Abstract
The genome of Tetrahymena thermophila contains 39 loci encoding NIMA-related kinases (NRKs), an extraordinarily large number for a unicellular organism. Evolutionary analyses grouped these sequences into several subfamilies, some of which have orthologues in animals, whereas others are protist specific. When overproduced, NRKs of three subfamilies caused rapid shortening of cilia. Ultrastructural studies revealed that each NRK triggered ciliary resorption by a distinct mechanism that involved preferential depolymerization of a subset of axonemal microtubules, at either the distal or proximal end. Overexpression of a kinase-inactive variant caused lengthening of cilia, indicating that constitutive NRK-mediated resorption regulates the length of cilia. Each NRK preferentially resorbed a distinct subset of cilia, depending on the location along the anteroposterior axis. We also show that normal Tetrahymena cells maintain unequal length cilia. We propose that ciliates used a large number of NRK paralogues to differentially regulate the length of specific subsets of cilia in the same cell.
INTRODUCTION
How the size of organelles is determined is an important and largely unanswered question. Cilia have emerged as a model organelle to study the mechanism of size regulation (Marshall, 2002). The size of an organelle is determined by the balance between assembly pathways that deliver structural components and disassembly pathways that remove components. In cilia, the intraflagellar transport (IFT), a bidirectional motility system that operates inside cilia (Rosenbaum and Witman, 2002; Scholey, 2003), plays a key role in length regulation. The anterograde IFT supplies structural subunits during assembly (Piperno et al., 1996; Qin et al., 2005) and maintains the axoneme after assembly (Brown et al., 1999). The retrograde IFT recycles the IFT machinery (Signor et al., 1999) and removes ciliary structural components that turn over (Qin et al., 2004). This turnover is the basis of the disassembly pathway (Stephens, 1997; Marshall and Rosenbaum, 2001; Song and Dentler, 2001; Thazhath et al., 2004). Experimental data and mathematical modeling in Chlamydomonas reinhardtii indicate that the length of flagella is dependent on the rates of elongation (equivalent to the efficiency of anterograde IFT) and disassembly. As the flagellum grows, the rate of elongation decreases due to the limited supply of IFT components that need to travel longer distances to the tip (Marshall and Rosenbaum, 2001). According to the “balance point model,” the steady-state length is achieved when the rate of elongation equals the rate of disassembly (Marshall and Rosenbaum, 2001; Marshall et al., 2005).
The length of already assembled cilia can be reduced drastically using two mechanisms: autotomy or resorption (reviewed by Quarmby, 2004). During autotomy, the axoneme breaks off from the basal body, the cilium falls off, and the plasma membrane reseals over the basal body. Protists undergo autotomy in response to low pH (Lewin et al., 1982). It is unclear whether autotomy occurs naturally or is a pathological process (Quarmby, 2004). Partial or complete resorption is based on a retraction of cilia, by enhanced disassembly. In mammalian cells, sensory primary cilia are resorbed before mitosis (Rieder et al., 1979). Chlamydomonas cells resorb flagella during mating and before mitosis (Cavalier-Smith, 1974). All three types of ciliary length reduction processes: autotomy, resorption, and disassembly at steady state, have a common denominator—they all involve physical separation of α/β tubulin dimers from the assembled axoneme. Therefore, the three mechanisms of ciliary length reduction may be related phenomena. There is genetic evidence that autotomy and resorption share components (Parker and Quarmby, 2003).
Some molecular components of the ciliary length reduction pathways have already been identified and notably, all of them are kinases. An Aurora kinase of Chlamydomonas, CALK, is required for autotomy and resorption (Pan et al., 2004). A mutation in the mitogen-activated protein kinase encoded by the LF4 gene of Chlamydomonas led to excessive flagellar length (Berman et al., 2003). Two NimA-related kinases (NRKs) of Chlamydomonas have been implicated in autotomy and disassembly at steady state (Mahjoub et al., 2002; Parker and Quarmby, 2003; Bradley and Quarmby, 2005).
Tetrahymena thermophila is a cell type in which the regulation of ciliary length requires a high level of sophistication, due to the large number of cilia and the fact that a subset of cilia can be either resorbed or elongated in a localized manner. In contrast to Chlamydomonas where the length of the two flagella is subject to equalization (Rosenbaum et al., 1969; Coyne and Rosenbaum, 1970; Barsel et al., 1988), the ciliary length in Tetrahymena can be highly eccentric. For example, cilia of the undulating membranelle of the anterior oral apparatus of the dividing cell undergo rapid shortening, whereas other cilia located nearby do not resorb (Nelsen et al., 1981). We show here that in Tetrahymena, the anterior cilia are shorter compared with cilia at the posterior end. In contrast, in cells subjected to starvation, a single posterior cilium elongates that is associated with transition to the “fast swimmer” form (Nelsen, 1978; Nelsen and DeBault, 1978). The factors that cause such localized adjustments of ciliary length are unknown.
