake function. Studies by using site-directed mutagenesis in the carboxyl-terminal region of PBR demonstrated that amino acids Tyr-153 and Arg-156 are involved in the interaction of the receptor with cholesterol. Thus, we postulated the existence of a CRAC sequence at the carboxyl terminus of PBR (VLNYYVWR) (15). Both purified recombinant PBR and the chemically synthesized TAT-CRAC peptide, which contains the CRAC domain of PBR, were photolabeled by [3H]promegestone, indicating that CRAC is the binding site for promegestone in PBR. The specificity of the crosslinking at the CRAC domain was further confirmed by using the TAT-mCRAC peptide, mutated at Tyr-153 and Arg-156 as in our previous studies (15); binding of [3H]promegestone to TAT-mCRAC was dramatically reduced as compared with TAT-CRAC. Computational modeling of the peptides indicated the presence of a pocket in the TAT-CRAC peptide that is absent in mutated peptide, suggesting that this pocket may be at least part of the cholesterol docking site. These results were confirmed and extended by using computational docking simulations, which indicated that indeed cholesterol docked in the 153YYWR156 amino acid sequence of the CRAC. Interestingly, these are the same amino acids found to interact with cholesterol in our previous mutagenesis study (15).
The TAT-CRAC peptide was able to compete with recombinant PBR protein for [3H]promegestone photolabeling, suggesting that under the conditions used, the peptide acquires a conformation allowing promegestone binding. [3H]Promegestone photolabeling could be competed by unlabeled promegestone and cholesterol but not by other steroids tested, such as progesterone, pregnenolone, testosterone, and 17β-estradiol. The finding that the IC50 for cholesterol inhibition of [3H]promegestone binding, in the in vitro system tested, is 1 μM, suggests that in the right protein/lipid microenvironment, the CRAC domain of PBR may be a high-affinity binding site for cholesterol. The observation that promegestone displaced [3H]promegestone with an IC50 of 200 μM indicates that promegestone has much lower affinity for this steroid binding site of PBR as compared with cholesterol. Moreover, the observations that the dissociation constant of [3H]promegestone for TAT-CRAC is 192 μM (data not shown), close to the IC50 reported above, [3H]promegestone does not bind to mutated TAT-CRAC, cholesterol docking occurred in TAT-CRAC, and PBR mutated at the CRAC site does not bind and uptake cholesterol (15) suggest that promegestone is modeling cholesterol in binding to the CRAC site. Comparison of the chemical structures of the steroids tested suggests that the C17 side chain of promegestone and cholesterol may be responsible for the binding specificity to PBR. It is evident that, at present, we cannot exclude the possibility that other lipids, chemicals, or endogenous entities may bind to the CRAC domain of PBR.
To further demonstrate the physiological role of the CRAC domain of PBR, we introduced the synthetic peptide containing the cholesterol recognition domain of PBR (CRAC) into Leydig cells in an effort to generate an intracellular competing binding site for the cholesterol targeted to the mitochondrial PBR. Leydig cells contain high levels of mitochondrial PBR involved in the transport of cholesterol into mitochondria (16), a rate-determining step in steroid biosynthesis (9). Thus, introduction of high levels of CRAC into Leydig cells may be able to divert cholesterol from its route to the mitochondrial PBR and steroidogenesis (dominant-negative phenotype). To achieve the rapid transduction of high levels of the CRAC domain of PBR into the MA-10 Leydig cells, we used the TAT domain of the HIV TAT protein. In 1988, Green and Loewenstein (25) and Frankel and Pabo (26) independently discovered that the HIV TAT protein is able to cross cell membranes. In 1994, Fawell et al. (27) demonstrated that chemical crosslinking a 36-amino acid domain of TAT to heterologous proteins conferred the ability to transduce these proteins into the cells. This approach was recently applied to various proteins (17, 28). In this study, we took advantage of the ability of the HIV TAT protein to cross cell membranes and transduce proteins into the cells (17). However, instead of making recombinant fusion protein, we chemically synthesized a TAT-CRAC fusion peptide containing the 11 amino acids from the TAT protein (YGRKKRRQRRR), one glycine, and the 13 amino acids of the CRAC domain of the 18-kDa PBR protein (ATVLNYYVWRDNS). Peptides where the Val, Tyr, and Arg amino acids were replaced by Gly, Ser and Leu, respectively, were also chemically synthesized (TAT-mCRAC, YGRKKRRQRRRGATGLNSSVWLDNS). These amino acid replacements were based on our previous studies where we demonstrated their importance in the uptake of cholesterol by bacteria expressing the PBR protein (15).
