Summary
A 45 kb DNA sequencing analysis from Streptomyces hygroscopicus 5008 involved in validamycin A (VAL-A) biosynthesis revealed 16 structural, 2 regulatory, and 5 genes related to transport, transposition/integration, tellurium resistance, and another 4 genes with no obvious identity. The VAL-A biosynthetic pathway was proposed, with assignment of the required genetic functions confined in the sequenced region. A cluster of eight reassembled genes was found to support the VAL-A synthesis in a heterologous host, S. lividans 1326. In vivo inactivation of the putative glycosyltransferase gene (valG) abolished the final attachment of glucose for VAL production, and resulted in the accumulation of the VAL-A precursor, validoxylamine, while the normal production of VAL-A could be restored by complementation with valG. The role of ValG in the glycosylation of validoxylamine to VAL-A was demonstrated in vitro by enzymatic assay.
Keywords: biosynthetic genes, aminocyclitol antibiotics, validamycin, glycosyltransferase, validoxylamine
Abbreviations: aa, amino acid; NDP, nucleoside-diphosphate; PLP, pyridoxal 5′-phosphate; tsr, thiostrepton resistance gene; aac(3)IV, apramycin resistance gene; oriT, origin of transfer; PermE*, up-mutated promoter of erythromycin resistance gene; kDa, Kilo-dalton; Rf, retardation factor; ESI-MS, electrospray ionization-mass spectrometry; IPTG, isopropyl-β-D-thiogalactopyranoside; DTT, dithiothreitol; VAL-A, VAL-A
Introduction
VAL-A (validamycin A, Figure 1A), a weakly basic C7N-aminocyclitol-containing antibiotic, was first isolated from Streptomyces hygroscopicus var. limoneus, and later from Streptomyces hygroscopicus var. jinganggensis 5008 (hereafter S. hygroscopicus 5008 or strain 5008). It is widely used, especially in Asia to control sheath blight disease of rice plants and dumping-off of cucumber seedlings caused by the fungus Pellicularia sasakii (Rhizoctonia solani). In the presence of VAL-A, normal extension of the main fungal hyphae is switched to an abnormal branching at the tips and further development of those growing fungi is repressed. VAL-A was also found to have insecticidal activity, most likely resulting from its strong inhibition of trehalase, the trehalose-degrading enzyme. Trehalose is a primary storage carbohydrate in fungi (8 to 10% of the dry cell weight of P. sasakii) and is recognized as a characteristic blood sugar of insects. The enzyme trehalase appears to play an important metabolic role in these organisms in generating glucose for energy supply or for other physiological purposes. Its inhibition results in disruption of the glucose supply system of the fungus leading to growth abnormality and death. Although in vitro experiments revealed that the pseudo-disaccharide, validoxylamine A, is more active than VAL-A against trehalase, results of in vivo experiments suggest the opposite. It is proposed that the presence of the glucose moiety on VAL-A is essential for its efficient entry into fungal mycelia, in which it is hydrolyzed by an α-glucosidase to yield the active pharmacophore, validoxylamine A. VAL-A is also used as the source of valienamine, a pharmaceutically important precursor for the production of the antidiabetic drug, voglibose (Basen®), whose mechanism of action is to inhibit α-glucosidase in the intestine, comparable to another antidiabetic agent acarbose.
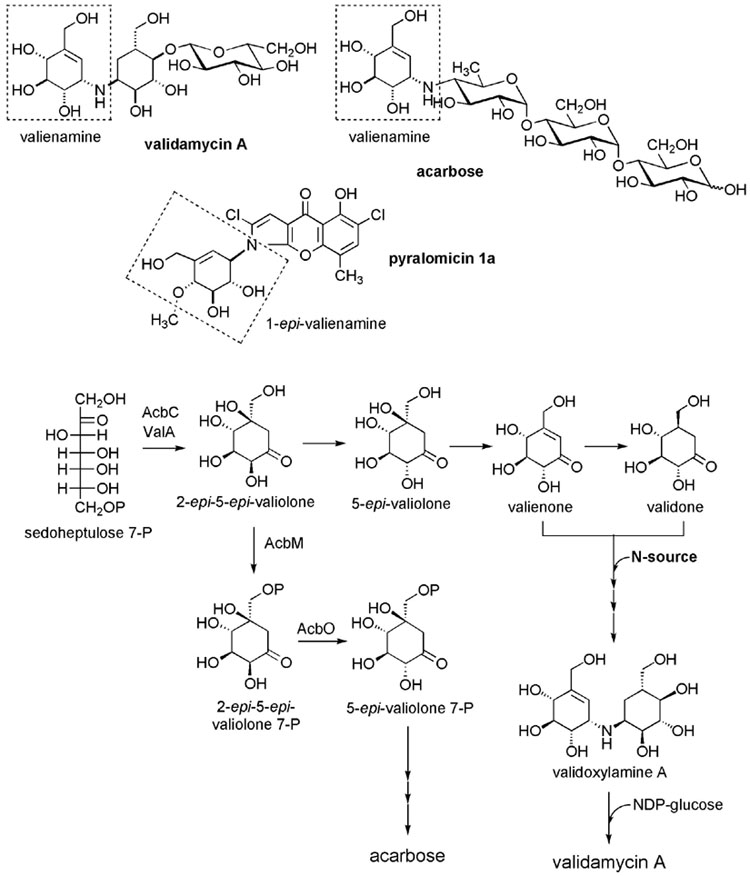
Figure 1.
Valienamine-containing C7N Aminocyclitols and Proposed Biosynthetic Pathways to VAL-And Acarbose. (A) Chemical structures of VAL-A, acarbose and pyralomicin 1a. Dashed-boxed regions show the cyclitol moiety shared by all three compounds. (B) Previously proposed biosynthetic pathways to VAL-A [9] and acarbose [14, 15].
The biosynthesis of the C7N-aminocyclitol moiety has been investigated to some extent in Actinoplanes sp. SE50/110 (acarbose-producer), in S. hygroscopicus var. limoneus (VAL-A producer), and more recently in Microtetraspora spiralis (pyralomicin-producer). Feeding experiments with a number of isotopically labelled potential intermediates to S. hygroscopicus var. limoneus demonstrated that 2-epi-5-epi-valiolone, 5-epi-valiolone, valienone, and validone were incorporated into VAL-A, leading to a proposed biosynthetic pathway to VAL-A as shown in Figure 1B. Similar feeding experiments were also performed with Actinoplanes sp. SE50/110 and Microtetraspora spiralis but with surprisingly different incorporation patterns. Only 2-epi-5-epi-valiolone and 5-epi-valiolone were found to be efficiently incorporated into pyralomicin, whereas 2-epi-5-epi-valiolone was the only one of these compounds incorporated into acarbose.
The gene cluster for acarbose biosynthesis has been cloned and a number of the proteins involved have been characterized, resulting in a proposed biosynthetic pathway to acarbose. The pathway is initiated by a cyclization of sedoheptulose 7-phosphate, a C7 sugar phosphate involved in the pentose phosphate pathway, catalyzed by the dehydroquinate synthase-like protein, AcbC, to give 2-epi-5-epi-valiolone (Figure 1B). The latter compound is then phosphorylated to its 7-phosphate derivative catalyzed by the kinase AcbM. Furthermore, the product of acbO, which was found in the same operon as acbC and acbM, was identified as a 2-epi-5-epi-valiolone 7-phosphate 2-epimerase. Efficient incorporation of 2-epi-5-epi-valiolone into both VAL and acarbose strongly suggested that both biosynthetic pathways share the same cyclization step. However, further downstream the pathways to the two compounds may be different, as feeding experiments with the producing strains gave two different patterns. This leaves many open questions surrounding the biosynthesis of acarbose and VAL-A, which cannot be addressed by conventional feeding experiments alone.
Using the acbC gene from the acarbose cluster as a heterologous probe, we have successfully cloned and identified the VAL-A biosynthetic genes from a genomic library of S. hygroscopicus 5008. Sequencing of a 6 kb BamHI fragment (Figure 2A), in vivo gene inactivation and in vitro biochemical characterization resulted in the identification of 2-epi-5-epi-valiolone synthase (ValA), a homolog of AcbC, paving the way for further sequencing of the entire cluster.