Cells could achieve a localized effect on a subpopulation of cilia by using isoforms of a rate-limiting regulatory protein that are restricted in subcellular localization. We show here that NRKs form a large protein family in Tetrahymena and negatively regulate the length of cilia in a localized manner. The founding member of the NRK family, the Never-in-Mitosis A (NimA) kinase of Aspergillus nidulans, is required for cell entry into mitosis (Osmani et al., 1988). Some NRKs participate in ciliary functions (reviewed by Quarmby and Mahjoub, 2005). Mutations in the vertebrate NRKs, Nek1 and Nek8, cause polycystic kidney disease in mice and zebrafish (Upadhya et al., 2000; Liu et al., 2002). The FA2 gene of Chlamydomonas encodes an NRK that is essential for flagellar autotomy and normal rate of disassembly (Mahjoub et al., 2002). Another NRK of Chlamydomonas, Cnk2p, negatively regulates the steady-state length of flagella (Bradley and Quarmby, 2005). Here, we show that the activity of NRKs closely related to Cnk2p reduces the length of cilia in Tetrahymena. Furthermore, we show that overexpressed members of two additional subtypes of NRKs also shorten cilia but use a distinct mechanism. We also show that the responses of individual cilia of Tetrahymena to the activity of specific NRKs are dependent on the subcellular location.
MATERIALS AND METHODS
Analyses of Genomes
Sequences of annotated NRK loci were obtained from the following databases: C. reinhardtii, fungi and humans, National Center for Biotechnology Information; Drosophila melanogaster, FlyBase; Ceanorhabditis elegans, Sangers Institute; Paramecium tetraurelia, Genoscope; Volvox carteri, Joint Genome Institute (http://jgi.doe.gov). The T. thermophila NRK1 sequence was published by Christensen et al. (2003) (AAN76826) as “protein tyrosine kinase 1, TtPTK1.” Our phylogenetic analyses grouped TtPTK1 with NRKs and TtPTK1 has motifs diagnostic of the NRK family. Therefore, we will refer to this gene as NRK1, which is consistent with the name of the orthologue found earlier in Tetrahymena pyriformis (Wang et al., 1998). The NRK2 gene was identified from an expressed sequence tag (EST) clone from the PEPdb database (http://amoebidia.bcm.umontreal.ca/public/pepdb/agrm.php). We used the sequences of Nrk1p, Nrk2p to identify all NRK loci of T. thermophila, by tBLASTn searches of the macronuclear genome (http://tigrblast.tigr.org/er-blast/index.cgi?project=ttg). We defined a sequence as encoding a putative NRK, if an established NRK sequence of another organism was returned as the best match using BLASTx. Gene accession numbers of sequences used for phylogenetic analyses are listed in the legend of Supplemental Figure S6. The predicted kinase domain sequences were aligned using Clustal X 1.82 and corrected manually in SeaView (Galtier et al., 1996). A tree was calculated using the PHYLIP package (Felsenstein, 1997). One hundred replicates of the sequence set were created using SEQBOOT. The distances were calculated in PROTDIST, and trees were reconstructed using NEIGHBOR. The Jones–Taylor–Thorton substitution model was used. A consensus tree was obtained using CONSENSE, and the tree was plotted using DRAWGRAM.
Strains and Culture
Tetrahymena cells were grown in 1% proteose-peptone, 0.2% glucose, 0.1% yeast extract, and 0.003% ferric-sodium:EDTA (SPP) supplied with an antibiotic-antimycotic mix (Invitrogen, Carlsbad, CA). The CU522 strain (from Dr. Donna Cassidy-Hanley, Cornell University, Ithaca, NY) was used to insert transgenes as described previously (Gaertig et al., 1999), and such strains were maintained in SPP with 20 μM paclitaxel. To overexpress a transgene under MTT1 promoter (Shang et al., 2002), cells were grown without paclitaxel overnight, diluted to 105–2 × 105 cells/ml, and incubated with 2.5 μg/ml CdCl2.
