Abstract
Although the Ran GTPase-activating protein RanGAP mainly functions in the cytoplasm, several lines of evidence indicate a nuclear function of RanGAP. We found that Schizosaccharomyces pombe RanGAP, SpRna1, bound the core of histone H3 (H3) and enhanced Clr4-mediated H3-lysine 9 (K9) methylation. This enhancement was not observed for methylation of the H3-tail containing K9 and was independent of SpRna1–RanGAP activity, suggesting that SpRna1 itself enhances Clr4-mediated H3-K9 methylation via H3. Although most SpRna1 is in the cytoplasm, some cofractionated with H3. Sprna1ts mutations caused decreases in Swi6 localization and H3-K9 methylation at all three heterochromatic regions of S. pombe. Thus, nuclear SpRna1 seems to be involved in heterochromatin assembly. All core histones bound SpRna1 and inhibited SpRna1–RanGAP activity. In contrast, Clr4 abolished the inhibitory effect of H3 on the RanGAP activity of SpRna1 but partially affected the other histones. SpRna1 formed a trimeric complex with H3 and Clr4, suggesting that nuclear SpRna1 is reciprocally regulated by histones, especially H3, and Clr4 on the chromatin to function for higher order chromatin assembly. We also found that SpRna1 formed a stable complex with Xpo1/Crm1 plus Ran-GTP, in the presence of H3.
INTRODUCTION
The concentration gradient of Ran-GTP from the nucleus to the cytoplasm (Kalab et al., 2002) is important to carry out the Ran-mediated cellular processes, such as nucleocytoplasmic transport of macromolecules, mitotic spindle formation, and postmitotic nuclear envelope assembly (Moore, 2001; Dasso, 2002; Hetzer et al., 2002; Weis, 2003; Mattaj, 2004). It is maintained by the cytoplasmic RanGAP (Becker et al., 1995; Bischoff et al., 1995a) and the chromosomal Ran-GDP/GTP exchange factor RCC1 (Kai et al., 1986; Ohtsubo et al., 1989; Bischoff and Ponstingl, 1991). Although RCC1 possesses only the nuclear localization signal (NLS) (Seino et al., 1992), RanGAP possesses a nuclear export signal (NES), in addition to NLS. For example, the Saccharomyces cerevisiae RanGAP homologue ScRna1p possesses a novel type of NLS and two of the classical NES signals, indicating that RanGAP is localized in the nucleus and exported, depending on the nuclear export receptor Xpo1/Crm1 (Feng et al., 1999). In Drosophila melanogaster, a naturally occurring meiotic drive system of the Segregation Distorter (SD) (Lyttle, 1991), which shows a preferential transmission of the SD chromosome from SD/SD+ heterozygous males, is caused by a mutated RanGAP, referred to as Sd-RanGAP (Merrill et al., 1999). This is enzymatically active but lacks a functional NES, so Sd-RanGAP accumulates in the nucleus (Kusano et al., 2001). Yrb1p, an S. cerevisiae homologue of mammalian RanBP1 that enhances RanGAP activity (Bischoff et al., 1995b; Noguchi et al., 1997; Seewald et al., 2003), shuttles between the nucleus and cytoplasm (Kunzler et al., 2000). We have also found that disruption of the S. cerevisiae YRB2 gene encoding a homologue of mammalian RanBP3, another RanGAP activator in the nucleus (Welch et al., 1999), is synthetically lethal with a temperature-sensitive mutant of S. cerevisiae RanGAP, rna1-1 (Noguchi et al., 1997). These reports suggest a hitherto unsuspected role of RanGAP in the nucleus.
We previously isolated a series of the temperature-sensitive (ts) mutants of the Sprna1+ gene encoding the Schizosaccharomyces pombe homologue of mammalian RanGAP. Sprna1ts shows a defect in chromosome segregation rather than in mitotic spindle formation or nucleocytoplasmic transport (Kusano et al., 2004). Interestingly, the temperature sensitivity of Sprna1ts is suppressed by overexpression of Clr4, a histone methyltransferase (HMTase) specific for histone H3 (H3)-K9 that is essential for heterochromatin assembly (Rea et al., 2000; Bannister et al., 2001; Nakayama et al., 2001), and is synthetically enhanced by a deletion of the clr4+ gene. Consistently, Sprna1ts shows a centromeric gene-silencing defect (Kusano et al., 2004). Thus, the phenotype of Sprna1ts suggests that RanGAP might have an unsuspected nuclear function related to heterochromatin assembly. In this context, it is intriguing how RanGAP may be functionally related with Clr4-HMTase. Here, Clr4 and its substrate H3 were found to play an important role in regulating a nuclear RanGAP.
MATERIALS AND METHODS
Yeast Media and Strains
S. pombe strains were grown in rich medium (YE5S) or Edinburgh minimal medium (EMM) with appropriate supplements. The strains used in this experiment are listed in Table 1.
Table 1.
S. pombe strains used in this study
| Strainsa | Genotype |
|---|---|
| AK4 | h−ura4-D18 leu1-32 |
| FY2267 | h+ura4-D18 leu1-32 ade6-m210 clr4::ura4+otr1R(dg-glu)Sph::ade6 |
| FY498 | h+ura4-DS/E leu1-32 ade6-m210 imr1R(NcoI)::ura4+ |
| FY648 | h+ura4-DS/E leu1-32 ade6-m210 otr1R(NcoI)::ura4+ |
| FY336 | h−ura4-DS/E leu1-32 ade6-m210 cnt1/TM(NcoI)::ura4+ |
| HN1 | h−ura4-D18 leu1-32 [pREP3X] |
| HN2 | h−ura4-D18 leu1-32 [pREP3X-hht1] |
| HN3 | h−ura4-D18 leu1-32 [pREP3X-NES-hht1] |
| HN4 | h+ura4-D18 leu1-32 ade6-m210 clr4::ura4+otr1R(dg-glu)Sph::ade6 [pREP3X] |
| HN5 | h+ura4-D18 leu1-32 ade6-m210 clr4::ura4+otr1R(dg-glu)Sph::ade6 [pREP3X-hht1] |
| HN6 | h+ura4-D18 leu1-32 ade6-m210 clr4::ura4+otr1R(dg-glu)Sph::ade6 [pREP3X-NES-hht1] |
| HN7 | h+ura4-DS/E leu1-32 imr1R(NcoI)::ura4+ |
| HN8 | h−ura4-DS/E leu1-32 imr1R(NcoI)::ura4+sprna1-1ts |
| HN9 | h−ura4-DS/E imr1R(NcoI)::ura4+sprna1-47ts |
| HN10 | h−ura4-DS/E leu1-32 imr1R(NcoI)::ura4+sprna1-86ts |
| HN11 | h+ura4-DS/E leu1-32 imr1R(NcoI)::ura4+sprna1-87ts |
a Strains FY498, FY648, and FY336 are described in Nakagawa et al. (2002). FY2267 is described in Bannister et al. (2001). Strains starting with AK are described in Kusano et al. (2004). Strains starting with HN are generated in this study.
Recombinant Protein Preparation
Clr4: S. pombe clr4+ that was isolated previously (Kusano et al., 2004) was fused with 6xHis in-frame using pRSETc (Table 2) and was expressed in Escherichia coli. Clr4 was purified using Ni-NTA agarose (QIAGEN, Hilden, Germany) and MonoQ (GE Healthcare, Piscataway, NJ) as reported previously (Nakayama et al., 2001).
Table 2.
Plasmids used in this study
| Plasmid | Marker | Description | Reference |
|---|---|---|---|
| For yeast | |||
| pREP3X | LEU2 | Yeast-inducible expression vectors | Maundrell (1993) |
| pREP3X-hht1 | LEU2 | pREP3X with hht1+ fragment at BamHI/SmaI site | This study |
| pREP3X-NES-hht | LEU2 | pREP3X with NES fused hht1+ fragment at XhoI/BamHI site | This study |
| For E. coli | |||
| pRSETc-Clr4 | pRSETc with Clr4+ at BamHI/EcoRI site | Nakayama et al. (2001) | |
| pQE31-Rna1 | pQE31 with Rna1+ at SalI/PstI site | This study | |
| pQE31-Rna1-8 | pQE31 with Rna1-8ts at SalI/PstI site | This study | |
| pQE31-Rna1-15 | pQE31 with Rna1-15ts at SalI/PstI site | This study | |
| pQE31-Rna1-87 | pQE31 with Rna1-87ts at SalI/PstI site | This study | |
| pET3b-hht1FL | pET3b with hht1+ (1-136) at NdeI/BamHI site | This study | |
| pET3b-hht1N | pET3b with hht1 (1-40) at NdeI/BamHI site | This study | |
| pET3b-hht1C | pET3b with hht1 (41-136) at NdeI/BamHI site | This study | |
| pET8c-Ran | pET8c with Ran at NcoI/BamHI site | Dasso et al. (1994) | |
| pET3b-RCC1 | pET3b with RCC1 at NdeI/BamHI site | Dasso et al. (1994) | |
| pGEX-CS-RanBP1 | pGEX-CS with RanBP1 at NcoI/XhoI site | Hayashi et al. (1995) |
SpRna1: S. pombe Sprna1+ and Sprna1ts genes were amplified from the S. pombe genomic DNA by PCR using as primers Rna1-N (5′-AAC GCG TCG ACA TGT CGC GTT TTT CAA TAG AAG GG) and Rna1-C (5′-AAA ACT GCA GCA TCC CTA AAT ATG AGC TTT TGA TAG CTC). Amplified DNA fragments were inserted into pQE31 (QIAGEN) (Table 2). Resulting 6xHis-fused SpRna1 was expressed in E. coli and purified using Ni-NTA agarose and MonoQ.