Figure 2.
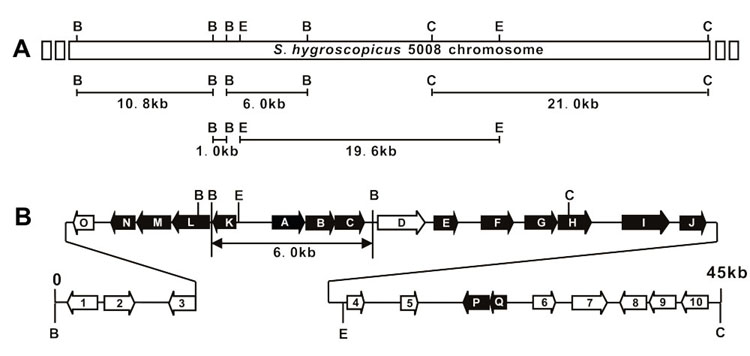
Cloning Strategy for Sequencing and Genetic Organization of val Gene Cluster. (A) Cloning strategy for sequencing of the 45 kb region. B: BamHI; C: ClaI; E: EcoRI. (B) Genetic organization of the val gene cluster. Genes proposed to be involved in validamycin biosynthesis are indicated as black arrows. The previously sequenced 6.0 kb BamHI fragment was indicated by double-arrowed line.
Here we describe the sequencing and functional analysis of 45 kb of DNA, which led to the identification of eight out of 27 genes as essential for validamycin biosynthesis through reconstituted heterologous gene expression in S. lividans 1326. We also demonstrate the glycosyltransferase function of the identified valG gene, first by its in vivo inactivation and functional complementation in strain 5008, secondly by in vitro enzymatic characterization of the ValG protein expressed in E. coli, and thirdly by heterologous expression of the val gene cluster, with or without valG, to yield VAL-A and validoxylamine A, respectively, in S. lividans 1326.
Results
Overall Architecture of the Sequenced Region
A total of 45,168 bp of DNA sequence (deposited in GenBank under accession number DQ164098) including the previously reported 5,877 bp of DNA (AY753181) was determined using overlapping or linking fragments (Figure 2A). Overall GC content of this region is 68.7%, somewhat lower than the typical GC content as exemplified by S. coelicolor A3(2) and S. avermitilis (ca. 72%). In silico analysis of the sequence revealed the presence of 27 complete open reading frames (ORFs), whose organization is shown in Figure 2B and proposed functions are listed in Table 1. Structural and regulatory genes likely involved in VAL-A biosynthesis, designated as valA-Q and indicated as black arrows in Figure 2B, are distributed in the middle part of the sequenced region, among which subcluster valA – valJ and valK – valN seem to be transcribed in opposite directions.
Table 1.
Deduced Functions of the Open Reading Frames In the val Gene Cluster
| Protein | aa | Proposed Function | Homologs, origin | Identity, similarity | Accession No. |
|---|---|---|---|---|---|
| ORF1 | 400 | Integrase | Urd-Int, Streptomyces fradiae | 88%, 94% | AAF00216 |
| ORF2 | 391 | Transposase | SAV1204, Streptomyces avermitilis | 90%, 92% | BAC68914 |
| ORF3 | 276 | Transposase | SCO7080, Streptomyces ceolicolor | 91%, 93% | CAB89752 |
| ValO | 221 | Phosphatase/phosphohexomutase | COG0637, Thermobifida fusca | 39%, 58% | ZP_00293893 |
| ValN | 331 | Cyclitol reductase | Glr0369, Gloeobacter violaceus | 29%, 39% | BAC88310 |
| ValM | 424 | Aminotransferase | DRA0029, Deinococcus radiodurans | 39%, 54% | NP_285353 |
| ValL | 492 | Validoxylamine A 7′-phosphate synthase | Rxy102001033, Rubrobacter xylanophilus | 30%, 47% | ZP_00187712 |
| ValK | 324 | Epimerase/dehydratase | APE1179, Aeropyrum pernix | 31%, 51% | BAA80165 |
| ValA | 414 | 2-epi-5-epi-valiolone synthasea | AcbCa, Actinoplanes sp. 50/110 | 48%, 65% | CAA77208 |
| ValB | 373 | Valienol-1-phosphate nucleotidylyltransferase | GlgC, Bacillus halodurans | 33%, 50% | BAB04806 |
| ValC | 351 | Cyclitol kinase | GlcK, Bacillus cereus | 29%, 44% | ZP_00238533 |
| ValD | 451 | Glyoxalase | NocaDRAFT_3188, Nocardioides sp. JS614 | 40%, 49% | ZP_00657213 |
| ValE | 331 | Oxidoreductase | SAV4805, Streptomyces avermitilis | 45%, 56% | BAC72517 |
| ValF | 401 | Oxidoreductase | SAV1825, Streptomyces avermitilis | 80%, 87% | BAC69536 |
| ValG | 422 | Glycosyltransferasea | KfoCa, Escherichia coli | 35%, 49% | BAC00523 |
| ValH | 398 | Transport protein | SAV6900, Streptomyces avermitilis | 52%, 66% | BAC74611 |
| ValI | 611 | Glycosyl hydrolase | SAV6503, Streptomyces avermitilis | 69%, 81% | BAC74214 |
| ValJ | 305 | Oxidoreductase | SAV4805, Streptomyces avermitilis | 43%, 55% | BAC72517 |
| ORF4 | 204 | Unknown | Mll4476, Mesorhizobium loti | 39%, 55% | NP_105339 |
| ORF5 | 347 | Unknown | SMc00549, Sinorhizobium meliloti | 28%, 47% | NP_385900 |
| ValP | 293 | Regulatory protein | SCO3723, Streptomyces coelicolor | 49%, 63% | CAD55330 |
| ValQ | 163 | Regulatory protein | SCO3723, Streptomyces coelicolor | 45%, 59% | CAD55330 |
| ORF6 | 191 | Tellurium resistance protein | SAV896, Streptomyces avermitilis | 92%, 97% | BAC68606 |
| ORF7 | 451 | Unknown | SCO3725, Streptomyces coelicolor | 72%, 81% | CAB76972 |
| ORF8 | 290 | Unknown | SAV7268, Streptomyces avermitilis | 52%, 68% | BAC74979 |
| ORF9 | 308 | Esterase | Bcepa03001004, Burkholderia cepacia | 47%, 59% | ZP_00218391 |
| ORF10 | 349 | Cytochrome P450 hydroxylase | P450-SU2, Streptomyces griseolus | 51%, 61% | AAA26825 |
with confirmed function
The valA – valJ Subcluster
valA, whose deduced protein sequence shows significant homology to AcbC from the acarbose biosynthetic gene cluster, was confirmed to encode 2-epi-5-epi-valiolone synthase. The deduced aa sequence of ValB shows 33% identity with GlgC, a glucose-1-phosphate adenylyltransferase from Bacillus halodurans C-125. ValC shows 30% identity with AcbM, the 2-epi-5-epi-valiolone 7-kinase from the acarbose cluster, whose demonstrated function is to phosphorylate 2-epi-5-epi-valiolone to form 2-epi-5-epi-valiolone 7-phosphate. Additionally, ValC also shows a weaker homology to glucokinase from a Bacillus species (29% identity/44% similarity).
Full length ValD (451 aa) is homologous to a putative glyoxalase/bleomycin resistance protein/dioxygenase domain from Nocardioides sp. JS614. Two domains of ValD, one at Gly35-Glu194 and the other at Gly220-Ala382, are showing 35.5% identity/50% similarity each other.
Both ValE (331 aa) and ValJ (305 aa) show homology to 2-oxoglutarate (2-OG) and Fe(II)-dependent oxygenases, whose enzymatic functions are known to catalyze a variety of reactions typically involving the oxidation of an organic substrate using an oxygen molecule, such as hydroxylation, desaturation, or oxidative ring closure. Noticeably, ValE and ValJ showed significant homology (79.8% identity at nt level and 66.3% identity at the protein level), and mainly differed from each other at the C-terminus.