Green Fluorescent Protein (GFP) Tagging
The IFT52 coding region of pMTT1-IFT52-GFP plasmid (Brown et al., 2003) was replaced with the NRK2 coding region, which was amplified from genomic DNA with HindIII and BclI sites at the 5′ and MluI site at the 3′ end, respectively, and cloned using the same sites of pMTT1-IFT52-GFP, to produce pMTT1-NRK2-GFP. The NRK1 coding region was amplified from genomic DNA with addition of BclI and MluI sites at 5′ and 3′ ends, respectively, and cloned into the same restriction sites of pMTT1-NRK2-GFP, to give pMTT1-NRK1-GFP. For overexpression of fusion proteins with an N-terminal GFP, we replaced the NRK2-GFP coding region of pMTT1-NRK2-GFP with the GFP coding region, which was amplified with an addition of a HindIII site at 5′ and MluI-BamHI sites at the 3′ end, respectively. The MluI and BamHI sites were separated by a TGA stop codon. The fragment was digested with HindIII and BamHI and inserted into the same sites of pMTT1-NRK2-GFP to create the pMTT1-GFP. To overexpress Nrk17p and Nrk30p tagged with an N-terminal GFP, the coding regions were amplified with addition of MluI and BamHI sites at 5′ and 3′ ends, respectively, and inserted into pMTT1-GFP to create pMTT1-GFP-NRK17 and pMTT1-GFP-NRK30. The following primers were used to amplify the coding regions: 5′-AATAAACGCGTCATGCTTAATAATGGATTC-3′ and 5′-TTATTGGATCCGCTAAAATCATGCTGGCAATA-3′ for NRK17 and 5′-AATAAACGCGTCATGAGTAAATAGTCTAAATTA-3′ and 5′-TTAATTGGATCCCGATTATAGAATAATTTCTTCCA-3′ for NRK30.
To overexpress the transgenes in Tetrahymena, the backbones of the above-mentioned plasmids were separated from the inserts using ApaI and SacI or SacII. The total digested plasmids were used to biolistically transform CU522 cells, and transformants were selected in SPP with 20 μM paclitaxel. To prepare “kinase-dead” variants, plasmids with NRK1-GFP and NRK2-GFP coding regions were subjected to in vitro mutagenesis, to change the conserved lysine codon of the kinase domain to arginine at positions 40 and 35, respectively (Fry et al., 1995), using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Immunofluorescence
A standard fixation method was used to analyze cells expressing either Nrk1p-GFP or Nrk2p-GFP (Thazhath et al., 2002). Secondary antibodies were conjugates to either fluorescein isothiocyanate or Cy3 (Zymed Laboratories, South San Francisco, CA) and were used at 1:150 dilution. The following primary antibodies were used: rabbit polyclonal anti-GFP (Clontech, Mountain View, CA) at 1:200 dilution, mouse monoclonal anti-GFP (Clontech) at 1:600, TAP952 monoclonal anti-monoglycylated tubulin at 1:50, polyclonal anti-total Tetrahymena tubulins, SG at 1:300, monoclonal anti-tubulin polyglutamylation antibodies ID5 at 1:30, monoclonal anti-α-tubulin, 12G10 at 1:20, and anti-centrin 20H5 at 1:100. Nrk17p-GFP was not retained in cells using the standard fixation procedure. Therefore, Nrk17p-GFP cells were fixed with 15 μl of 2% paraformaldehyde in PHEM buffer (Schliwa and van Blerkom, 1981) and after 10–15 s permeabilized with 10 μl of 0.5% Triton X-100 in the same buffer. We could not retain Nrk30p-GFP inside cells under fixation conditions, which included detergent permeabilization. Thus, these cells were only fixed with 2% paraformaldehyde for microscopic analyses. Cells were viewed using a Leica TCS SP2 spectral confocal microscope with coherent Ti:sapphire multiphoton laser. To measure the length of cilia, cells were labeled by immunofluorescence with a mixture of 12G10 and SG anti-tubulin antibodies. Cilia length was measured on confocal images using NIH Image 1.62 (http://rsb.info.nih.gov/nih-image/). The transmission electron microscopy (TEM) analysis was done as described previously (Jerka-Dziadosz et al., 2001).