Hht1: S. pombe hht1+ (S. pombe gene, no. SPAC1834.04), encoding a mammalian H3 homologue, was amplified from the S. pombe genomic DNA using as primers Hht1-N (5′-CCG CAT ATG GCT CGT ACT AAA CAA AC), Hht1-M (5′-CCG CAT ATG CGT TAT CGT CCT GGT ACT GT), Hht1-Ccom (5′-CGG GAT CCT TAT GAG CGT TCG CCA CGG A), and Hht1-Mcom (5′-CCG CAT ATG CGT TAT CGT CCT GGT ACT GT). Amplified DNA fragments were inserted into pET3b vectors (Table 2). A full-sized, core, and tail Hht1 were expressed in E. coli and purified as described previously (Luger et al., 1999). Purified proteins were conjugated to the beads, NHS-activated Sepharose 4FF (GE Healthcare), at 2 mg protein/ml Sepharose.
Analysis of Clr4-mediated HMTase Activity
Recombinant Clr4 (80 nM) was mixed with commercially available calf H3 (Roche Diagnostics, Mannheim, Germany) (8 μM), S-adenosyl-l-[methyl-14C]methionine ([14C]SAM) (80 μM) as the methyl donor, and the indicated proteins in 40 μl of HMTase buffer (50 mM Tris, pH 8.0, 20 mM KCl, 10 mM MgCl2, 250 mM sucrose, and 0.5 mM dithiothreitol [DTT]). After incubation for 1 h at 30°C, each sample was given SDS sample buffer and boiled. Boiled samples were separated by 17% SDS-PAGE and visualized by Coomassie staining. 14C-Labeled H3 was detected and analyzed using Bio-Imaging analyzer BAS-2500 (Fujifilm, Tokyo, Japan).
Methylation of the H3/Hht1-tail (ARTKQTARKSTGGKAPRKQL) and the Hht1-core was carried out in the same condition. After incubation with [14C]SAM, the sample was spotted onto the P81 phosphocellulose filter paper (catalog no. 3698023; Whatman, Maidstone, United Kingdom) and washed four times by incubating each time for 10 min in 50 mM NaHCO3, pH 9.0. The radioactivity incorporated into each substrate was calculated by liquid scintillation counter as described previously (Nakayama et al., 2001).
Analysis of RanGAP Activity
[γ-32P]GTP was loaded on Ran by incubating for 10 min at 30°C in loading buffer (25 mM Tris, pH 7.5, 50 mM NaCl, 10 mM EDTA, and 1 mM DTT). The reaction was stopped with the addition of 20 mM MgCl2 and the resulting Ran-[γ-32P]GTP molecules were collected through a PD10 column (GE Healthcare) that had been equilibrated with GAP buffer (25 mM Tris, pH 7.5, 50 mM NaCl, 20 mM MgCl2, 1 mM DTT, and 0.05% gelatin [catalog no. G-7765; Sigma-Aldrich, St. Louis, MO]). Fifty nanomolar Ran-[γ-32P]GTP were incubated for 10 min at 30°C in 100 μl of GAP buffer containing various concentrations of SpRna1 and the indicated proteins. The reaction was stopped by addition of ice-cold stop buffer {20 mM Tris, pH 7.5, 25 mM MgCl2, and 100 mM NaCl). Resulting reaction mixtures were spotted onto the nitrocellulose membrane (0.45 μm, NC45; Whatman Schleicher and Schuell, Dassel, Germany) and washed with ice-cold stop buffer. The radioactivity of the [γ-32P]GTP remaining on Ran is calculated by a liquid scintillation counter.
To separate GTP and Pi, the reaction was stopped by the addition of EDTA (final concentration 72 mM), and the reaction mixtures were boiled. Samples (0.3 μl) were spotted on a thin layer chromatography plate (PEI matrix; Sigma-Aldrich) and then [γ-32P]GTP and 32P-labeled inorganic phosphate (32Pi) were separated with 1 M LiCl and 1 M formic acid for 60 min. 32P-labeled reagents were detected and analyzed using BAS-2500. By setting the sum of a radioactivity of [γ-32P]GTP and 32Pi measured by BAS-2500 to 100%, the amount of GTP molecule hydrolyzed per second by Ran was calculated.
Surface Plasmon Resonance Analysis
Measuring was done in a Biacore 2000 (BIAcore, Uppsala, Sweden) instrument. Purified Clr4, and calf H3, were immobilized separately onto the biosensor chip CM5 (BIAcore) with an amine coupling kit (BIAcore). SpRna1 suspended in HBS-EP buffer (BIAcore) was injected for 180 s. The response of each flow cell from which the response of a blank flow cell was subtracted is indicated. The sensorgrams obtained were evaluated by BIAevaluation software (BIAcore) to estimate the value of ka and kd.
Nucleosome Purification
The procedure described by Edmondson et al. (1996) was modified as following. Cells in 1 liter of YE5S culture (1 × 107 cells/ml) were grown at 30°C and harvested. Cell pellets were washed with sterile water and then suspended in 50 ml of buffer (0.1 mM Tris, pH 8.5, and 10 mM DTT). After incubation for 10 min at 30°C with gentle shaking, cells were washed with PEMS buffer (100 mM PIPES, pH 6.9, 1 mM EDTA, 1 mM MgCl2, and 1.2 M sorbitol) and suspended in PEMS buffer supplemented with 1.0 mg/ml zymolyase 100T (Seikagaku, Tokyo, Japan). After incubation at 30°C for 30 min with gentle shaking, the reaction was stopped by addition of ice-cold PEMS buffer. Resulting spheroplasts were washed three times with ice-cold PEMS buffer. Cell pellets were suspended in 50 ml of ice-cold NIB buffer (0.25 M sucrose, 60 mM KCl, 14 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM PIPES, pH 6.9, and 0.8% Triton X-100) supplemented with a mixture of protease inhibitors (phenylmethylsulfonyl fluoride [code. no. 273-27; Nacalai Tesque, Kyoto, Japan], pepstatin A [code no. 4039; Peptide Institute, Osaka, Japan], leupeptin (code no. 4041; Peptide Institute), aprotinin [code no. 016-11836; Wako Pure Chemicals, Osaka, Japan], and benzamidine (code no. 04036-72; Nacalai Tesque]) on ice for 20 min. After incubation, the insoluble fraction was spun down. Resulting precipitates were washed five times with washing buffer A (10 mM Tris, pH 7.5, 0.5% NP-40, 75 mM NaCl, and a mixture of protease inhibitors) and then incubated in washing buffer B (10 mM Tris, pH 7.5, 0.4 M NaCl, and a mixture of protease inhibitors) for 10 min on ice. After centrifugation, pellets were washed five times with washing buffer B. Both precipitated fractions, P1 and P2, shown in Figure 3A, were digested with 6 U/ml micrococcal nuclease (MNase; catalog no. N3755; Sigma-Aldrich) in MNase buffer {20 mM Tris, pH 7.5, 100 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5% glycerol, and 0.1% Triton X-100) at 30°C for 1 h. After treatment with MNase, samples were centrifuged to fractionate into the supernatants and the precipitates. The antibodies to SpRna1 were prepared, and other antibodies were obtained as follows: anti-Pim1 and anti-Spi1 antibodies were from Dr. Shelley Sazer (Baylor College of Medicine, Houston, TX) (Matynia et al., 1996), anti-histone H3 antibodies were from Abcam (ab1791; Abcam, Cambridge, United Kingdom), and the monoclonal antibody (mAb) to nucleoporins, mAb414, was from Covance {catalog no. MMS-120R; Covance, Berkeley, CA: Davis and Blobel, 1986).
Figure 3.
Chromatin-localization of SpRna1. (A) Schematic of fractionation of the cell extracts derived from spheroplasts of exponentially growing S. pombe wild-type AK4. The spheroplasts were treated with Triton X-100. Resulting cell extracts (total) were divided into supernatant (soluble) and precipitate (insoluble) fractions by centrifugation. Resulting insoluble fractions were treated as indicated. At each step, fractions were divided into the supernatants (S) and precipitates (P) by centrifugation. Precipitated fractions that had been treated with MNase were divided into the supernatants (Sup.) and precipitates (Pellet) by centrifugation. (B) Total cell extracts and indicated fractions were resolved in 5–20% gradient SDS-PAGE and analyzed by immunoblotting with antibodies to SpRna1, Pim1, Hht1, and Spi1 and with mAb414 as indicated. Based on the molecular mass, *1, *2, and *3 may include Nup189 (SPAC1885.12c), Nup124 (SPAC30D11.04c), and p65 (SPAC18B5.08c), respectively.