ValF is highly homologous with putative oxidoreductases from S. avermitilis and S. coelicolor (76% identity/82% similarity). In addition, a ValF-related glucose-fructose oxidoreductase from Zymomonas mobilis is known to catalyze the reductive conversion of fructose to sorbitol, suggesting that ValF might be involved in the keto reduction of intermediate compounds in VAL-A biosynthesis.
ValG exhibits considerable homology to chondroitin polymerase KfoC from E. coli K4, which is known to catalyze the polymerization of the chondroitin backbone in the assembly of capsule polysaccharide K4 antigen. However, ValG (422 aa) is 264 aa shorter than KfoC (686 aa), but 250 aa residues of its N-terminal domain, which is required for sugar donor binding, are very similar to known glycosyltransferases. In addition, one conserved short motif of DGS and two DXD motifs, which have been proposed to facilitate NDP-sugar binding and Mn2+ ion binding, respectively, are present in ValG.
The deduced product of valH is homologous to putative transport protein SAV6900 from S. avermitilis and the fucose permease FucP from Mesorhizobium sp. BNC1 (33% identity and 50% similarity), whose functions are known to be co-transport of fucose, glucose or galactose with H+. Analysis of ValH using the TMHMM Server v. 2.0 suggests that 11 transmembrane helices span the length of the protein.
ValI seems to be closely related to the putative glucoamylase SAV6503 from S. avermitilis, which belongs to the glycosyl hydrolase family 15. Glucoamylases are known to catalyze the hydrolysis of α-1,4 and α-1,6 glucosidic linkages to release β-D-glucose from the non-reducing ends of starch and related poly- and oligosaccharides. However, no obvious function could be assigned in the pathway for VAL-A biosynthesis, except that D-glucose could be a precursor for VAL-A.
The valK – valN Subcluster
The valK – valN genes are predicted to be transcribed in the opposite direction to the valA – valJ genes. BLAST analysis of ValK showed significant homology with dTDP-4-dehydrorhamnose reductase (RmlD) from Aeropyrum pernix, a member of the reductase/epimerase/dehydrogenase/dehydratase protein superfamily putatively involved in the biosynthesis of L-rhamnose from glucose-1-phosphate. ValK contains a strictly conserved (P-X3-Y-X3-K-X3-E) motif that is characteristic for this superfamily and another conserved (STDNVF) motif that is unique for RmlD-type proteins. Interestingly, the conserved NAD(P)-binding Wierenga motif (G-X2-G-X2-G) is missing from the N-terminus of ValK. NAD(P)+-independent sugar epimerases from E. coli and other microorganisms have been reported previously. The reaction mechanism involves substrate to metal binding and a cis enediolate-stabilized intermediate. However, no significant sequence similarity was found between ValK and the NAD(P)+-independent sugar epimerases.
ValL shows significant homology to the putative trehalose 6-phosphate synthase Rxyl021033 from Rubrobacter xylanophilus and lower homology to its counterpart OtsA in E. coli. In E. coli, the trehalose 6-phosphate synthase (OtsA) and the trehalose 6-phosphatase (OtsB) are responsible for the biosynthesis of trehalose from glucose-6-phosphate and UDP-glucose with net retention of the anomeric configuration of the donor sugar.
ValM is closely related to pyridoxal 5′-phosphate-dependent (PLP) aminotransferases, such as the putative 4-aminobutyrate aminotransferase from Deinococcus radiodurans and the acetylornithine aminotransferase from Methanosarcina mazei. Consistent with other aminotransferases, the conserved Lys268 of ValM is believed to be the PLP-binding residue for the Schiff base linkage. In addition, other conserved residues important for PLP-binding (Asp239, Gln242, and Thr322) and catalysis (Glu206 and Arg349) are also present.
ValN shows significant homology to the zinc-dependent threonine dehydrogenase Krad06004313 from Kineococcus radiotolerans and AcbL from the acarbose pathway of Actinoplanes sp. SE50/110 (29% identity/39% similarity). AcbL, a putative cyclitol dehydrogenase was proposed to be involved in the reduction of the C-1 keto group of valienone 7-phosphate to give 1-epi-valienol 7-phosphate.
Putative Regulatory Genes
ValP and valQ are predicted to encode a two-component regulatory system consisting of a separate histidine kinase (sensor) and a Sigma B PP2C-like phosphatase (response regulator), as ValP showed the highest homology with the response regulator phosphatase domain and ValQ with the sensor kinase domain of SCO3723 from S. coelicolor A3(2). The existence of valP and valQ as two independent genes separated by only 28 bp seems unique in the val gene cluster since most of the counterpart proteins, exemplified by SCO3723, are known to exist as single peptides with two domains conforming to ValP and ValQ, respectively. It is not known yet whether the independent organization of ValP and ValQ could have anything to do with the induction of Sigma B activity and subsequent validamycin production in response to environmental stress, such as increased temperature and/or high salt concentration. However, we have consistently observed an apparently increased production of VAL-A and enhanced transcriptions of both valP and valQ in parallel, when the producer was grown at 37°C rather than at 30°C (Lei Li and Linquan Bai, unpublished data).
Boundaries of the Cluster and Other Most Likely Unrelated Genes
Beyond valN, 3 transposase-like or integrase-like ORFs (ORF1 to 3) and valO, which exhibit homology to a predicted phosphatase/phosphohexomutase from Thermobifida fusca, were detected. To determine the left boundary of the biosynthetic gene cluster, valO, which is located 764 bp downstream of valN and most likely monocistronically transcribed, was inactivated. Through double cross-over recombination mediated by the 2.5 kb left flanking sequence and 4.0 kb right flanking sequence, an internal 588 bp fragment was replaced by aac(3)IV via ReDirect methodology. The mutant ZYR-5, confirmed by PCR amplification, was found to produce a comparable amount of VAL-A as the wild-type, suggesting that valO is not involved in the biosynthesis of VAL-A. Thus, the left-border of the val gene cluster was defined.
Of the seven ORFs flanking the putative two-component regulatory genes valP and valQ, ORF 4, 5, 7 and 8 seem to encode proteins with unknown biological functions. Moreover, none of the detected homologies of ORF 6, 9 and 10 with a tellurium resistance protein, a group of esterases, and a cytochrome P450 seem to be relevant to validamycin biosynthesis. It is thus likely that all of the genes required for VAL-A biosynthesis are located between valN and valQ in a ca. 30 kb region (Figure 2B).
Heterologous Production of Validoxylamine A and VAL-A in S. lividans 1326
valABC seems to transcribe as one operon and valKLMN as another operon transcribed in the opposite direction (Figure 2B). The two operons were reassembled to have seven genes in the same orientation under the control of a constitutive PermE* promoter (Figure 3A), and a construct (pJTU757) was introduced into a heterologous host, S. lividans 1326, to see if the biosynthetic intermediate, validoxylamine A, can be produced. Since the 251 bp fragment to the N-terminus of valK may still contain its own promoter, the recombinant, XH-6, was thus initially fermented at 37°C, instead of at 30°C as commonly set for S. lividans.
Figure 3.
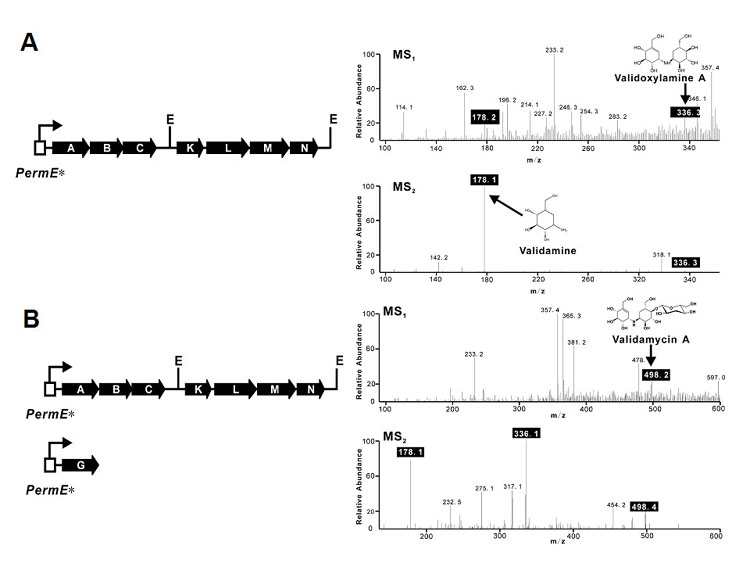
Heterologous Production of Validoxylamine A and VAL-A (A) Reassembly of valABCKLMN and MS analysis (MS1). PermE*, the un-mutated promoter of the erythromycin resistance gene; E, EcoRI; Fragmentation of validoxylamine A (m/z 336.1) yields validamine (m/z 178.1) (MS2). (B) Co-expression of valABCKLMN and valG and MS detection (MS1). Fragmentation of VAL-A (m/z 498.2) generates validoxylamine A (m/z 336.1) and validamine (m/z 178.1) (MS2).