Western Blots
To obtain the whole cytoskeleton fraction, 107 cells were extracted as described previously (Williams, 2000). Eight micrograms of proteins were separated on a 10% SDS-PAGE gel. Proteins were transferred to nitrocellulose, blocked in 5% skim milk in tris-buffered saline (TBS) for 1 h, and incubated overnight at 4°C with polyclonal anti-GFP antibodies (Torrey Pines Biolabs, Houston, TX) diluted 1:2000 in 1% skim dry milk in TBS. Next, filters were washed 3 × 10 min in TBS and incubated for 2 h in secondary antibodies (anti-rabbit IgG-horseradish peroxidase conjugated). After washing (3 × 15 min), proteins were visualized using either ECL system (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) on CL-Xposure TM film (Pierce Chemical, Rockford, IL).
RESULTS
NRK Loci in the Genome of T. thermophila
Wang et al. (1998) reported the first identification of a ciliate NRK in T. pyriformis named TpNrk, as a protein whose mRNA level changed under stress and during the cell cycle (Wang et al., 1998). An orthologous gene of T. thermophila was later identified by Christensen et al. (2003) and will be referred to as NRK1. The NRK2 sequence was identified in the EST database. By searching the macronuclear genome of T. thermophila, we identified 37 additional NRK sequences (Eisen, Wu, Wu, Thiagarajan, Wortman, Badger, Ren, Amedeo, Jones, Tallon, Delcher, Salzberg, delToro, Ryder, Williamson, Barbeau, Silva, Haas, Majoros, Farzad, Carlton, Garg, Pearlman, Karrer, Sun, Smith, Jr., Manning, Elde, Turkewitz, Asai, Wilkes, Wang, Cai, Collins, Wilamowska, Ruzzo, Weinberg, Stewart, Lee, Wloga, Rogowski, Frankel, Gaertig, Tsao, Gorovsky, Keeling, Waller, Patron, Cherry, Stover, Krieger, Hamilton, Orias, and Coyne, unpublished results). The kinase domains of the 39 predicted NRKs of T. thermophila have between 36 and 59% of amino acid sequence identity to the kinase domains of human NRKs (Neks). In all predicted Tetrahymena NRKs, the kinase domain is located near the N-terminal end and is followed by a nonconserved noncatalytic region.
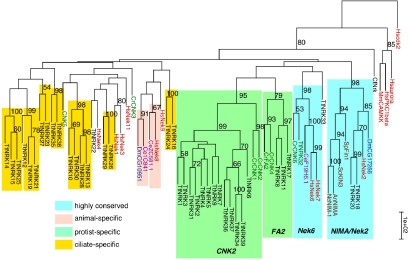
A neighbor-joining phylogenetic tree based on the kinase domains (Figure 1) indicates that NRKs of Tetrahymena and several model organisms form four well supported clades. We defined subfamilies based on these clades, using names of the best characterized members. Two subfamilies (NimA/Nek2 and Nek6) are conserved between animals and protists. Nrk18p and Nrk20p are homologues of the mammalian Nek2, implicated in the splitting of duplicated centrosomes (Fry et al., 1998; Faragher and Fry, 2003). The second conserved subfamily includes human Nek6 and Nek7, which have been implicated in the metaphase–anaphase transition (Yin et al., 2003), and a single Tetrahymena homologue, Nrk32p.
Figure 1.
Phylogeny of NRKs. Amino acid sequences of kinase domains of NRKs and other serine-threonine kinases were aligned and a neighbor-joining tree prepared using HsCdk2 as an outgroup. Numbers above branches represent bootstrap support values above 50%. The multiple sequence alignment is shown in Supplemental Figure S6. Novel motifs found downstream of the kinase domain of Cnk2p type NRKs are shown in Supplemental Figure S7.
Two additional well supported subfamilies do not have animal homologues, but they include NRKs of Chlamydomonas (Bradley et al., 2004) and Tetrahymena and may be protist specific. The FA2 subfamily includes Fa2p and Cnk4p NRKs of Chlamydomonas and Tetrahymena Nrk8p, Nrk11p, and Nrk17p. The CNK2 subfamily contains an astonishing cluster of 13 Tetrahymena sequences (including Nrk1p and Nrk2p), along with Cnk1p and Cnk2p of Chlamydomonas. Based on reverse transcription-PCR, it seems that most if not all paralogues in this subfamily are expressed, because we could detect mRNAs in vegetative cells for each of eight NRK sequences tested (Supplemental Figure S5). Within the CNK2 subfamily, we identified two conserved amino acid signature motifs, LL[Q/N]TI[K/R]xP, and LPxxY located downstream of the kinase domain. These motifs are also present in the closely related NRKs of Paramecium tetraurelia and V. carteri (Supplemental Figure S7). The motifs do not match any known protein domains or phosphorylation sites. The phylogenetic relationships among the remaining NRKs, which include 19 Tetrahymena sequences, were not well resolved.