As noted above, TAT-CRAC, but not TAT-mCRAC, was photolabeled by [3H]promegestone. [3H]Promegestone binding was efficiently displaced by cholesterol, and TAT-CRAC could compete with recombinant PBR for [3H]promegestone binding. These properties made TAT-CRAC and TAT-mCRAC excellent tools to examine the role of the CRAC domain in Leydig cell steroidogenesis in situ.
In agreement with previous studies by using TAT-fusion proteins and peptides (17, 27, 28), TAT-CRAC peptides were rapidly transduced into almost all of the exposed cells in a concentration-dependent manner. Thus, the HIV TAT domain represents a superior technique to perform in situ functional studies on an entire cellular population in precise time intervals. Transduction of the TAT-CRAC peptide inhibited the hormone- and cAMP-stimulated progesterone formation by MA-10 Leydig cells in a dose-dependent manner. TAT-mCRAC did not exert any effect on Leydig cell steroid synthesis even at 30 μM, the highest concentration tested. These data indicate that the inhibition seen with TAT-CRAC is because of the Tyr and Arg amino acids, shown to interact with cholesterol, present in the CRAC and not to a nonspecific or toxic effect of the peptide. Steroidogenesis begins with the transport of the intracellular free cholesterol to the mitochondria where, by interacting/binding to PBR, cholesterol is entering into the mitochondria. Thus, the inhibition seen with TAT-CRAC is probably because of the fact that transduced TAT-CRAC peptide presents a binding site for cholesterol that competes with the endogenous PBR. Thus, increasing concentrations of TAT-CRAC result in increased inhibition of cholesterol levels bound to endogenous PBR leading to inhibition of steroid synthesis. The specificity of the effect of TAT-CRAC at the level of cholesterol uptake and movement into mitochondria is indicated by the findings that TAT-CRAC did not affect the hormone-stimulated cAMP synthesis and the 22R-hydroxycholesterol-supported steroidogenesis. 22R-hydroxycholesterol is a cholesterol derivative, substrate of the P450scc, that can cross freely the mitochondrial membranes and load onto the enzyme in the inner mitochondrial membrane (16). Thus, this steroid bypasses the mitochondrial transport mechanism of the substrate cholesterol. Taken together, these results show that the effect of TAT-CRAC is localized at the level between cAMP formation and cholesterol metabolism, where the cholesterol transport is taking place.
In conclusion, these results demonstrate that (i) PBR is a cholesterol-binding protein, (ii) the carboxyl-terminal cytosolic CRAC domain of the receptor is responsible for cholesterol binding, (iii) TAT-CRAC transduced peptides create a dominant-negative phenotype resulting in diversion of the cholesterol flow from the mitochondria, and (iv) PBR mediates the hormone-stimulated cholesterol transport into the mitochondria and subsequent steroid formation.
Acknowledgments
We are grateful to Dr. S. F. Dowdy (Howard Hughes Medical Institute and Division of Molecular Oncology, Departments of Pathology and Medicine, Washington University School of Medicine) for advice on the use of the HIV TAT peptide, Dr. B. B. Wolfe (Georgetown University, Washington, DC) for helpful discussions, Dr. P. Scotney (University of Melbourne, Melbourne, Australia) for assistance in porting autodock to Linux, and Dr. M. Culty (Georgetown University, Washington, DC) for critical review of the manuscript. This work was supported by National Institute of Environmental Health Sciences, National Institutes of Health, Grant ES-07747.