Even though bioassay with chloroform-extracted fermentation broth of XH-6 showed almost no inhibitory activity against P. sasakii and HPLC analysis gave no obvious peak corresponding to validoxylamine A because of the lower yield, mass spectrometry analysis through direct injection of the sample did reveal the presence of validoxylamine A (Figure 3A). The typical fragmentation of validoxylamine A (m/z 336.1) under positive ion mode gives validamine (m/z 178.1). The isolated compound with m/z of 336.3 (MS1) from XH-6 was indeed fragmented to give a peak with m/z of 178.1 (Figure 3A, MS2). These data demonstrate that only seven genes, valABCKLMN, from the two original operons are required for the synthesis of validoxylamine A. Fermentation of XH-6 at 30°C followed by MS analysis also revealed the production of validoxylamine A at low yield (data not shown).
Since valG is the only gene found in the sequenced region encoding a glycosyltransferase, it was tempting to co-express this gene with valABCKLMN for the possible production of VAL-A in S. lividans. The new recombinant XH-9 harboring valABCKLMNG showed inhibitory activity in bioassay after fermentation at 37°C for 7 days, or at 30°C for 11 days (data not shown), suggesting the accumulation of VAL-A. The typical fragmentation pattern of VAL-A (m/z 498.2) under positive ion mode is the loss of glucose to give validoxylamine A (m/z 336.1), which is further fragmented to validamine (m/z 178.1). Detailed analysis by mass spectrometry of a 7-day fermented broth of XH-9 at 37°C clearly indicated the presence of VAL-A with m/z of 498.2, which was sequentially fragmented into validoxylamine A (m/z 336.1) and validamine (m/z 178.1) in the MS/MS experiment (Figure 3B). The 11-day fermentation broth of XH-9 at 30°C was also analyzed by MS/MS, and an obviously similar fragmentation pattern for VAL-A could be observed (data not shown).
The successful heterologous production of validoxylamine A and VAL-A in S. lividans 1326 not only confirmed the necessity for VAL-A biosynthesis of the eight structural genes from the 27 ORFs found in the sequenced region, but also revealed the identity of valG as a glycosyltransferase that transfers activated glucose to validoxylamine A as the final step in VAL-A biosynthesis.
Targeted Replacement of valG Abolishes VAL-A Biosynthesis
LL-1 (Figure 4B) was one of the confirmed mutant with an internal 806 bp region of valG (from nt number 112 to 918) replaced by an aac(3)IV from the strain 5008 (Figure 4A). Bioassay indicated that the LL-1 fermentation broth had retained inhibitory activity, albeit at a reduced capacity (Figure 4C), but the presence of VAL-A (retention time of 9.7 minute) could not be detected by HPLC analysis. Despite the fact that under normal conditions the wild-type strains also produced validoxylamine A (retention time of 6.5 minute), a significant increased accumulation of validoxylamine A was observed in LL-1 (Figure 4D). The abolished production of VAL-A and the increased accumulation of validoxylamine A in LL-1 strongly suggest that ValG catalyzes the glucosylation of validoxylamine A to yield VAL-A. The observed weak inhibitory activity agrees with the previous finding that validoxylamine A has much lower in vivo inhibitory activity than VAL-A.
Figure 4.
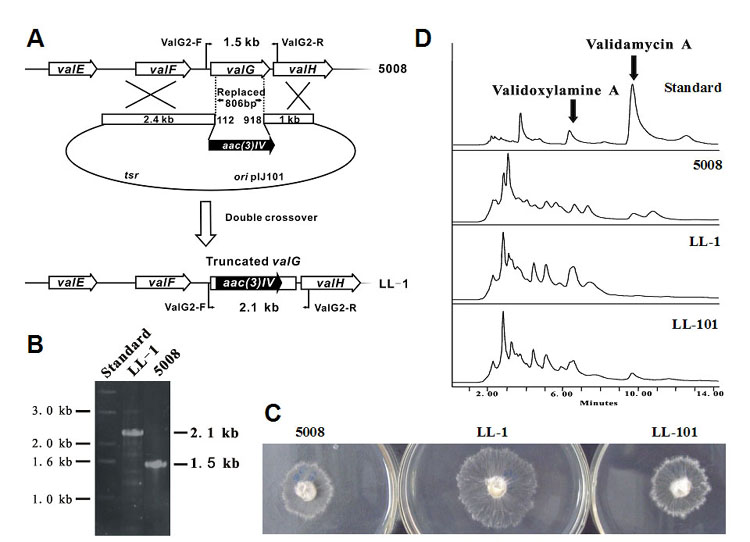
Inactivation and complementation of glycosyltransferase valG. (A) Schematic representation of the replacement of an 806 bp internal fragment of valG with the 1.4 kb aac(3)IV. In shuttle plasmid pJTU609, aac(3)IV was inserted between the 2.4 kb and 1.0 kb genomic fragments originally flanking the deleted 806 bp region. While wild-type S. hygroscopicus should give a 2.1 kb PCR-amplified product, mutant LL-1 should yield a 1.5 kb product using a pair of primers, ValG2-F and ValG2-R. (B) PCR analysis of wild-type S. hygroscopicus and mutant LL-1. (C) Bioassay comparison between the wild-type (left), LL-1 (middle) and LL-101 (right). LL-101 is the derivative of LL-1 harboring shuttle plasmid pJTU612 with valG. (D) HPLC profiles of the standards, wild-type, LL-1 and LL-101. The retention time of VAL-A is 9.7 min and that of validoxylamine A is 6.5 min.
Complementation by Cloned valG Restores VAL-A Production
When pJTU612, a pIJ101-derived plasmid with an intact valG, was introduced into mutant LL-1, the culture broth of the thiostrepton-resistant exconjugant (LL-101) was found to have regained the inhibitory activity (Figure 4C) in bioassay. HPLC analysis unambiguously demonstrated the presence of VAL-A with the retention time of 9.7 minute. However, the presence of valG under the strong constitutive expressed PermE* promoter in LL-101 was not sufficient for the conversion of all validoxylamine A to VAL-A, as a peak with the retention time of 6.5 minute corresponding to validoxylamine A was also observed (Figure 4D).
In Vitro Glycosylation of Validoxylamine A to VAL-A Using Recombinant ValG
Further proof to support the glycosyltransferase function of valG was obtained from its heterologous expression in E. coli, which gave rise to a 49 kDa soluble polyhistidine-tagged protein. Affinity purification on a Ni-NTA spin column (QIAGEN) or a BD TALON™ column gave a protein that was > 75% pure as judged by SDS-polyacrylamide gel electrophoresis (Figure 5A). To test the catalytic activity of ValG, the enzymatic reaction was carried out using validoxylamine A and UDP-, GDP-, ADP- or TDP-glucose as substrate. The reactions gave rise to a product, which has the same Rf value as authentic VAL-A on TLC (Figure 5B). On the other hand, incubation with cell-free extracts of E. coli harboring empty pRSET-B vector gave no product (data not shown). The product was confirmed to be VAL-A by ESI-MS and NMR. UDP-glucose was the most efficient glycosyl donor for the ValG reaction as judged by TLC (Figure 5B), whereas GDP-glucose was somewhat and ADP-glucose much less efficient. To our surprise, TDP-glucose was the least efficient donor for the reaction among the activated sugars tested.
Figure 5.