NRK Proteins of Tetrahymena Colocalize with Cilia and Microtubular Cortical Structures
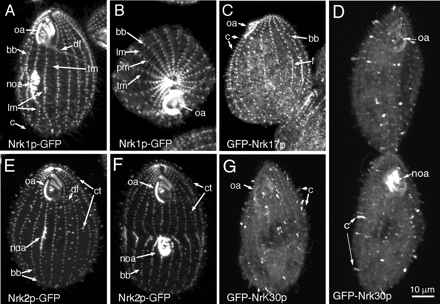
We overexpressed Nrk1p and Nrk2p as GFP fusions using the cadmium-inducible MTT1 promoter (Shang et al., 2002). Nrk1p-GFP was detected 2 h after cadmium addition, and the levels remained high for at least 24 h (Supplemental Figure S1, A). At 3–4 h, the Nrk1p-GFP–overexpressing cells greatly decreased the rate of proliferation (Supplemental Figure S1, C). Nrk1p-GFP localized to a large set of microtubular organelle types, including cilia, basal bodies (BBs), three classes of cortical microtubules (longitudinal, transverse, and postciliary), oral deep fiber (Figures 2, A and B, and 3, A–C), and contractile vacuole pores (CVPs) (our unpublished data). In dividing cells, the level of Nrk1p-GFP was enhanced in BBs and cilia of the newly formed oral apparatus (Figure 2A) and in the newly assembled locomotory cilia (Figure 3, A–C).
Figure 2.
Overexpressed NRKs colocalize with cilia and cortical microtubular organelles. GFP fluorescent confocal images in cells overproducing Nrk1p-GFP (A and B), GFP-Nrk17p (C), GFP-Nrk30p (D and G), and Nrk2p-GFP (E and F). Expression of transgenes was induced with 2.5 μg/ml CdCl2 for 2–4 h. oa, oral apparatus; noa, new oral apparatus; bb, basal bodies; df, deep fiber; tm, transverse microtubules; c, cilia; ct, ciliary tips, f, cytoplasmic fibers, lm, longitudinal microtubule bundles, pm, postciliary microtubules.
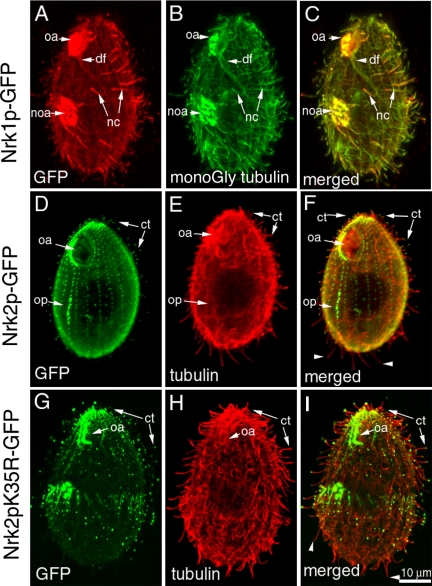
Figure 3.
Nrk1p and Nrk2p GFP fusion proteins preferentially localize to cilia and cortical microtubules. Immunofluorescent confocal images of cells overproducing Nrk1p-GFP (A–C), Nrk2p-GFP (D–F), and Nrk2p-K35R-GFP (G–I). Fluorescence of GFP is shown in A (red) and D and G (green). Top row, an Nrk1p-GFP–overproducing cell was processed for double immunofluorescence using anti-GFP antibodies (red in A) and TAP952 antibodies against monoglycylated tubulins (B). The TAP 952 antibody labels strongly new assembling cilia. Note that Nrk1p-GFP accumulates preferentially in TAP952-positive new cilia (C). Middle row, an Nrk2p-GFP–overproducing cell was subjected to double immunofluorescence with anti-GFP antibodies (D) and anti-total tubulin antibodies SG (E). Note an accumulation of Nrk2p-GFP at the tips of a subset of cilia located mainly in the ventral and anterior region of the cell (see F). Arrowheads indicate the absence of GFP label at tips of posterior cilia (F). Bottom row, an Nrk2p-K35R-GFP–overproducing cell labeled by anti-GFP antibodies (G) and total tubulin antibodies (H). The arrows point out a similar localization of Nrk2p-K35R-GFP at ciliary tips (ct) compared with Nrk2p. However, cilia of Nrk2p-K35R-GFP do not undergo shortening (I).