Chromatin Immunoprecipitation (ChIP) Assay
The procedure described by Hecht et al. (1996) was modified as follows. Cells in 100 ml of EMM with supplements culture (1 × 107 cells/ml) grown at 26°C were fixed by incubating with formaldehyde (final concentration 1%) for 15 min at 30°C and then on ice for 50 min. Fixed cells were washed four times with Tris-buffered saline (25 mM Tris, pH 7.5, and 150 mM NaCl). Resulting cells were suspended in 500 μl of extraction buffer (50 mM Tris, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and protease inhibitors) and disrupted with glass beads. Chromatin DNA was fragmented to an average length of 0.8 kb by sonication. Seventy microliters of cell extracts was mixed with antibodies to K9-methylated H3 (catalog no. 07-441; Upstate Biotechnology, Lake Placid, NY), SpRna1, Swi6 (Sadaie et al., 2004), K4-methylated H3 (ab7766; Abcam), Pim1, or as a control, to mouse immunoglobulin (code no. Z0109; DakoCytomation, Glostrup, Denmark). The immune complexes were purified using protein G-Sepharose beads (GE Healthcare), washed five times with extraction buffer and two times with LiCl buffer (10 mM Tris, pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP-40, and 0.5% Na-deoxycholate), and then with TE buffer (10 mM Tris, pH 8.0, and 1 mM EDTA). Whole cell extracts (WCE) and the chromatin DNA immunoprecipitated with antibodies were treated with ChEB buffer (10 mM Tris, pH 8.0, 300 mM NaCl, 5 mM EDTA, and 0.5% SDS) for 13 h at 65°C and digested with 10 μg/ml RNase A (Nacalai Tesque) for 30 min at 37°C and then with 80 μg/ml proteinase K (Merck, Darmstadt, Germany) for 1 h at 55°C. Resulting supernatants were given 50 μg of yeast tRNA (catalog no. 109495; Roche Diagnostics) and treated with phenol/chloroform. Purified DNA was precipitated by ethanol in the presence of Na-acetate. Immunoprecipitated DNA and the DNA from WCE were amplified by PCR using the indicated primers (Table 3) in the presence of [α-32P]dCTP. PCR products were separated on 5.0% nondenaturing polyacrylamide gel to be analyzed using BAS-2500.
Table 3.
Primers used in ChIP assay
| Locus | Name | Sequence | Reference |
|---|---|---|---|
| ura4 | ura4FW | 5′-GAGGGGATGAAAAATCCCAT-3′ | Ekwall et al. (1997) |
| ura4RV | 5′-TTCGACAACAGGATTACGACC-3′ | ||
| act1 | act1FW | 5′-GAAGTACCCCATTGAGCACGG-3′ | Noma et al. (2001) |
| act1RV | 5′-CAATTTCACGTTCGGCGGTAG-3′ | ||
| dg223 | dg223FW | 5′-TGGTAATACGTACTAGCTCTCG-3′ | Nakagawa et al. (2002) |
| dg223RV | 5′-AACTAATTCATGGTGATTGATG-3′ | ||
| E12 | B15E1-2490 | 5′-CGATGCTCTCGACAAAGCCGTTCT-3′ | Sadaie et al. (2003) |
| B15E1-3010 | 5′-CCATCTCAAACTTCTGTTCAACATT-3′ | ||
| matinga | mat107FW | 5′-TAATATGCTGGTATGGACATAGC-3′ | This study |
| mat648RV | 5′-AGTGGAGATGCGTATTTGGGAAC-3′ |
a Between the IR-L and the mat 2P.
Preparation of hht1+ for Expressing in S. pombe
The hht1+ gene was amplified from S. pombe genomic DNA by PCR using the primers H3-REPN (5′-CGG GAT CCA TGG CTC GTA CTA AAC AAA C) and H3-REPC (5′-CGC TCG AGT TAT GAG CGT TCG CCA CGG A). Resulting DNA fragments were introduced into pREP3X (Table 2). To construct the FLAG-NES fused hht1+ gene, two oligonucleotides—FLAG-NES (5′-CTA GAC TCG AGA TGG ACT ATA AAG ATG ACG ATG ACA AGG GGC TTG CGC TAA AAC TCG CCG GCC TCG ATA TCC A) and FLAG-NESr (5′-TAT GGA TAT CGA GGC CGG CGA GTT TTA GCG CAA GCC CCT TGT CAT CGT CAT CTT TAT AGT CCA TCT CGA GT) (Stade et al., 1997)—were annealed and then digested by the restriction enzymes XbaI and NdeI. Resulting DNA fragments were inserted into pET3b-hht1 (Table 2). The XhoI/BamHI DNA fragments of the resulting plasmid were inserted into pREP3X (Table 2), resulting in pREP3X-NES-hht1. Constructed plasmids (Table 2) were introduced into the clr4+ and clr4Δ strains with electroporation.
RESULTS
SpRna1 Enhances the HMTase Activity of Clr4 through Histone H3
Previously, we found a genetic interaction of SpRna1 with Clr4-HMTase (Kusano et al., 2004). To confirm this interaction biochemically, recombinant Clr4 and SpRna1 were prepared and incubated with H3, a specific substrate of the Clr4-HMTase (Rea et al., 2000; Nakayama et al., 2001). Methylated H3 was detected by the radioactivity of H3 labeled with [14C]SAM. Recombinant Clr4 proteins methylated H3 (Figure 1A, lane 3), as reported previously (Nakayama et al., 2001). The level of 14C-labeled H3 was apparently increased by the addition of SpRna1 (Figure 1A, compare lane 4 with lane 3), whereas SpRna1 itself had no activity to methylate H3 (Figure 1A, lane 2). To confirm this finding, increasing doses of SpRna1, and the controls, RanGEF-RCC1, Ran-GTP, or Ran-GDP, were mixed with H3 and Clr4 in the presence of [14C]SAM. As shown in Figure 1B, the amount of labeled H3 increased in a dose-dependent manner with the addition of SpRna1. In contrast, RanGEF-RCC1, Ran-GTP, and Ran-GDP, in addition to the boiled SpRna1, showed no effect on Clr4-mediated H3 methylation.
Figure 1.
Enhancement of Clr4-HMTase with SpRna1. (A) Clr4 (80 nM), SpRna1 (2 μM), and H3 (8 μM) were mixed as indicated and incubated in the presence of [14C]SAM (80 μM) as a methyl donor in 30 μl of HMTase buffer. After incubation for 1 h at 30°C, reaction mixtures were given SDS sample buffer, boiled, and separated by 17% SDS-PAGE and visualized by Coomassie staining. The methylated protein was analyzed using BAS-2500. (B) Clr4 (80 nM) was mixed with H3 (8 μM), [14C]SAM (80 μM), and the increasing amount (20 nM, 200 nM, 2 μM, and 20 μM) of RCC1, Ran that had bound GTP or GDP as indicated, and SpRna1 in 30 μl of HMTase buffer. Boiled SpRna1 was also added as a control. After incubation for 1 h at 30°C, reaction mixtures were given SDS sample buffer, boiled, and separated by 17% SDS-PAGE and visualized by Coomassie staining. The methylated H3 was analyzed using BAS-2500.
The interactions of SpRna1 with Clr4 and H3 were then examined using surface plasmon resonance analysis. Clr4 and H3 were immobilized separately onto a biosensor chip. When SpRna1 was injected at the indicated concentration, SpRna1 bound H3, but not Clr4, in a dose-dependent manner (Figure 2A). The calculated ka and kd values of SpRna1 to H3 were 9.8 × 104 and 4.4 × 10−4, respectively (equilibrium constant KD = 4.5 nM). H3 consists of the C-terminal core and the N-terminal tail that contains lysine 9 (K9), which is methylated by Clr4 (Nakayama et al., 2001; Khan and Hampsey, 2002). To determine which part of H3 binds SpRna1, an S. pombe hht1+ gene encoding a mammalian H3 homologue was cloned and divided into tail and core regions (Figure 2B, top). The full-sized (Hht1-FL), tail (Hht1-tail), and core (Hht1-core) Hht1 produced in E. coli were purified and conjugated with Sepharose beads. When these were mixed with SpRna1, SpRna1 coprecipitated with the core of Hht1 and with the full-sized Hht1, but not with the tail (Figure 2B). The amount of SpRna1 coprecipitated with the full-sized Hht1 was similar to the amount coprecipitated with the core (Figure 2B, compare lanes 4 and 6), indicating that SpRna1 binds to H3 through its core region.
Figure 2.