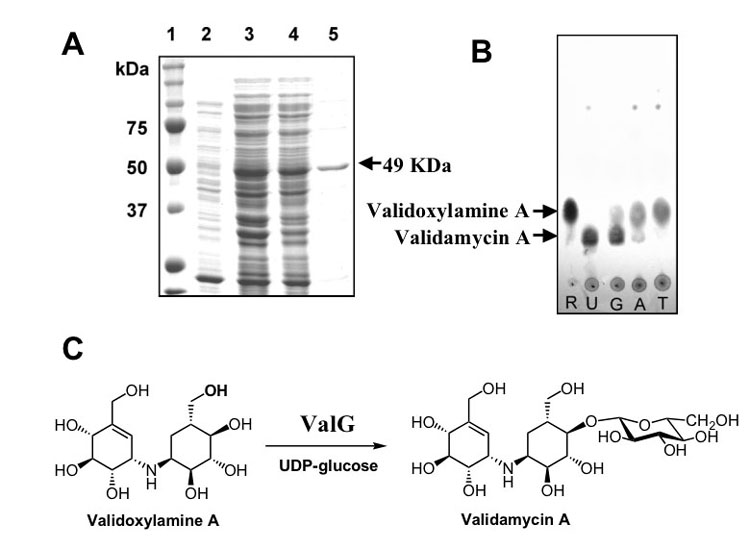
Heterologous Expression of valG and Characterization of the Glycosyltransferase. (A) SDS-PAGE analysis of ValG protein: 1. molecular weight marker, 2. soluble protein of extract of E. coli BL21Gold(DE3)pLysS/valG before induction, 3. total protein of extract of E. coli BL21Gold(DE3)pLysS/valG after induction with IPTG, 4. soluble protein of line 3, 5. purified his-tagged ValG. (B) TLC analysis of ValG reaction (silica gel, solvent system: nPrOH:AcOH:H2O = 4:1:1); R: validoxylamine A standard, U: ValG reaction using UDP-glucose as glucosyl donor, G: ValG reaction using GDP-glucose as glucosyl donor, A: ValG reaction using ADP-glucose as glucosyl donor, T: ValG reaction using TDP-glucose as glucosyl donor. (C) Conversion scheme of validoxylamine A to VAL-A catalyzed by ValG.
Discussion
Earlier feeding experiments with isotopically labeled potential intermediates led to a proposal that the biosynthesis of VAL-A is initiated by the cyclization of D-sedoheptulose 7-phosphate to form 2-epi-5-epi-valiolone, which subsequently epimerizes at C-2 to give 5-epi-valiolone, followed by dehydration between C-5 and C-6 to generate valienone. The conversion of valienone to validone takes place by anti-addition of hydrogen followed by transamination, with the α-nitrogen of glutamate as the probable nitrogen source, to form validamine. Analysis of a ca. 45 kb region of the S. hygroscopicus 5008 chromosome revealed 16 potential structural genes, which include genes deduced or proven to encode 2-epi-5-epi-valiolone synthase (ValA), epimerase/dehydratase (ValK), reductase (ValN), transaminase (ValM) and glucosyltransferase (ValG) (Table1). These enzymes fit nicely with the proposed scheme of VAL-A biosynthesis resulting from the feeding experiments. Despite some discrepancies between the biosynthesis of acarbose and VAL-A, cloning of the acarbose biosynthetic gene cluster and subsequent biochemical characterization of several enzymes involved in the biosynthesis has provided additional information for the elucidation of VAL-A biosynthesis. In the acarbose case, 2-epi-5-epi-valiolone, the cyclization product of AcbC, is phosphorylated by the kinase AcbM to give 2-epi-5-epi-valiolone 7-phosphate. Further conversion to 5-epi-valiolone 7-phosphate is catalyzed by the epimerase AcbO. The presence of ValC, a homolog of AcbM, in the val gene cluster suggests the involvement of phosphorylated intermediates in VAL-A biosynthesis as well. However since 2-epi-5-epi-valiolone, 5-epi-valiolone, valienone and validone were efficiently incorporated into VAL-A, the kinase (ValC) may utilize valienone and/or validone as substrate. Alternatively, the kinase might have broad substrate specificity, activating 2-epi-5-epi-valiolone, 5-epi-valiolone, valienone and validone and making it possible for them to be incorporated into the biosynthetic pathway.
In analogy to the proposed function of the dehydrogenase AcbL in acarbose biosynthesis, ValN is predicted to reduce the C-1 keto group of valienone-7-phosphate to give either valienol 7-phosphate or its epimer, 1-epi-valienol 7-phosphate (Figure 6). If valienol 7-phosphate is formed, a downstream coupling reaction, most likely catalyzed by the trehalose 6-phosphate synthase homolog (ValL), would occur with net retention of the C-1 configuration of the unsaturated cyclitol. However, if 1-epi-valienol 7-phosphate is formed the coupling reaction must take place with inversion of the C-1 configuration. In acarbose biosynthesis, it was proposed that reduction of valienone 7-phosphate yields 1-epi-valienol 7-phosphate and further phosphorylation of the latter compound by an unidentified kinase gives 1-epi-valienol 1,7-diphosphate. However, while this hypothesis is attractive, there is no evidence for the presence of a second kinase in the validamycin cluster. The presence of valO, which encodes a protein homologous to phosphatase/phosphohexomutase, in the validamycin cluster, conversely suggests the involvement of a phosphomutation step from the C-7 to the C-1 hydroxyl group, as commonly observed in sugar biosyntheses. However, inactivation of valO did not abolish the production of VAL-A, which leaves this part of the pathway unclear.
Figure 6.
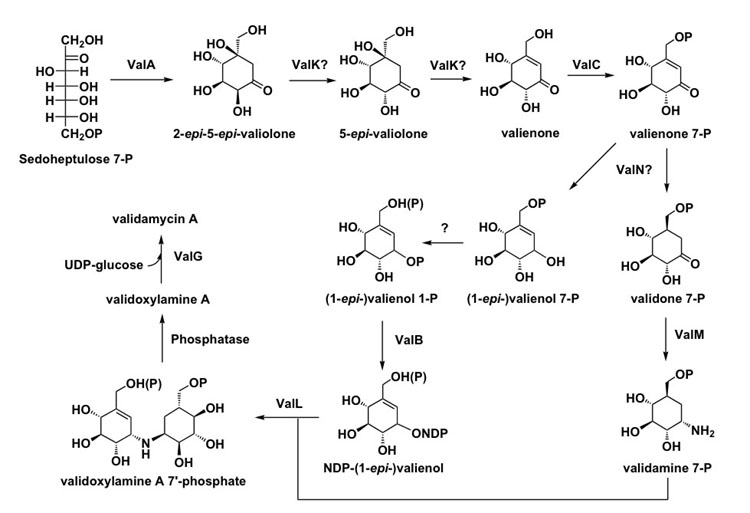
Proposed Biosynthetic Pathway to VAL-A.
Nevertheless, the availability of the genetic information reported here as well as results from our previous feeding experiments and biochemical analysis of several related enzymes, allows us to propose a more comprehensive pathway to VAL-A (Figure 6). In vivo inactivation and in vitro characterization of ValA confirmed the initiation step from D-sedoheptulose 7-phosphate to the cyclic product, 2-epi-5-epi-valionone. Subsequently, epimerization at C-2 and dehydration at C-5/C-6 of 2-epi-5-epi-valionone by the proposed bi-functional ValK would give valienone. ValK has significant homology with the sugar epimerase/dehydratase and it appears to be the only plausible candidate enzyme for the epimerization and dehydration reactions among the eight proteins known to be involved in VAL-A biosynthesis. In addition, ValK may not require NAD(P)+ for catalytic activity, which is similar to the L-ribulose-5-phosphate 4-epimerase and the dTDP-4-dehydrorhamnose 3,5-epimerase. However, no significant homology was found between the primary structures of ValK and the NAD+-independent sugar epimerases. This raises questions regarding the function and reaction mechanism of ValK, which prompts further investigations.