Although the kinase domain sequence of NRK2 is closely related to that of NRK1, the pattern of expression and localization of Nrk2p-GFP was not identical. Within 2 h of cadmium exposure, the Nrk2p-GFP cells grew more slowly (Supplemental Figure S1, C) and many cells were arrested in cytokinesis displaying a highly disorganized new oral apparatus (Supplemental Figure S2). The level of Nrk2p-GFP decreased after 4 h (Supplemental Figure S1, B). Nrk2p-GFP localized to cilia, BBs, LMs (weakly) (Figures 2, E and F, and 3D) and CVPs (our unpublished data). Strikingly, in cilia, Nrk2p-GFP accumulated at the tips (Figure 3, D–F, arrowheads). However, a subset of posterior and dorsal cilia did not accumulate Nrk2p-GFP. The distinct temporal patterns of expression and localization of Nrk2p and Nrk1p, suggests that the fast-evolving nonkinase domains determine the quantitative regulation and subcellular localization. We also localized Nrk1p and Nrk2p by adding a GFP sequence to the 3′ end of the coding regions inside native loci so that the tagged genes are produced using native promoters. Using immunofluorescence, we detected a GFP signal above background only in cilia (Supplemental Figure S3). Like in overproducing cells, Nrk1p-GFP was found preferentially in assembling locomotory and oral cilia (Supplemental Figure S3, B and C). Nrk2p-GFP accumulated at the tips of assembling oral cilia and anterior locomotory cilia in dividing cells. It is possible that Nrk1p and Nrk2p, when present at normal levels are exclusively ciliary proteins. The colocalization of Nrk1p-GFP and Nrk2p-GFP with nonciliary microtubules seen in overproducing cells may therefore reflect a microtubule-binding ability of these proteins.
We also overexpressed NRKs from two distinct subfamilies. Nrk17p is related to Chlamydomonas Fa2p (Figure 1A), implicated in severing of axonemal microtubules during deflagellation (Mahjoub et al., 2002), whereas Nrk30p belongs to a Tetrahymena-specific clade. The localization of overproduced GFP-Nrk17p was far more restricted, because this protein was mainly observed in association with BBs or the most proximal segment of cilia and some fibrous structures in the cell body (Figure 2C). Overexpression of GFP-Nrk17p led to a loss of cell motility in 2 h, and cells ceased to proliferate within 3 h (Supplemental Figure S1, C). Interestingly, ∼20% of cells were arrested in cytokinesis, often with a cleavage furrow shifted to the anterior end (Supplemental Figure S2). Apart from diffuse staining of the cell body, GFP-Nrk30p localized exclusively inside cilia (Figure 2, D and G). GFP-Nrk30p cells ceased to move within 3–4 h but continued to proliferate at decreased rate (Supplemental Figure S1, C).
Overexpression of Three Distinct NRKs Shortens Cilia
We noticed that in NRK-overproducing cells, the length of cilia decreased rapidly. We verified that CdCl2, at the concentration used to activate the MTT1 promoter, did not affect ciliary length. Wild-type cells treated with cadmium maintained normal length cilia of ∼6 μm (Figure 4, A, F, and G). Also, the ciliary length in cells carrying an NRK transgene grown without cadmium was normal (Figure 4F, zero time point). Overexpression of Nrk1p-GFP kinase had a mild reducing effect on ciliary length (Figure 4, B and F). After 4 h of induction, cilia of Nrk1p-GFP cells had ∼77% of original length (4.27 ± 0.69 μm; n = 260). Overexpression of Nrk2p-GFP caused a rapid and more extensive shortening of cilia (Figure 4, C and F) from 6.18 ± 0.51 μm (n = 32) to 2.09 ± 0.72 μm (n = 161) after 2 h and 1.84 ± 0.74 μm (n = 133) after 4 h (note that we underestimated the extent of shortening because we did not include completely resorbed cilia). The extent of shortening depended on the subcellular location. The shortest cilia were observed at the anterior and ventral side (Figure 5, A–E), and all affected cilia had Nrk2p-GFP at the tips (Figure 3, D–F). The unaffected cilia lacked Nrk2p-GFP at the tips and were mostly scattered within the posterior and dorsal region. In these areas, affected and unaffected cilia were often seen side by side (Figure 3, D–F).