SpRna1 enhanced the Clr4-HMTase activity through H3. (A) Clr4 and H3 were immobilized separately onto the biosensor chip CM5. The various concentrations of SpRna1 (1:0, 2:20, 3:60, 4:100, and 5:150 nM) were injected for 180 s (from on to off) in HBS-EP buffer. The responses of flow cells conjugated with H3 and Clr4, from which the response of a blank flow cell had been subtracted, are shown on the vertical line. (B) Top, schematic of S. pombe H3, Hht1. Indicated proteins conjugated to Sepharose 4FF beads were incubated with 2 nM 6xHis-SpRna1 in 1 ml of GAP buffer supplemented with 1 mM CHAPS. After incubation for 1 h at 4°C, beads were spun down, washed five times, and proteins coprecipitated with beads were separated by 5–20% gradient SDS-PAGE and blotted with the mAb to 6xHis (catalog no. 8916-1, Clontech, Mountain View, CA). An arrowhead indicates the position of 6xHis-SpRna1. Input indicated a total amount of 6xHis-SpRna1 used in this experiment. (C) Clr4 (80 nM) was mixed in 30 μl of HMTase buffer, with [14C]SAM (80 μM), 8 μM full-sized H3 (●), H3/Hht1-tail* (○), or Hht1-core (□), and an increasing concentration (100, 300, or 900 nM) of SpRna1 (horizontal line). After incubation for 1 h at 30°C, the reaction mixtures were spotted onto the P81 phosphocellulose filter papers that were washed four times in 50 mM NaHCO3, pH 9.0. The radioactivity incorporated into each substrate was calculated by liquid scintillation counter. The percentage of radioactivity of each substrate labeled with 14C is calculated by setting the radioactivity of the reaction possessing H3, but lacking SpRna1, to 100% and plotted on the vertical line. The same experiment was repeated three times and error bars were marked. Asterisk (*) indicates that mammalian H3-tail and Hht1-tail possess the same amino acid sequences.
As reported previously (Nakayama et al., 2001), Clr4 specifically methylated the H3/Hht1-tail (Figure 2C, open symbols). Under the same conditions, SpRna1 enhanced the methylation of the full-sized H3, but not the H3/Hht1-tail (Figure 2C). Therefore, we conclude that SpRna1 enhances Clr4-mediated H3 methylation by binding to the H3-core.
SpRna1 Is Localized on Chromatin
Besides SpRna1 binding to H3, SpRna1 enhanced Clr4-HMTase that is required for heterochromatin assembly via H3-K9 methylation. This raises the question of whether SpRna1 is localized in the nucleus. To address this issue, spheroplasts of exponentially growing wild-type cells were lysed with Triton X-100 to fractionate them into soluble and insoluble fractions (Figure 3A). The insoluble fractions containing chromatin were treated with NP-40 and then with 0.4 M NaCl (Figure 3A). Finally, both precipitated fractions, P1 and P2 (Figure 3A), were digested with MNase. The resulting supernatants and precipitates were analyzed by immunoblotting using the indicated antibodies. Although most SpRna1 was dissolved after treatment with Triton X-100 as described previously (Feng et al., 1999; Dasso, 2002), some SpRna1 molecules were fractionated into the insoluble fraction containing chromatin (Figure 3B, lane 3). They were rendered soluble after digestion with MNase (Figure 3B, lanes 6 and 10), like Hht1, the S. pombe homologue of mammalian H3 used as a control for chromatin-bound protein (Figure 3B, compare SpRna1 with Hht1). In contrast, Pim1, another chromosomal protein, dissolved after treatment with 0.4 M NaCl, as reported previously for RCC1 (Ohtsubo et al., 1989). To confirm our fraction assay, we examined the behavior of nucleoporins by immunoblotting with mAb414, which stains S. pombe nucleoporins (Tange et al., 2002). Although p65 (designated as *3 in Figure 3B) was soluble, some nucleoporins were insoluble (Figure 3B, mAb414). Among these, proteins designated as *2 dissolved in 400 mM NaCl (Figure 3B, lane 8). In contrast, the nucleoporin designated as *1 partially fractionated into the P2 fraction (Figure 3B, lane 9). However, this was not dissolved by MNase digestion (Figure 3B, compare lane 10 with 11), in contrast to SpRna1 and Hht1. These results suggest that a nuclear SpRna1 binds chromatin in a manner similar to H3.
To determine where SpRna1 associates with chromatin, ChIP assay was carried out on S. pombe Sprna1+ and Sprna1ts cells, both of which contain the ura4+ gene inserted at the innermost repeat of the centromere (imr1R::ura4+), and the ura4 minigene (ura4DS/E). DNA was immunoprecipitated with antibodies to SpRna1. As controls, we used antibodies to the methylated H3-K9 peptide Swi6 (S. pombe homologue of human HP1) and the methylated H3-K4 peptide or Pim1. DNA immunoprecipitated with the antibodies, and DNA of WCE as a control, were subjected to PCR amplification using the primer sets shown in Table 3. These amplified the centromere (ura4+ and dg223), telomere (E12) or mating-type regions. In addition, a primer pair to amplify the act1+ gene was included as an internal control to verify the enrichment. A set of ura4 primers amplified ura4DS/E, which can also be used as an internal control, in addition to the ura4+ genes inserted in the centromere. The relative enrichment of heterochromatic regions was calculated based on the radioactivity incorporated into PCR products (Noma et al., 2001). Autoradiographs of PCR products are shown in Figure 4, A–D. Unfortunately, we could not detect any PCR products in SpRna1-ChIP.
Figure 4.
Figure 4 (facing page). SpRna1 was required for hetrochromain assembly. (A–D) H3-K9 methylation at CEN, TEL, and MAT was reduced in Sprna1ts. DNA isolated from the immunoprecipitated chromatin fractions using the indicated antibodies or from the WCE was used as a template for PCR-amplifying ura4 gene (A), centromeric region (B), telomeric region (C), or mating-type locus (D). The DNA samples were prepared from Sprna1+ or Sprna1ts cells cultured at 26 or 34°C for 5 h. The relative enrichment of ura4+ to ura4DS/E (A) and of cen-dg223 (B), tel-E12 (C), or mating type locus (D) to act1 was calculated as reported previously (Noma et al., 2001), and its ratio to that of WCE is shown at the bottom of each lane. (E) Centromeric gene silencing activity of Sprna1ts mutants. Top, schematic showing the cen1 and the ura4+ insertion within imr1R. Sprna1+ and Sprna1ts strains possessing the ura4+ gene at the imr1R domain that were spotted. Each strain was grown to 1.0 × 107 cells/ml in EMM supplemented with uracil. Serial dilution (1:5) of the indicated cultures were spotted onto nonselective (+ura) or selective (−ura) plates and incubated at 26°C, the permissive temperature, for 6 d. The highest density spots contained 1 × 104 cells.
SpRna1 Is Required for H3-K9 Methylation in All Heterochromatic Regions of S. pombe
Figure 4 also shows the effect of Sprna1ts mutation on H3-K9 methylation and the association of Swi6 with heterochromatin. Even at 26°C, a permissive temperature for Sprna1ts, the level of H3-K9 methylation was reduced in all three heterochromatic regions (centromeres, telomere, and the mating-type locus) of Sprna1ts compared with Sprna1+. In particular, H3-K9 methylation was very low at the imr1R::ura4+ region of Sprna1ts (Figure 4A). After incubation at 34°C, a nonpermissive temperature, H3-K9 methylation was further reduced in all three heterochromatic regions of Sprna1ts (Figure 4, A–D). In contrast, the levels of H3-K4 methylation at both the act1+ gene and the ura4DS/E minigene were not affected by incubation at 34°C. Thus, a defect of SpRna1 inhibited H3-K9 methylation in all three heterochromatic regions. In parallel with the reduction of H3-K9 methylation, the level of Swi6 binding the methylated H3-K9 (Bannister et al., 2001; Nakayama et al., 2001) was reduced at all three heterochromatic regions of Sprna1ts (Figure 4, A–D, Swi6).
It is notable that the level of Swi6 associated with heterochromatin was lower than that of H3-K9 methylation even at 26°C, the permissive temperature (Figure 4, A–D, relative enrichment). Consistent with the fact that the association of Swi6 with methylated H3-K9 is essential for the establishment of heterochromatin, the silencing of the ura4+ gene inserted into the centromeric region as shown in Figure 4E, top, was abolished in Sprna1ts, even at 26°C (Figure 4E).