Prior to any further processing, valienone may be phosphorylated, presumably by the kinase, ValC, to give valienone 7-phosphate. Valienone 7-phosphate may serve as the branching point to be reduced to validone 7-phosphate on the one hand and to valienol 7-phosphate or 1-epi-valienol 7-phosphate on the other by the reductase ValN and/or other reductases/dehydrogenases. There are three oxidoreductase genes present in the cluster, valE, valF and valJ. However, the results of the heterologous expression experiments in S. lividans suggest that these genes are not necessary for the biosynthesis of VAL-A. Even so, it may be that they are involved in the biosynthesis of validamycins B and/or G, the hydroxylated analogs of VAL-A. Transamination of the keto group of validone 7-phosphate catalyzed by the aminotransferase ValM would give validamine 7-phosphate. On the other hand, valienol 7-phosphate or its 1-epimer is either phosphorylated at C-1 by an unidentified kinase to generate the 1,7-diphosphate derivative or transformed into valienol 1-phosphate through catalysis by a phosphoglucomutase. The product is subsequently converted to an NDP-valienol derivative by the nucleotidyl transferase ValB. Condensation of validamine 7-phosphate with the NDP-valienol derivative would give phosphorylated validoxylamine A, whose conversion to validoxylamine A requires a dephosphorylation activity. The corresponding phosphatase has not been located in the sequenced region. However, an internal trehalose 7-phosphate phosphatase or a non-specific sugar phosphatase may be able to carry out this hydrolytic reaction. Finally, validoxylamine A is converted to VAL-A biosynthesis through catalysis by the glucosyltransferase ValG using UDP-glucose as the sugar donor.
Significance
VAL-A is a strong crop protectant produced by S. hygroscopicus and is widely used for the treatment of the sheath blight disease of rice plants and the vegetable dumping-off caused by the fungus P. sasakii. In addition, its degradation product, valienamine, is a pharmaceutically important precursor for the production of the antidiabetic agent, voglibose. Despite its significant and wide applications in the biomedical and agricultural sectors, little is known about its biosynthesis. Sequencing and bioinformatics analysis of the validamycin biosynthetic gene cluster, as well as in vivo and in vitro characterization of a number of enzymes involved in VAL-A biosynthesis, provide important insights into the biosynthesis of this antibiotic. The data presented will also aid in the elucidation of the biosynthesis of other related aminocyclitol-containing natural products. In combination with knowledge about the acarbose biosynthetic gene cluster, it may now be possible to design unique primers for the screening and cloning of other aminocyclitol biosynthetic gene clusters from various sources. Exemplified by the significant accumulation of validoxylamine A in the glycosyltransferase mutants, in-depth knowledge about VAL-A biosynthesis could potentially be used to establish a platform for the production of drug precursors, e.g., valienamine, validamine, valiolamine and hydroxyvalidamine. Functional analysis of validamycin biosynthesis also sets the stage for creating novel analogs of VAL-A via combinatorial biosynthesis and yield improvement through metabolic engineering. And finally, with all the genes necessary for the synthesis of this important crop protectant at hand, one can contemplate the possibility of expressing them in plant systems to endow crop plants with their built-in protection against pathogens such as P. sasakii.
Experimental Procedures
Bacterial Strains, Plasmids, Culture Techniques and Media
S. hygroscopicus 5008 and its derivatives were grown either on solid SFM medium or in modified liquid TSB medium (supplemented with 1% yeast extract and 10.3% sucrose). Fermentation was carried out in FM-II medium at 37°C for 6 days. DH10B (GIBCO-BRL), ET12567(pUZ8002) and BL21Gold(DE3)pLysS were used as E. coli hosts for plasmid construction, E. coli-Streptomyces conjugation and protein over-expression, respectively. Pellicularia sasakii was used as indicator strain for VAL-A bioassay. pHZ1358 was the cosmid vector used for constructing the 5008 genomic library and for gene inactivation. pJTU472 contains aac(3)IV with HindIII sites on both sides, which was originally cloned from pHZ1070. pIJ2925 and pUC18 was E. coli vectors for plasmid construction or DNA sequencing. pRSET-B (Invitrogen) was used as vector for protein over-expression in E. coli.
Shuttle vector pJTU695 for complementation study was constructed as follows. The 1628 bp region between SphI and NruI of the pET15b (Novagen) was replaced first by a 868 bp SphI-BalI fragment containing oriT from pSET152. A 163 bp NdeI-BglII region of the resulting plasmid was further replaced by a 4365 bp fragment containing the replication origin of pIJ101 and tsr, creating pJTU676. A region of pJTU676 with the PermE* promoter, ribosomal binding site (RBS) and NdeI site was replaced with a corresponding fragment from pIB139 after digestion with EcoRI and HindIII. The distance between the RBS and the start codon of the downstream gene is 7 bp in the final construct, pJTU695, while that in pJTU676 is 23 bp.
DNA Sequencing and Analysis
DNA sequencing was performed using a set of five fragments directly isolated from cosmids 4G8 and 17F2, or cloned onto pBluescript SK(+) (Stratagene) or pSL301 (Invitrogen), which were originally connected or overlapped to cover a ca. 45 kb region with the validamycin biosynthetic gene cluster (Figure 2A). pHZ2232 with the 1.0 kb BamHI fragment was directly sequenced with T3 and T7 primers. Sequencing analysis of the 21 kb ClaI fragment, a 19.6 kb EcoRI fragment (pHZ2239), a 6.0 kb BamHI fragment (pHZ2229), and 7.0 kb and 3.8 kb BamHI fragments (pJTU464) were as described in.
Inactivation of valG
A 2.4 kb EcoRI-HindIII fragment flanking the left of the nt 112 of valG and a 1.0 kb HindIII-EcoRI fragment flanking the right of the nt 918 of valG were co-ligated into EcoRI-digested pIJ2925 to generate pJTU600. A HindIII fragment carrying the 1.4 kb aac(3)IV cassette from pJTU472 was inserted into the HindIII site between the 2.4 kb and 1.0 kb fragments to generate pJTU607, from which a 4.8 kb BglII fragment was excised for insertion into the BamHI-digested pHZ1358 to construct a vector (pJTU609) for mediating gene replacement (Figure 4) to obtain a valG mutant (LL-1). The mutation was confirmed by PCR amplification using forward primer valG2-F (5′-AGAGCCGATCTGGTGGTGAG-3′) and reverse primer valG2-R (5′-GGTGATGATTAGCCCTTCTCG-3′).
Complementation of Mutant LL-1 With Cloned valG
PCR-amplified valG was cleaved as a 1.2 kb NdeI-EcoRI fragment and cloned into pJTU695 digested with the same restriction enzymes. The new plasmid, pJTU612, was introduced from E.coli into LL-1 through conjugation as described and exconjugants were selected with 25 μg/ml thiostrepton. Further confirmation was carried out through plasmid isolation from 2-day old mycelia, transformation into E. coli, and comparison between the newly purified pJTU612 and the original one by restriction enzymes digestions. An exconjugant having the correct plasmid, named LL-101, was investigated through fermentation, bioassay and HPLC analysis as described.
Targeted disruption of valO
The valO gene was disrupted using ReDirect Technology. The aac (3)IV and oriT cassette was amplified from the pIJ773 disruption cassette using the primers ValO-PCR-F (5′-ATGTATAAGGTCGCACTTTTCGATCTGGACGGCACGTTAattccggggatccgtcgacc-3′) and ValO-PCR-R (5′-TCACGTGACGTCGTGATTCGAATCGGAGGGAGGAACGATtgtaggctggagctgcttc-3′) (pIJ773 homologous sequence is in lowercase letters). The resulting PCR product was used to replace valO first in pJTU714, a pHZ1358-derived plasmid carrying the 7.0 kb BamHI fragment containing valO, and then in strain 5008 as described [16]. Allelic replacement of valO in the ZYR-5 mutant was confirmed by PCR amplification with the primers of ValO-Det-F (5′-CGACGACCCTCAACCTC-3′) and ValO-Det-R (5′-CTGGCATCAAGCGACAC-3′). Fermentation of ZYR-5 and activity comparison between the wild-type and ZYR-5 through bioassay and HPLC were performed.