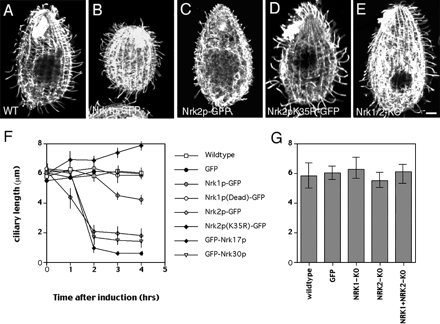
Figure 4.
Overproduction of NRKs causes ciliary resorption. Immunofluorescence confocal images of cells after 4 h of treatment with cadmium, which are either wild-type (A) or overproduce Nrk1p-GFP (B), Nrk2p-GFP (C), Nrk2p-K35R-GFP (D), or lack both NRK1 and NRK2 genes (E). (F) Graph that shows the average length of cilia as a function of time of treatment with cadmium either in wild-type or overproducing cells. Between 30 and 268 cilia were measured on multiple cells for each time point. Because many cilia were completely resorbed, and only visible cilia were measured, results at later time points are overestimated for GFP-Nrk17p, Nrk2p-GFP, and GFP-Nrk30p. (G) A histogram showing an average length of cilia in a growing population of cells that are either wild type, overproduce GFP (negative control), or lack NRK1, NRK2, or both genes. Between 150 and 260 cilia of multiple cells were measured for each strain.
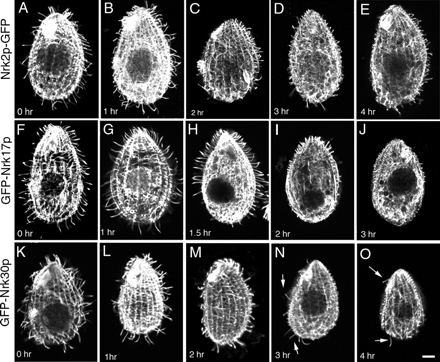
Figure 5.
A time-course study of the distribution of shortening cilia in cells overproducing Nrk2p-GFP (A–E), GFP-Nrk17p (F–J), and GFP-Nrk30p (K–O). Cells were treated with 2.5 μg/ml cadmium chloride for the time indicated and fixed and labeled by immunofluorescence with anti-total tubulin SG antibodies and anti-α-tubulin 12G10 antibodies.
In cells with GFP-Nrk17p, we noticed a rapid shortening of cilia, initially at the posterior end (Figure 5, F–J). At 1.5 h, cells had extremely short cilia in the posterior region and normal length cilia elsewhere (Figure 5H). The zone of shortening cilia expanded gradually toward the anterior end (Figure 5I), until 3–4 h after induction, when cells had become nonmotile with extremely short or completely resorbed cilia (0.63 ± 0.25 μm; n = 72) (Figures 4F and 5J).
In cells overexpressing GFP-Nrk30p, the ciliary length was also rapidly reduced (in 4 h down to 1.40 ± 0.47 μm; n = 218) (Figure 4F). Unlike in Nrk2p-GFP and GFP-Nrk17p cells, shortening of cilia in GFP-Nrk30p cells was observed throughout the cell (Figure 5, K–O), but a few anterior and posterior end cilia resisted the resorption (Figure 5, N and O, arrows).
Nrk2p, a CNK2-type NRK, Contributes to Length Regulation of Cilia at Steady State
Using germline targeting technology, we obtained cells lacking either NRK1 or NRK2 or both loci. However, single and double knockout cells had normal phenotype, including normal length cilia (Figure 4, E and G), normal growth rate, and conjugation. Given that NRK1 and NRK2 are in a group of 13 paralogues (Figure 1A), and most if not all of these paralogues are expressed (Supplemental Figure S5), one reasonable explanation of the lack of mutant phenotype is functional redundancy. We therefore attempted to produce a dominant-negative mutation. We mutated the highly conserved lysine residue to arginine to produce variants of Nrk1p and Nrk2p, which were predicted to lack kinase activity (Fry et al., 1995). Overexpression of Nrk1p-K40R-GFP did not affect the length of cilia (Figure 4F). However, overexpression of Nrk2p-K35R-GFP caused marked lengthening of cilia. The pattern of localization of Nrk2p-K35R-GFP was identical to its unmutated form (Figure 3, G–I). After 4 h of cadmium induction, cilia of Nrk2p-K35R-GFP cells had ∼132% their original length (from 5.99 ± 0.55 μm, n = 43 to 7.89 μm ±1.07 μm, n = 26) (Figure 4, D and F). These observations indicate that an ongoing resorption mediated by at least some NRKs regulates the length of cilia at steady state. As we were analyzing ciliary length of control cells, we noticed that cilia present in the anterior end are shorter compared with cilia in other parts of the cell (Figures 4A and 5, A and F). The average length of cilia at the anterior end in wild-type cells was 4.38 ± 0.49 μm (n = 50) compared with 6.45 ± 0.68 μm (n = 45) for the posterior cilia. Thus, normal Tetrahymena maintains unequal length cilia.