Abbreviations
- CRAC
cholesterol recognition/interaction amino acid consensus
- PBR
benzodiazepine receptor
- AChR
acetylcholine receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031461598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031461598
References
- 1.Snyder S H, McEnery M W, Verma A. Neurochem Res. 1990;15:119–123. doi: 10.1007/BF00972201. [DOI] [PubMed] [Google Scholar]
- 2.Papadopoulos V. Endocr Rev. 1993;14:222–240. doi: 10.1210/edrv-14-2-222. [DOI] [PubMed] [Google Scholar]
- 3.Costa E, Auta J, Guidotti A, Korneyev A, Romeo E. J Steroid Biochem Mol Biol. 1994;49:385–389. doi: 10.1016/0960-0760(94)90284-4. [DOI] [PubMed] [Google Scholar]
- 4.Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Cancer Res. 1999;59:831–842. [PubMed] [Google Scholar]
- 5.Miettinen H, Kononen J, Haapasalo H, Helen P, Sallinen P, Harjuntausta T, Helin H, Alho H. Cancer Res. 1995;55:2691–2695. [PubMed] [Google Scholar]
- 6.Brown R C, Degenhardt B, Kotoula M, Papadopoulous V. Cancer Lett. 2000;156:125–132. doi: 10.1016/s0304-3835(00)00451-1. [DOI] [PubMed] [Google Scholar]
- 7.Venturini I, Alho H, Podkletnova I, Corsi L, Rybnikova E, Pellicci R, Baraldi M, Pelto-Huikko M, Helen P, Zeneroli M L. Life Sci. 1999;65:2223–2231. doi: 10.1016/s0024-3205(99)00487-7. [DOI] [PubMed] [Google Scholar]
- 8.Papadopoulos V, Amri H, Li H, Boujrad N, Vidic B, Garnier M. J Biol Chem. 1997;272:32129–32135. doi: 10.1074/jbc.272.51.32129. [DOI] [PubMed] [Google Scholar]
- 9.Simpson E R, Waterman M R. Can J Biochem Cell Biol. 1983;61:692–707. doi: 10.1139/o83-088. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch J D, Beyer C F, Malkowitz L, Beer B, Blume A J. Mol Pharmacol. 1989;35:157–163. [PubMed] [Google Scholar]
- 11.Larcher J C, Vayssiere J L, Le Marquer F J, Cordeau L R, Keane P E, Bachy A, Gros F, Croizat B P. Eur J Pharmacol. 1989;161:197–202. doi: 10.1016/0014-2999(89)90843-1. [DOI] [PubMed] [Google Scholar]
- 12.Carayon P, Portier M, Dussossoy D, Bord A, Petitpretre G, Canat X, Le Fur G, Casellas P. Blood. 1996;87:3170–3178. [PubMed] [Google Scholar]
- 13.Papadopoulos V, Dharmarajan A M, Li H, Culty M, Lemay M, Sridaran R. Biochem Pharmacol. 1999;58:1389–1393. doi: 10.1016/s0006-2952(99)00215-4. [DOI] [PubMed] [Google Scholar]
- 14.Papadopoulos V, Kapsis A, Li H, Amri H, Hardwick M, Culty M, Kasprzyk P G, Carlson M, Moreau J, Drieu K. Anticancer Res. 2000;20:2835–2848. [PubMed] [Google Scholar]
- 15.Li H, Papadopoulos V. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulos V, Mukhin A G, Costa E, Krueger K E. J Biol Chem. 1990;265:3772–3779. [PubMed] [Google Scholar]
- 17.Nagahara H, Vocero-Akbani A M, Snyder E L, Ho A, Latham D G, Lissy N A, Becker-Hapak M, Ezhevsky S A, Dowdy S F. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 18.Dudek M, Ramnarayan K, Ponder J W. J Comput Chem. 1998;19:548–573. [Google Scholar]
- 19.Morris G M, Goodsell D S, Halliday R S, Huey R, Hart W E, Belew R K, Olson A J. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Stromstedt P E, Berkenstam A, Jornvall H, Gustafsson J A, Carlstedt-Duke J. J Biol Chem. 1990;265:12973–12977. [PubMed] [Google Scholar]
- 22.Sadler S E, Maller J L. J Biol Chem. 1982;257:355–361. [PubMed] [Google Scholar]
- 23.Blanton M P, Xie Y, Dangott L J, Cohen J B. Mol Pharmacol. 1999;55:269–278. doi: 10.1124/mol.55.2.269. [DOI] [PubMed] [Google Scholar]
- 24.Rankin S E, Addona G H, Kloczewiak M A, Bugge B, Miller K W. Biophys J. 1997;73:2446–2455. doi: 10.1016/S0006-3495(97)78273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green M, Loewenstein P M. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 26.Frankel A D, Pabo C O. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 27.Fawell S, Seery J, Daikh Y, Moore C, Chen L L, Pepinsky B, Barsoum J. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gius D R, Ezhevsky S A, Becker-Hapak M, Nagahara H, Wei M C, Dowdy S F. Cancer Res. 1999;59:2577–2580. [PubMed] [Google Scholar]