Cloning and Heterologous Over-expression of Recombinant His6-tagged ValG
The valG gene was amplified by PCR with Platinum® Pfx DNA polymerase (Invitrogen) using the cosmid 3G8 as template and primers ValGF3 ( 5′-GAAGATCTGCATATGCCCGGTGCGACATCCCATG-3′, engineered BglII and NdeI sites are in italic) and ValAR2 (5′-GGAATTCTCAGTCACCGCGAAGAGACGGCTCG-3′, engineered EcoRI site is in italic). The PCR amplification was done in a thermocycler (Eppendorf, Mastercycler gradient) under the following conditions: 33 cycles of 90 s at 95°C, 45 s at 60°C, and 45 s at 72°C. The PCR products were digested with BglII and EcoRI, and subsequently ligated into BamHI/EcoRI-digested pRSET-B. The constructs were transformed into E. coli XL-1 Blue and plated on LB agar plates containing 100 μg/ml ampicillin. The plasmid DNA was isolated and introduced by heat-pulse transformation into E. coli BL21 Gold(DE3)/pLysS (Stratagene), which was then plated onto LB agar plates containing 100 μg/ml ampicillin and 25 μg/ml chloramphenicol. The transformants were grown in 20 ml LB medium containing ampicillin and chloramphenicol at 37°C to an OD600 of 0.6. IPTG was added to a final concentration of 0.2 mM and the incubation was continued at 28°C for 24 h. The cells were harvested by centrifugation at 3500 rpm for 15 min and stored frozen at −80°C until further use.
Preparation of Cell-free Extracts and Purification of His6-tagged ValG
Cells were thawed and resuspended in disruption buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), the suspension was sonicated three times for 25 s each and cell debris were removed by centrifugation at 10,000 rpm for 10 min. The protein solution was applied to a BD TALONspin™ column (BD Biosciences) and centrifuged at 500 rpm for 2 min. The column was washed with washing buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0, twice, and 50 mM NaH2PO4, 300 mM NaCl, 100 mM imidazole, pH 8.0). The His6-tagged protein was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, pH 8.0) and dialyzed for 24 h against 1 liter of dialysis buffer (25 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 20 mM NH4Cl and 0.5 mM DTT). Protein concentration was measured by the Bradford protein microassay with bovine serum albumin as standard.
Enzyme Assay
The enzyme assay was carried out at 30°C for 3–12 h in a 100 μl volume of 25 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 20 mM NH4Cl, 15 mM NDP-glucose, 10 mM validoxylamine A and 50 μl of protein solution (2.4 mg/ml protein). The reaction progress was monitored by TLC analysis. For a scale-up experiment, 3.35 mg of validoxylamine A was used. The reaction mixture was applied to a Microcon YM-10 (Amicon) spin column. The flow through was subjected to a cation-exchange (Dowex 50Wx8-200 [H+]) chromatography column. The column was washed with 20 ml of water and the reaction product was eluted with 0.5M aqueous NH4OH. Fractions containing the desired product were pooled and lyophilized to give 4.1 mg of VAL-A as a white powder.
Heterologous Production of Validoxylamine A and VAL-A
The valABC operon was first cloned downstream of the PermE* promoter by inserting a 623 bp NdeI-FspI DNA fragment of valA and a 3280 bp FspI-BamHI fragment from pHZ2229 into pSL301 digested with NdeI and BamHI. Then, a 3964 bp NdeI-EcoRI fragment containing the complete valABC was cleaved from the resulting plasmid, pJTU755, and ligated into pIB139 digested with the corresponding enzymes, generating pJTU756. Subsequently a 5611 bp EcoRI fragment, containing valKLMN from cosmid 17F2, was inserted into EcoRI-digested pJTU756. Clones with valABCKLMN transcribed in the same orientation were selected and named pJTU757.
Plasmid pJTU757 was integrated into the chromosome of S. lividans 1326 by protoplast transformation and selection with apramycin to create a recombinant XH-6. Introduction of pJTU612 carrying valG into XH-6 by conjugation resulted in another recombinant XH-9, which was selected with thiostrepton and apramycin. Fermentation and bioassay analysis of XH-6 and XH-9 were done as previously described [16]. Tandem MS analysis was performed with fermentation broth extracted once with chloroform through directed injection on an Agilent 1100 series LC/MSD Trap system. The iontrap mass spectrometer was operated with the electrospray ionization source in the positive ion mode. Drying gas flow was 10 l/min, and nebulizer pressure was 50 psi. Drying gas temperature was 325°C. The fragmentation amplitude was varied between 1.0 and 1.8 V.
Acknowledgments
This work received support from the Ministry of Science and Technology (2003CB114205), the National Science Foundation of China, the Ph.D. Training Fund of the Ministry of Education and the Shanghai Municipal Council of Science and Technology. Work at Oregon State University was supported by NIH Grant RAI061528A. KM received postdoctoral training funds from Shionogi & Co., LTD, Osaka, Japan. The authors thank Dr. Patricia M. Flatt for helpful comments.
Footnotes
Data deposition: The complete DNA and deduced protein sequences of the val gene cluster reported in this paper have been deposited in GenBank under the accession number DQ164098.
References
- 1.Iwasa T, Yamamoto H, Shibata M. Studies on validamycins, new antibiotics. I. Streptomyces hygroscopicus var. limoneus nov. var., validamycin-producing organism. Jpn J Antibiot. 1970;23:595–602. [PubMed] [Google Scholar]
- 2.Iwasa T, Higashide E, Yamamoto H, Shibata M. Studies on validamycins, new antibiotics. II. Production and biological properties of validamycins A and B. J Antibiot (Tokyo) 1971;24:107–113. doi: 10.7164/antibiotics.24.107. [DOI] [PubMed] [Google Scholar]
- 3.Asano N, Takeuchi M, Kameda Y, Matsui K, Kono Y. Trehalase inhibitors, validoxylamine A and related compounds as insecticides. J Antibiot (Tokyo) 1990;43:722–726. doi: 10.7164/antibiotics.43.722. [DOI] [PubMed] [Google Scholar]
- 4.Kameda Y, Asano N, Yamaguchi T, Matsui K. Validoxylamines as trehalase inhibitors. J Antibiot (Tokyo) 1987;40:563–565. doi: 10.7164/antibiotics.40.563. [DOI] [PubMed] [Google Scholar]
- 5.Asano N, Tanaka K, Kameda Y, Matsui K. All eight possible mono-beta-D-glucosides of validoxylamine A. II. Biological activities. J Antibiot (Tokyo) 1991;44:1417–1421. doi: 10.7164/antibiotics.44.1417. [DOI] [PubMed] [Google Scholar]
- 6.Stratmann A, Mahmud T, Lee S, Distler J, Floss HG, Piepersberg W. The AcbC protein from Actinoplanes species is a C7-cyclitol synthase related to 3-dehydroquinate synthases and is involved in the biosynthesis of the alpha-glucosidase inhibitor acarbose. J Biol Chem. 1999;274:10889–10896. doi: 10.1074/jbc.274.16.10889. [DOI] [PubMed] [Google Scholar]
- 7.Mahmud T, Tornus I, Egelkrout E, Wolf E, Uy C, Floss HG, Lee S. Biosynthetic studies on the alpha-glucosidase inhibitor acarbose in Actinoplanes sp.: 2-epi-5-epi-valiolone is the direct precursor of the valienamine moiety. J Am Chem Soc. 1999;121:6793–6983. [Google Scholar]
- 8.Arakawa K, Bowers SG, Michels B, Trin V, Mahmud T. Biosynthetic studies on the alpha-glucosidase inhibitor acarbose: the chemical synthesis of isotopically labeled 2-epi-5-epi-valiolone analogs. Carbohydr Res. 2003;338:2075–2082. doi: 10.1016/s0008-6215(03)00315-x. [DOI] [PubMed] [Google Scholar]