SpRna1 Enhances the HMTase Activity of Clr4 Independently of Its RanGAP Activity
Because SpRna1 seems to be involved in heterochromatin assembly, we tested whether the ability of SpRna1 to enhance the Clr4-mediated H3 methylation could be further increased by the RanGAP activity of SpRna1. Given that RCC1 showed no effect on the Clr4-mediated H3 methylation (Figure 1B), we constructed a system where Ran-GTP is continuously supplied via RanGEF-RCC1 in the presence of high amounts of GTP, because Ran-GTP is hydrolyzed rapidly to Ran-GDP by the RanGTPase in the presence of SpRna1–RanGAP (for details, see Materials and Methods and legend to Figure 5A legend). In this assay condition, upon adding nonradioactive GTP, the amount of residual [γ-32P]GTP increased (Figure 5A, a), indicating that the exchange Ran-GTP ↔ Ran-GDP occurred continuously in the reaction mixture in which Ran-GTP was mixed with SpRna1, RCC1, RanBP1, [γ-32P]GTP, and the increasing doses of nonradioactive GTP. The rates of hydrolysis of GTP in the presence of 50 nM RCC1 (open squares) or 500 nM RCC1 (closed circles), were shown as representative results (Figure 5A, b). The optimal reaction mixture in this experiment contained 1000 nM Ran, 500 nM RCC1, 800 nM RanBP1, and 5 mM GTP. In this system, 3.7 pmol of GTP was hydrolyzed per second on average at 250 nM SpRna1. Even under this Ran-GTP supplying system, the kinetics of the SpRna1-mediated enhancement of Clr4-HMTase activity was unchanged (Figure 5B, compare −Ran with +Ran). After incubation with increasing doses of SpRna1 (Figure 5B, horizontal line), a sufficient amount of Ran-GTP was still present (our unpublished data), revealing that SpRna1 enhances Clr4-mediated H3 methylation independent of RanGAP activity. Indeed, this Clr4-mediated H3 methylation was enhanced by the mutated SpRna1ts proteins, which did not show any detectable RanGAP activity (Figure 5, B and C).
Figure 5.
SpRna1 enhanced H3-K9 methylation independently of its RanGAP activity. (A) Construction of the continuous supplying system of Ran-GTP. (a) 1000 nM Ran-GTP was incubated with SpRna1 (250 nM), RCC1 (500 nM), RanBP1 (800 nM), and [γ-32P]GTP in the presence of the various concentrations of GTP (0, 5, 50, 500, or 5000 μM) in 30 μl of HMTase buffer for 1 h at 30°C. After incubation, the reaction was stopped by the addition of EDTA (final concentration 72 mM) and boiled. Resulting [γ-32P]GTP and 32Pi were separated by thin layer chromatography with 1 M LiCl and 1 M formic acid. 32P-labeled reagents were detected and analyzed using BAS-2500. (b) GTP hydrolysis carried out in the mixture containing of 1000 nM Ran, 800 nM RanBP1, an indicated concentration of GTP, 250 nM SpRna1, and 50 nM (□) or 500 nM RCC1 (●). 32Pi derived by hydrolysis of [γ-32P]GTP that were detected and analyzed using BAS-2500 are shown on the vertical line. Radioactivity of [γ-32P]GTP and 32Pi was measured using BAS-2500 for setting the sum of the radioactivity of [γ-32P]GTP and 32Pi to 100%. By estimating what percentage of [γ-32P]GTP is hydrolyzed in each reaction mixture, how many molecules of GTP were hydrolyzed in the reaction was calculated. (B) Enhancement of Clr4-HMTase with SpRna1 in the absence or presence of Ran-GTP. Clr4 (80 nM) was mixed in 30 μl of HMTase buffer with H3 (8 μM) and [14C]SAM (80 μM) in the presence of increasing amount (0, 50, 250, or 1250 nM) of SpRna1+ (○), SpRna1-8ts (●), SpRna1-15ts (▴), or SpRna1-87ts (■) under the continuous supply of Ran-GTP (+Ran) or not (−Ran). After incubation for 1 h at 30°C, the reaction mixtures were boiled in SDS sample buffer and resolved by 17% SDS-PAGE. The bands of H3 were visualized by Coomassie staining. Methylated H3 was analyzed using BAS-2500. The percentage of enzyme activity is plotted, setting the radioactivity in the reaction lacking SpRna1 to 100% (vertical line). (C) RanGAP activity of SpRna1ts. Fifty nanomolar Ran-[γ-32P]GTP was incubated in 100 μl of GAP buffer with various concentrations (0, 0.1, 0.3, 0.9, and 2.7 nM) of SpRna1+ (○), SpRna1-8ts (●), SpRna1-15ts (▴), or SpRna1-87ts (■). The reaction mixture was stopped by addition of ice-cold stop buffer. Resulting reaction mixtures were spotted onto the nitrocellulose membrane and washed with stop buffer. The radioactivity of the [γ-32P]GTP remaining on Ran was calculated by a liquid scintillation counter, which was shown as a ratio (percentage) by setting the radioactivity of a sample that was not incubated to 100% on the vertical line. The same experiments were repeated three times and error bars were marked in B and C.
Histones Inhibit the RanGAP Activity of SpRna1
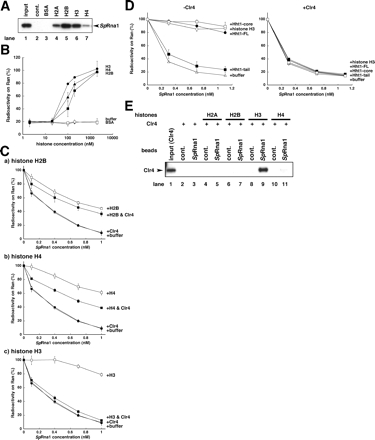
The nuclear localization of RanGAP led us to test how nuclear RanGAP activity could be inhibited to keep the concentration gradient of Ran-GTP from the nucleus to the cytoplasm. Histone H2, binding RCC1, enhances the RanGEF activity of RCC1 (Nemergut et al., 2001). Based on this report, we studied the effects of core histones on SpRna1–RanGAP activity. First, we examined whether the core histones, including H3, bound SpRna1. The indicated histones and bovine serum albumin (BSA) as a control were conjugated with Sepharose beads and then incubated with His-tagged SpRna1. When the beads were spun down, SpRna1 coprecipitated significantly with all of the histones, compared with BSA and with beads alone (Figure 6A). Among the histones, H3 and H2B efficiently coprecipitated with SpRna1. Based on these results, H2B and H4, in addition to H3, were chosen to investigate the effects of histones on the RanGAP activity of SpRna1. When an increasing dose of histones was incubated with a fixed amount of SpRna1 (0.5 nM), H3 most efficiently inhibited the RanGAP activity of SpRna1 (Figure 6B); this was expected because H3 bound SpRna1 strongly. However, H4 bound SpRna1 less strongly than H2B but inhibited the RanGAP activity of SpRna1 more efficiently than H2B. The same results were obtained when an increasing dose of SpRna1 was incubated with a fixed amount (100 nM) of histones (Figure 6C, open symbols). These results raised the question whether the ability of histones to bind SpRna1 is important for inhibiting the RanGAP activity of SpRna1. To study this, S. pombe H3, Hht1 (Hht1-FL), and its core (Hht1-core) or tail (Hht1-tail) was incubated with SpRna1. The Hht1-core, which binds SpRna1, inhibited the RanGAP activity like the full-sized Hht1 (Figure 6D, −Clr4), but the Hht1-tail, which does not bind SpRna1, could not. Thus, the binding ability of histones to SpRna1 was important to inhibit SpRna1–RanGAP activity, but it may not be sufficient, because H2B did not inhibit the SpRna1–RanGAP activity significantly. This might explain why a large molar excess of H3 was required to inhibit SpRna1–RanGAP activity, which was unexpected from the KD of SpRna1 to H3 calculated from surface plasmon resonance analysis (Figure 2A). In general, the value of KD means the binding affinity itself, but it does not always indicate the enzymatic Km or Ki values.
Figure 6.