- 9.Dong H, Mahmud T, Tornus I, Lee S, Floss HG. Biosynthesis of the validamycins: identification of intermediates in the biosynthesis of VAL-A by Streptomyces hygroscopicus var. limoneus. J Am Chem Soc. 2001;123:2733–2742. doi: 10.1021/ja003643n. [DOI] [PubMed] [Google Scholar]
- 10.Mahmud T, Lee S, Floss HG. The biosynthesis of acarbose and validamycin. Chem Rec. 2001;1:300–310. doi: 10.1002/tcr.1015. [DOI] [PubMed] [Google Scholar]
- 11.Mahmud T, Xu J, Choi YU. Synthesis of 5-epi-[6-(2)H(2)]valiolone and stereospecifically monodeuterated 5-epi-valiolones: exploring the steric course of 5-epi-valiolone dehydratase in VAL-A biosynthesis. J Org Chem. 2001;66:5066–5073. doi: 10.1021/jo0101003. [DOI] [PubMed] [Google Scholar]
- 12.Naganawa H, Hashizume H, Kubota Y, Sawa R, Takahashi Y, Arakawa K, Bowers SG, Mahmud T. Biosynthesis of the cyclitol moiety of pyralomicin 1a in Nonomuraea spiralis MI178-34F18. J Antibiot (Tokyo) 2002;55:578–584. doi: 10.7164/antibiotics.55.578. [DOI] [PubMed] [Google Scholar]
- 13.Kameda Y, Horii S, Yamano T. Microbial transformation of validamycins. J Antibiot (Tokyo) 1975;28:298–306. doi: 10.7164/antibiotics.28.298. [DOI] [PubMed] [Google Scholar]
- 14.Wehmeier UF, Piepersberg W. Biotechnology and molecular biology of the alpha-glucosidase inhibitor acarbose. Appl Microbiol Biotechnol. 2004;63:613–625. doi: 10.1007/s00253-003-1477-2. [DOI] [PubMed] [Google Scholar]
- 15.Zhang CS, Podeschwa M, Altenbach HJ, Piepersberg W, Wehmeier UF. The acarbose-biosynthetic enzyme AcbO from Actinoplanes sp. SE 50/110 is a 2-epi-5-epi-valiolone-7-phosphate 2-epimerase. FEBS Lett. 2003;540:47–52. doi: 10.1016/s0014-5793(03)00221-7. [DOI] [PubMed] [Google Scholar]
- 16.Yu Y, Bai L, Minagawa K, Jian X, Li L, Li J, Chen S, Cao E, Mahmud T, Floss HG, Zhou X, Deng Z. Gene Cluster Responsible for Validamycin Biosynthesis in Streptomyces hygroscopicus subsp. jinggangensis 5008. Appl Environ Microbiol. 2005;71:5066–5076. doi: 10.1128/AEM.71.9.5066-5076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 18.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henner DJ, Band L, Shimotsu H. Nucleotide sequence of the Bacillus subtilis tryptophan operon. Gene. 1985;34:169–177. doi: 10.1016/0378-1119(85)90125-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhang CS, Stratmann A, Block O, Bruckner R, Podeschwa M, Altenbach HJ, Wehmeier UF, Piepersberg W. Biosynthesis of the C(7)-cyclitol moiety of acarbose in Actinoplanes species SE50/110. 7-O-phosphorylation of the initial cyclitol precursor leads to proposal of a new biosynthetic pathway. J Biol Chem. 2002;277:22853–22862. doi: 10.1074/jbc.M202375200. [DOI] [PubMed] [Google Scholar]
- 21.Prescott AG, Lloyd MD. The iron(II) and 2-oxoacid-dependent dioxygenases and their role in metabolism. Nat Prod Rep. 2000;17:367–383. doi: 10.1039/a902197c. [DOI] [PubMed] [Google Scholar]
- 22.Kingston RL, Scopes RK, Baker EN. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure. 1996;4:1413–1428. doi: 10.1016/s0969-2126(96)00149-9. [DOI] [PubMed] [Google Scholar]
- 23.Ninomiya T, Sugiura N, Tawada A, Sugimoto K, Watanabe H, Kimata K. Molecular cloning and characterization of chondroitin polymerase from Escherichia coli strain K4. J Biol Chem. 2002;277:21567–21575. doi: 10.1074/jbc.M201719200. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9:1287–1296. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 25.Gunn FJ, Tate CG, Sansom CE, Henderson PJ. Topological analyses of the L-fucose-H+ symport protein, FucP, from Escherichia coli. Mol Microbiol. 1995;15:771–783. doi: 10.1111/j.1365-2958.1995.tb02384.x. [DOI] [PubMed] [Google Scholar]
- 26.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Ullman CG, Perkins SJ. A classification of mucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–942. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiba S. Molecular mechanism in alpha-glucosidase and glucoamylase. Biosci Biotechnol Biochem. 1997;61:1233–1239. doi: 10.1271/bbb.61.1233. [DOI] [PubMed] [Google Scholar]
- 29.Rocchetta HL, Burrows LL, Lam JS. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graninger M, Nidetzky B, Heinrichs DE, Whitfield C, Messner P. Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-L-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J Biol Chem. 1999;274:25069–25077. doi: 10.1074/jbc.274.35.25069. [DOI] [PubMed] [Google Scholar]
- 31.Wierenga R, De Maeyer M, Hol W. Interaction of pyrophosphate moieties with α-helices in dinucleotide binding proteins. Biochemistry. 1985;24:1346–1357. [Google Scholar]
- 32.Johnson AE, Tanner ME. Epimerization via carbon-carbon bond cleavage. L-ribulose-5-phosphate 4-epimerase as a masked class II aldolase. Biochemistry. 1998;37:5746–5754. doi: 10.1021/bi972984j. [DOI] [PubMed] [Google Scholar]
- 33.Samuel J, Luo Y, Morgan PM, Strynadka NC, Tanner ME. Catalysis and binding in L-ribulose-5-phosphate 4-epimerase: a comparison with L-fuculose-1-phosphate aldolase. Biochemistry. 2001;40:14772–14780. doi: 10.1021/bi011252v. [DOI] [PubMed] [Google Scholar]
- 34.Lee LV, Poyner RR, Vu MV, Cleland WW. Role of metal ions in the reaction catalyzed by L-ribulose-5-phosphate 4-epimerase. Biochemistry. 2000;39:4821–4830. doi: 10.1021/bi9928952. [DOI] [PubMed] [Google Scholar]
- 35.Storici P, Capitani G, De Biase D, Moser M, John RA, Jansonius JN, Schirmer T. Crystal structure of GABA-aminotransferase, a target for antiepileptic drug therapy. Biochemistry. 1999;38:8628–8634. doi: 10.1021/bi990478j. [DOI] [PubMed] [Google Scholar]
- 36.Delumeau O, Dutta S, Brigulla M, Kuhnke G, Hardwick SW, Volker U, Yudkin MD, Lewis RJ. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J Biol Chem. 2004;279:40927–40937. doi: 10.1074/jbc.M405464200. [DOI] [PubMed] [Google Scholar]
- 37.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia TH, Jiao RS. Studies on glutamine synthetase from Streptomyces hygroscopicus var. jinggangensis. Sci Sin [B] 1986;29:379–388. [PubMed] [Google Scholar]
- 39.Kieser, T., Bibb, M.J., Chater, K.F., Butter, M.J., and Hopwood, D.A. (2000). Practical Streptomyces genetics. A Laborratory Manual (John Innes Foundation, Norwich, UK).
- 40.Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moffatt BA, Studier FW. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell. 1987;49:221–227. doi: 10.1016/0092-8674(87)90563-0. [DOI] [PubMed] [Google Scholar]
- 42.Janssen GR, Bibb MJ. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y, Zhou X, Liu J, Bao K, Zhang G, Tu G, Kieser T, Deng Z. ‘Streptomyces nanchangensis’, a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology. 2002;148:361–371. doi: 10.1099/00221287-148-2-361. [DOI] [PubMed] [Google Scholar]
- 44.Chen S, Huang X, Zhou X, Bai L, He J, Jeong KJ, Lee SY, Deng Z. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem Biol. 2003;10:1065–1076. doi: 10.1016/j.chembiol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 47.Oliynyk M, Stark CB, Bhatt A, Jones MA, Hughes-Thomas ZA, Wilkinson C, Oliynyk Z, Demydchuk Y, Staunton J, Leadlay PF. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol Microbiol. 2003;49:1179–1190. doi: 10.1046/j.1365-2958.2003.03571.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Zhou X, Dong H, Tu G, Wang M, Wang B, Deng Z. A Complete Gene Cluster from Streptomyces nanchangensis NS3226 Encoding Biosynthesis of the Polyether Ionophore Nanchangmycin. Chem Biol. 2003;10:431–441. doi: 10.1016/s1074-5521(03)00092-9. [DOI] [PubMed] [Google Scholar]