Interaction of SpRna1 with histones and Clr4. (A) Indicated histones and as control, BSA, that were conjugated to Sepharose 4FF beads, and beads alone (cont.), were incubated with 2 nM 6xHis-SpRna1 in 1 ml of GAP buffer supplemented with 1 mM CHAPS. After incubation for 2 h at 4°C, beads were spun down, washed five times, and proteins coprecipitated with beads were separated by 4–20% gradient SDS-PAGE and blotted with the anti-SpRna1 antibody. An arrowhead indicates the position of 6xHis-SpRna1. Input included a half amount of total 6xHis-SpRna1 used in this experiment. (B) Fifty nanomolar Ran-[γ-32P]GTP was incubated in GAP buffer for 10 min at 30°C with 0.5 nM SpRna1 in the presence of various concentration of indicated histones H3 (●), H4 (▴), and H2B (■) and as controls, BSA (□) and buffer alone (○) (horizontal line). The reaction was stopped by addition of ice-cold stop buffer. Resulting reaction mixtures were spotted on a nitrocellulose membrane. The radioactivity of [γ-32P]GTP remaining on Ran was calculated by a liquid scintillation counter. The vertical line showed the ratio (percentage) of radioactivity remaining on Ran after incubation, compared with the radioactivity without incubation. (C) Fifty nanomolar Ran-[γ-32P]GTP was incubated in GAP buffer for 10 min at 30°C with various concentrations of SpRna1 (horizontal line) in the presence of 100 nM indicated histones H2B (a), H4 (b), and H3 (c) (□), 20 nM Clr4 (●), 100 nM indicated histones plus 20 nM Clr4 (■), or buffer alone (○). The reaction was stopped by the addition of ice-cold stop buffer. Reaction mixtures were then spotted on a nitrocellulose membrane. The radioactivity of [γ-32P]GTP remaining on Ran was calculated by a liquid scintillation counter. The vertical line showed the ratio (percentage) of radioactivity remaining on Ran after incubation, compared with the radioactivity without incubation. (D) Fifty nanomolar Ran-[γ-32P]GTP was incubated in GAP buffer for 10 min at 30°C with various concentrations of SpRna1 (horizontal line) in the presence of 100 nM H3 (○), Hht1-FL (●), Hht1-tail (■), Hht1-core (□), or buffer (Δ) alone, containing 20 nM Clr4 (+Clr4) (right) or not (−Clr4) (left) as indicated. The reaction was stopped by the addition of ice-cold stop buffer. Resulting reaction mixtures were spotted on a nitrocellulose membrane. The radioactivity of [γ-32P]GTP remaining on Ran was calculated by a liquid scintillation counter. The vertical line showed the ratio (percentage) of radioactivity remaining on Ran after incubation, compared with the radioactivity without incubation. The same experiments were repeated three times, and error bars are marked in B, C, and D. (E) SpRna1 conjugated to Sepharose 4FF beads were mixed with 250 nM 6xHis-tagged Clr4 (lanes 1–11) in the presence of 100 nM indicated histones in GAP buffer supplemented with 1 mM CHAPS. After incubation for 2 h at 4°C, beads were spun down, washed, and boiled in SDS sample buffer. Proteins coprecipitated with SpRna1 beads were resolved in 4–20% gradient SDS-PAGE, transferred to polyvinylidene diflouride (PVDF) membrane, and blotted with the mAb to 6xHis (Clr4). An arrowhead indicates the position of 6xHis-tagged Clr4. Input showed 20% of a total amount of Clr4 used in this experiment.
Because many positively charged amino acid residues were retained in the H3/Hht1-tail, compared with the Hht1-core, positive charge per se might not cause histones to bind and inhibit SpRna1; the mechanism is presently unknown. Regardless, we conclude that the RanGAP activity of a nuclear SpRna1 is inhibited by core histones.
Clr4 Abolishes H3-mediated RanGAP Inhibition
The effect of Clr4 on histone-mediated RanGAP inhibition was examined, because overexpression of Clr4 suppresses Sprna1ts (Kusano et al., 2004). When Clr4 was added to the mixture of SpRna1 and histones, the inhibitory effects of H2B and H4 on the RanGAP activity of SpRna1 were reduced partially but only that of H3 was abolished (Figure 6, C and D, +Clr4). Consistent with these results, Clr4 was spun down with SpRna1-conjugated Sepharose beads only in the presence of H3 (Figure 6E). Because Clr4 itself showed no ability to enhance the RanGAP activity of SpRna1 (Figure 6C, closed circles), we then determined how Clr4 could compromise the H3-mediated RanGAP inhibition. A simple idea is that Clr4 released SpRna1 from H3 by competing for binding H3. To test this, Sepharose beads conjugated with H3 were mixed with SpRna1 and increasing doses of Clr4. After incubation on ice for 60 min, beads were spun down. Consistent with the result shown in Figure 6E, both Clr4 and SpRna1 coprecipitated with H3 (Figure 7A, lane 6). However, the amount of SpRna1 coprecipitated with H3 was not reduced by the addition of an increasing dose of Clr4 (Figure 7A, lane 7), indicating that Clr4 did not release SpRna1 from H3. This result suggests that Clr4 makes a trimeric complex by binding to the H3 and SpRna1, as shown in Figure 7C. To study this, SpRna1-conjugated Sepharose beads were initially mixed with H3 as indicated in Figure 7B, lanes 2–5, and then the beads were spun down. After washing, the precipitated beads were mixed with Clr4 (Figure 7B, lanes 6–9). When beads were again spun down after incubation, the amount of H3 associated with SpRna1 had not been reduced by the addition of Clr4 (Figure 7B, compare H3 of lane 5 with lane 9). Moreover, Clr4 bound the complex of H3 and SpRna1 (Figure 7B, lane 9). Thus, a trimeric complex of SpRna1, H3, and Clr4 (Figure 7C) may form on the chromatin and abolish H3-mediated RanGAP inhibition.
Figure 7.
Formation of a complex consisted of SpRna1, H3, and Clr4. (A) Five hundred pM of 6xHis-tagged SpRna1 was incubated with mock conjugated Sepharose beads (cont.) and H3-conjugated Sepharose 4FF beads (histone H3) in 100 μl of GAP buffer containing 1 mM CHAPS in the presence of an increasing amount of 6xHis-tagged Clr4 (lanes 3 and 6: 10 nM or lanes 4 and 7: 100 nM). After incubation for 1 h, beads were spun down and boiled in SDS sample buffer. Precipitated proteins were resolved by 5–20% gradient SDS-PAGE and transferred to PVDF membrane. 6xHis-tagged SpRna1 and Clr4 were detected with the mAb to 6xHis. Input showed a position of SpRna1 and Clr4 used in this experiment. No proteins were precipitated in the lanes of control (mock-conjugated Sepharose beads). (B) Initially, SpRna1 conjugated to Sepharose 4FF beads was mixed with or without 250 nM H3 alone in GAP buffer supplemented with 0.2% Tween 20 as indicated (lanes 2–9). After incubation for 1 h at 4°C, beads were spun down, washed, and then mixed with 80 nM 6xHis-tagged Clr4 (lanes 6–9) or not (lanes 2–5) as indicated. After incubation for 1 h at 4°C, beads were spun down, washed, and boiled in SDS sample buffer. Proteins coprecipitated with SpRna1 beads were resolved in 5–20% gradient (for Clr4) or 17% (for H3) SDS-PAGE and then blotted with the mAb to 6xHis and the anti-H3 antibodies (catalog no. FL-136; Santa Cruz Biotechnology, Santa Cruz, CA). Arrowheads indicate the position of H3 and 6xHis-tagged Clr4, respectively. Input showed a total amount of Clr4 and H3 used in this experiment. (C) Model of interaction of Clr4 with H3 and SpRna1.
Xpo1/Crm1 Binds SpRna1 in the Presence of H3
Finally, we tested how the nuclear SpRna1 could be exported to the cytoplasm. Feng et al. (1999) suggested that S. cerevisiae ScRna1p could be exported to the cytoplasm depending on Xpo1p/Crm1p (Weis, 2003), which binds Ran-GTP and various intracellular cargoes, in this case, ScRna1p. To form an export complex containing Xpo1/Crm1 plus Ran-GTP for ScRna1p, there must be at least one mediator that inhibits the RanGAP activity of ScRna1p. Our results suggest that H3 plays such a role to export ScRna1p to the cytoplasm. To test this idea, glutathione S-transferase (GST)–fused Xpo1/Crm1 was mixed with SpRna1+, and as a control, with SpRna1-87ts, in the presence of Ran-GTP and H3 (Figure 8). When GST-Xpo1/Crm1 beads were spun down in the presence of Ran-GTP alone, SpRna1-87ts (with very little or no RanGAP activity as shown in Figure 5C) coprecipitated, but SpRna1+ did not (Figure 8, lanes 7 and 18). SpRna1+ coprecipitated with Xpo1/Crm1 in the presence of both Ran-GTP and H3 (Figure 8, lane 20). Thus, it seems that SpRna1 is exported to the cytoplasm with the aid of H3, in addition to Xpo1/Crm1 plus Ran-GTP. To confirm this, Clr4 was added to a mixture of SpRna1, Xpo1/Crm1, Ran-GTP and H3. As expected from the finding that Clr4 abolishes the H3-mediated inhibition of RanGAP activity, SpRna1+ could not interact with Xpo1/Crm1 even in the presence of Ran-GTP and H3, when Clr4 was added (Figure 8, lane 22).
Figure 8.
Interaction of SpRna1 with Xpo1/Crm1. 6xHis (800 pM)-fused SpRna1+ or SpRna1-87ts was mixed with Xpo1/Crm1, Ran-GTP, H3, and Clr4 as indicated in GAP buffer supplemented with 1 mM CHAPS. Final concentrations of GST-Xpo1/Crm1, Ran-GTP, H3, and Clr4 used in this assay were 40, 60, 200, and 20 nM, respectively. After 10-min incubation at 30°C, the reaction was stopped by ice-cold GAP buffer containing CHAPS. Glutathione Sepharose 4FF (code no. 17-5132; GE Healthcare) (GSH) or protein G-Sepharose 4FF as a control (cont.) was added to the reaction mixture. After incubation, beads were spun down. Proteins cofractionated with beads were resolved in 5–20% gradient gel and analyzed by blotting with the mAb to 6xHis and the mAb to GST (catalog no. sc-138; Santa Cruz Biotechnology). Input indicated a half amount of SpRna1+ and SpRna1-87ts proteins used in this experiment.
Overexpression of NES-Hht1 Inhibits the Growth of clr4Δ, but Not clr4+, Resulting in Chromosome Missegregation
To test how H3 and Clr4 might regulate SpRna1 in vivo reciprocally, the S. pombe H3 gene hht1+ was expressed from the nmt promoter in S. pombe clr4Δ and clr4+ strains, because our in vitro data suggested that overexpression of H3 could be toxic for the growth of S. pombe in the absence of Clr4. The hht1+ and NES-hht1+ genes, fused with NES in-frame, were conjugated with the thiamine-regulated nmt promoter in the pREP3X plasmid (Table 2) and then introduced into the clr4Δ and clr4+ strains. Transformants of clr4Δ and clr4+ containing pREP3X-hht1, pREP3X-NES-hht1, or pREP3X alone were cultivated on synthetic medium plates, with or without thiamine at 30°C. Five days later, the clr4Δ cells containing pREP3X-NES-hht1 could not make a clear colony in the absence of thiamine, whereas they papillated in the presence of thiamine. In contrast, clr4+ cells containing pREP3X-NES-hht1 papillated even in the absence of thiamine (Figure 9A).
Figure 9.
Overexpression of NES-fused S. pombe hht1+ was lethal for clr4Δ but not for clr4+. (A) clr4Δ and clr4+ cells expressing Hht1, NES-Hht1, or not (vector alone) were grown in EMM supplemented with uracil, adenine, and thiamine to 1.0 × 107 cells/ml. After fivefold serial dilution, cells were spotted onto thiamine additive (+thiamine) or thiamine-free (−thiamine) plates and incubated at 30°C for 5 d. The highest density spots contained 1 × 104 cells. (B) Cultures of clr4Δ cells containing pREP3X-NES-hht1 (NES-hht1+, rectangle) or pREP3X alone (vector, circle) were diluted to optical density (O.D.)595 = 0.02 and then incubated in the presence of thiamine (○, □) or not (●, ■). At the indicated time, O.D.595 values were measured and blotted (top left). After incubation for 24 h, cells were collected, fixed with 3.3% of paraformaldehyde in phosphate-buffered saline, and mounted in VECTASHIELD with 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) to visualize chromosome. Representative cells showing unequal chromosome segregation (arrow) and multinuclei (arrowhead) were indicated in the right photograph. Bar = 10 μm. Bottom left, to verify the expression of FLAG-NES-Hht1, cells (5.0 × 107) containing the indicated plasmid were harvested after incubation for 20 h in the absence of thiamine and then incubated in PEMS buffer supplemented with 1.0 mg/ml zymolyase 100T for 20 min at 30°C. Resulting spheroplasts were treated with 1% NP-40 and then centrifuged. Resulting supernatants and pellets were used as soluble and insoluble chromatin fractions, respectively. Fractions were separated in 17% SDS-PAGE and blotted with the mAb to FLAG M2 (catalog no. F-3165; Sigma-Aldrich). (C) Frequency of mitotic cells. More than 200 mitotic cells were examined three times to calculate the frequency (percentage) of cells showing unequal chromosome segregation (closed bar), multinuclei (hatched bar), and normal mitosis (open bar).
Twenty-four hours after thiamine depletion, the growth of clr4Δ cells containing pREP3X-NES-hht1, was retarded (Figure 9B, top left). At that time, NES-Hht1 accumulated in both the nucleus (insoluble) and the cytoplasm (soluble) (Figure 9B, bottom left). The calculated ratio of NES–Hht1 level between the nucleus and the cytoplasm was 2.5:1 (Figure 9B, bottom left), suggesting that both cytoplasmic and nuclear RanGAP could be inhibited. Consistently, cells showing abnormal chromosome segregation, accumulated, similar to Sprna1ts (Figure 9B, right, cells indicated by an arrow and an arrowhead). The calculated frequency of clr4Δ [pREP3X-NES-hht1] cells showing abnormal chromosomal segregation in total mitotic cells increased compared with clr4Δ possessing pREP3X vector alone after 24-h incubation without thiamine (Figure 9C).
DISCUSSION
Here, we found that S. pombe RanGAP is localized on the chromatin in addition to cytoplasm, and it seems likely to be involved in heterochromatin assembly.
An Unexpected Function of SpRna1 Enhances Clr4-mediated H3-K9 Methylation Independently of RanGAP Activity
It is very surprising that the recombinant SpRna1 enhanced HMTase activity of Clr4. Under the same conditions, recombinant Clr4 methylated both full-sized and tail-H3, but not the core of S. pombe H3, Hht1, as reported previously (Nakayama et al., 2001). In contrast, SpRna1 enhanced the methylation of full-sized H3 but not of the H3-tail alone. Thus, SpRna1 did not directly enhance Clr4-HMTase. From these results combined with SpRna1 bounding the core of S. pombe H3, Hht1, but not the tail of H3/Hht1, we conclude that SpRna1 enhances the Clr4-mediated H3-K9 methylation via the core region of Hht1. Because SpRna1 is the S. pombe RanGAP, Ran GTPase-activating protein, it was important to determine whether Ran-GTPase is involved in the SpRna1-mediated Clr4-HMTase enhancement. SpRna1ts-mutated proteins with no significant RanGAP activity enhanced the Clr4-mediated H3 methylation, similar to SpRna1+. In addition, we developed an in vitro system in which Ran-GTP was continuously supplied by RanGEF, RCC1, in the presence of a high dose of RanGAP, SpRna1. However, the ability of SpRna1 to enhance the Clr4-mediated H3-K9 methylation was unchanged, even in the presence of a sufficient amount of Ran-GTP. Thus, SpRna1 seems to enhance Clr4-mediated H3-K9 methylation independently of Ran-GTP in vitro.
The critical issue was whether the ability of SpRna1 to enhance the Clr4-HMTase activity could be observed in vivo. Cell fractionation analysis revealed that SpRna1 was present in the nucleus as well as the cytoplasm, as suggested by Feng et al. (1999). Although nucleoporins were fractionated into the insoluble fraction containing chromatin, they were not dissolved by MNase treatment. In contrast, the nuclear SpRna1 was dissolved by MNase treatment, similar to H3. Thus, a nuclear SpRna1 seems to bind chromatin in a manner similar to S. pombe H3, Hht1. The level of nuclear SpRna1 dissolved by MNase treatment seemed to be lower than that of Hht1, suggesting that nuclear SpRna1 might be localized in chromosomal regions resistant to MNase treatment. In vitro, SpRna1 bound the core histones, particularly H3 and H2B. Because H3 and H2B form dimers with H4 and H2A, respectively (Luger et al., 1997; Black et al., 2004), it is possible that SpRna1 is anchored to the chromatin through the core histones. Because SpRna1 binds to Clr4 in the presence of H3, a nuclear SpRna1 could make a trimeric complex with H3 and Clr4 to enhance the Clr4-HMTase activity that is essential for heterochromatin assembly. Consistently, Sprna1ts showed a defect in heterochromatin assembly; compared with H3-K4 methylation, H3-K9 methylation of Sprna1ts was strongly reduced after incubation at 34°C at all three heterochromatic regions of S. pombe. In parallel with the reduction of H3-K9 methylation, the association of Swi6 with chromatin was inhibited, consistent with the observation that Swi6 binds the methylated H3-K9 (Bannister et al., 2001; Nakayama et al., 2001). Thus, SpRna1 seems to be required for H3-K9 methylation in all heterochromatic regions of S. pombe. In this context, whether SpRna1 enhances the Clr4-HMTase activity independently of its RanGAP activity is now questionable, because the Sprna1ts mutation should affect the RanGAP activity of SpRna1. One possibility is that the ability of SpRna1 to enhance Clr4-HMTase might be affected by the Sprna1ts mutation in vivo, in a temperature-dependent manner by an unknown mechanism. In this context, it is notable that the crystal structure of SpRna1 is highly similar to the ribonuclease inhibitor and to the U2A′ small nuclear ribonucleoprotein (Hillig et al., 1999), because several lines of evidence support a role for RNA in the formation of heterochromatin (Maison et al., 2002; Grewal and Moazed, 2003). The other puzzle is that the Sprna1ts mutation shows a gene-silencing defect at the centromere but not at telomeres (Kusano et al., 2004), although all of the heterochromatic regions of S. pombe are affected by the Sprna1ts mutation. A similar centromere-specific silencing defect has been observed in RNA interference (RNAi) mutant cells (Volpe et al., 2002; Hall et al., 2003) and chp1-deleted cells (Thon and Verhein-Hansen, 2000), whereas both RNAi components and Chp1 are involved in heterochromatin assembly at all three heterochromatic regions of S. pombe (Sadaie et al., 2004; Kanoh et al., 2005; Miller et al., 2005). Taz1 (a telomere-associated factor in Schizosaccharomyces pombe) is specifically required for the establishment of telomeres but not for that of centromeres in S. pombe (Cooper et al., 1997; Kanoh et al., 2005; Miller et al., 2005). The centromere-specific silencing defect observed in Sprna1ts cells may reflect such a difference between centromeric and telomeric chromatin.
Histones and Clr4 Reciprocally Regulates Nuclear RanGAP Activity
Our finding that SpRna1 is localized on the chromatin raised the important general question of how the RanGAP activity of nuclear SpRna1 is inhibited, otherwise it abolishes the nucleocytoplasmic gradient of Ran-GTP concentration. In this context, it is notable that all core histones bound SpRna1 and inhibited its RanGAP activity. Among core histones, H3, which cooperates with H4 (Luger et al., 1997; Black et al., 2004), most strongly inhibited the RanGAP activity of SpRna1. Thus, we conclude that the RanGAP activity of nuclear SpRna1 is inhibited by core histones, particularly H3. In contrast, Clr4 abolished the H3-mediated inhibition of SpRna1–RanGAP activity. This finding raised another question of whether the SpRna1–RanGAP activity uncovered by Clr4 may play a role in the nucleus, independent of the ability of SpRna1 to enhance the Clr4-HMTase. It has been reported that Ran can bind to chromatin in manners dependent or independent of RCC1. In the RCC1-independent mode, Ran directly binds both H3 and H4 (Bilbao-Cortes et al., 2002). Chromatin-bound Ran, suggested to function for spindle formation and for nuclear envelope assembly, might cooperate with a nuclear RanGAP for Ran-mediated nuclear events. It remains to be determined whether a nuclear RanGAP functions for higher order chromatin assembly through the Ran cycle, as in microtubule assembly and nuclear membrane formation. In this context, it is notable that most Sprna1ts mutants do not show detectable defects in nucleocytoplasmic transport or in microtubule assembly. Because the disruption of clr4+ gene increased the temperature sensitivity of Sprna1ts, but it was not lethal for Sprna1ts (Kusano et al., 2001), a nuclear SpRna1 might function in an unknown pathway, other than the pathway including Clr4.
The RanGAP activity of a nuclear SpRna1 should be carefully regulated temporally and spatially to maintain the nucleocytoplasmic gradient of Ran-GTP concentration. After establishment of heterochromatin, a nuclear SpRna1 would be immediately inactivated or exported to the cytoplasm with the aid of its NES signal. Indeed, SpRna1 could make a stable complex with Xpo1/Crm1 plus Ran-GTP in the presence of H3. Accordingly, we could not detect any association of SpRna1 with chromatin by the ChIP assay, whereas a chromatin-bound SpRna1 was detected by immunoblotting analysis. In conclusion, we suggest that histones, particularly H3, and Clr4 regulate a nuclear SpRna1 reciprocally for heterochromatin assembly and for its nuclear export.
ACKNOWLEDGMENTS
We thank Drs. Shelley Sazer for the anti-Pim1 and anti-Spi1 antibodies, Kathy Wilson (Johns Hopkins University School of Medicine, Baltimore, MD) for helpful discussion, and Genevieve Almouzni (Institut Curie, Paris, France) for reading this manuscript and for helpful suggestions. This work was supported by grants-in-aid for specially promoted research from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0893) on March 15, 2006.
REFERENCES
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Becker J., Melchior F., Gerke V., Bischoff F. R., Ponstingl H., Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- Bischoff F. R., Krebber H., Kempf T., Hermes I., Ponstingl H. Human RanGTPase activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc. Natl. Acad. Sci. USA. 1995a;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Krebber H., Smirnova E., Dong W., Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995b;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bilbao-Cortes D., Hetzer M., Langst G., Becker P. B., Mattaj I. W. Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 2002;12:1151–1156. doi: 10.1016/s0960-9822(02)00927-2. [DOI] [PubMed] [Google Scholar]
- Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Cleveland D. W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Nimmo E. R., Allshire R. C., Cech T. R. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Dasso M. The Ran GTPase: theme and variations. Curr. Biol. 2002;12:R502–R508. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- Dasso M., Seki T., Azuma Y., Ohba T., Nishimoto T. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 1994;13:5732–5744. doi: 10.1002/j.1460-2075.1994.tb06911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. I., Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Edmondson D. G., Smith M. M., Roth S. Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Olsson T., Turner B. M., Cranston G., Allshire R. C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Feng W., Benko A. L., Lee J. H., Stanford D. R., Hopper A. K. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J. Cell Sci. 1999;112:339–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Hall I. M., Noma K., Grewal S. I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N., Yokoyama N., Seki T., Azuma Y., Ohba T., Nishimoto T. RanBP1, a Ras-like nuclear G protein binding to Ran/TC4, inhibits RCC1 via Ran/TC4. Mol. Gen. Genet. 1995;247:661–669. doi: 10.1007/BF00290397. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger S., Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Gruss O. J., Mattaj I. W. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 2002;4:E177–E184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- Hillig R. C., Renault L., Vetter I. R., Drell T., Wittinghofer A., Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol. Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- Kai R., Ohtsubo M., Sekiguchi T., Nishimoto T. Molecular cloning of a human gene that regulates chromosome condensation and is essential for cell proliferation. Mol. Cell. Biol. 1986;6:2027–2032. doi: 10.1128/mcb.6.6.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kanoh J., Sadaie M., Urano T., Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 2005;16:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Khan A. U., Hampsey M. Connecting the DOTs: covalent histone modifications and the formation of silent chromatin. Trends Genet. 2002;18:387–389. doi: 10.1016/s0168-9525(02)02746-4. [DOI] [PubMed] [Google Scholar]
- Kusano A., Staber C., Ganetzky B. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev. Cell. 2001;1:351–361. doi: 10.1016/s1534-5807(01)00042-9. [DOI] [PubMed] [Google Scholar]
- Kusano A., Yoshioka T., Nishijima H., Nishitani H., Nishimoto T. Schizosaccharomyces pombe RanGAP homolog, SpRna1, is required for centromeric silencing and chromosome segregation. Mol. Biol. Cell. 2004;15:4960–4970. doi: 10.1091/mbc.E04-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzler M., Gerstberger T., Stutz F., Bischoff F. R., Hurt E. Yeast Ran-binding protein 1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a CRM1 (XPO1)-dependent pathway. Mol. Cell. Biol. 2000;20:4295–4308. doi: 10.1128/mcb.20.12.4295-4308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D., Richmond T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T. J., Richmond T. J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Lyttle T. W. Segregation distorters. Annu. Rev. Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A. H., Quivy J. P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Matynia A., Dimitrov K., Mueller U., He X., Sazer S. Perturbations in the spi1p GTPase cycle of Schizosaccharomyces pombe through its GTPase-activating protein and guanine nucleotide exchange factor components result in similar phenotypic consequences. Mol. Cell. Biol. 1996;16:6352–6362. doi: 10.1128/mcb.16.11.6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 2004;5:65–69. doi: 10.1038/nrm1263. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Merrill C., Bayraktaroglu L., Kusano A., Ganetzky B. Truncated RanGAP encoded by the segregation distorter locus of Drosophila. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- Miller K. M., Ferreira M. G., Cooper J. P. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. D. The Ran-GTPase and cell-cycle control. Bioessays. 2001;23:77–85. doi: 10.1002/1521-1878(200101)23:1<77::AID-BIES1010>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Nakagawa H., Lee J. K., Hurwitz J., Allshire R. C., Nakayama J., Grewal S. I., Tanaka K., Murakami Y. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nemergut M. E., Mizzen C. A., Stukenberg T., Allis C. D., Macara I. G. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- Noguchi E., Hayashi N., Nakashima N., Nishimoto T. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-GTPase-activating protein. Mol. Cell. Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Allis D. A., Grewal S. S. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Okazaki H., Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Sadaie M., Iida T., Urano T., Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino H., Hisamoto N., Uzawa S., Sekiguchi T., Nishimoto T. DNA-binding domain of RCC1 protein is not essential for coupling mitosis with DNA replication. J. Cell Sci. 1992;102:393–400. doi: 10.1242/jcs.102.3.393. [DOI] [PubMed] [Google Scholar]
- Stade K., Ford C. S., Guthrie C., Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Seewald M. J., Kraemer A., Farkasovsky M., Korner C., Wittinghofer A., Vetter I. R. Biochemical characterization of the Ran-RanBP1-RanGAP system: are RanBP proteins and the acidic tail of RanGAP required for the Ran-RanGAP GTPase reaction? Mol. Cell. Biol. 2003;23:8124–8136. doi: 10.1128/MCB.23.22.8124-8136.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange Y., Hirata A., Niwa O. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 2002;115:4375–4385. doi: 10.1242/jcs.00135. [DOI] [PubMed] [Google Scholar]
- Thon G., Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155:551–568. doi: 10.1093/genetics/155.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., Martienssen R. A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Welch K., Franke J., Kohler M., Macara I. G. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by Importin-α3. Mol. Cell. Biol. 1999;19:8400–8411. doi: 10.1128/mcb.19.12.8400. [DOI] [PMC free article] [PubMed] [Google Scholar]