NRKs of Distinct Subtypes Resorb Cilia by Distinct Mechanisms
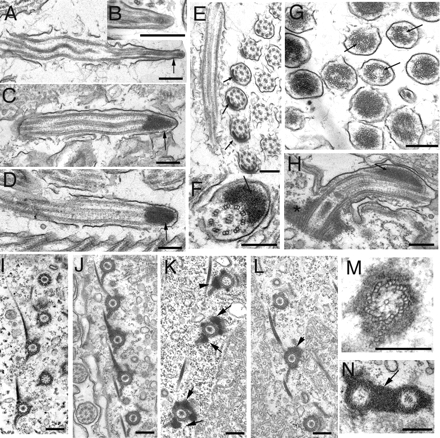
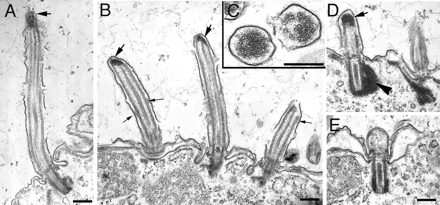
To shed light on the mechanism of cilia shortening, we analyzed the ultrastructure of NRK-overproducing cells. Cilia of wild-type cells treated with CdCl2 were ultrastructurally normal (Figure 6B), and so were cilia of overproducing cells grown without cadmium (Figure 6A). Examination of transverse ciliary sections of Nrk2p-GFP–overproducing cells showed normal 9 + 2 configuration of axonemal microtubules despite shortening (Figure 7C), suggesting that resorption is based on a coordinated disassembly of the entire axoneme. Prominent electron-dense deposits were present at the tips of cilia (Figures 6C and 7, A, B, and D, large arrow), which could correspond to foci of Nrk2p-GFP observed in immunofluorescence (Figure 3, D–F). These deposits are clearly distinct from naturally occurring axoneme-associated inclusions in one of the three rows of the adoral membranelle (Figure 6E), because they are larger and present in locomotory cilia. Most cilia that could be analyzed in their entirety were shorter than normal (Figure 7, A, B, and D) but completely resorbed cilia were seen rarely (Figure 7E), suggesting that the proximal part of cilia is more resistant to Nrk2p action. On cross-sections through the ciliary tips where the electron dense deposits were present, the doublet and central pair microtubules of the axoneme were displaced (Figure 6G, arrows). Electron-dense deposits were also observed in the cortex around BBs (Figures 6, J, K, M, and N, and 7D, arrowhead). Similar strong deposits were seen in cells overexpressing Nrk2p-K35R-GFP except that the deposits were often seen both at the tip as well as on the side of the axoneme (Figure 6, D, F, H, and L). Thus, the accumulation of deposits is not a sufficient cause of shortening, and kinase activity of Nrk2p is required. Strikingly, structures that closely resemble IFT particles described in Chlamydomonas (Kozminski et al., 1995) were frequently seen between the axonemes and the ciliary membranes of shortening cilia (Figure 7B, small arrows). Such IFT particle-like structures are rarely seen in cilia of normal or in cilia of Nrk2p-GFP cells without cadmium treatment (Figures 6A and 8A), which could be due to a slow rate of subunit exchange and low level of IFT components in already assembled cilia of Tetrahymena (Brown et al., 1999; Thazhath et al., 2004). This observation suggests that the resorption caused by Nrk2p-GFP is associated with an up-regulation of IFT. To test whether shortening cilia accumulate IFT components, we constructed strains that express a GFP-tagged subunit of IFT motor, kinesin-2, Kin1p (in the native locus), and an MTT1-driven transgene encoding 6xHis-Nrk2p. Overexpression of 6xHis-Nrk2p led to preferential shortening of anterior cilia as was earlier observed for GFP-Nrk2p. These shortening anterior cilia also showed increased signal of Kin1p-GFP in comparison with nonresorbing posterior cilia in the same cells (Supplemental Figure S4).
Figure 6.
Transmission electron microscopy of controls and cells overproducing NRKs. (A–D) Longitudinal sections of cilia of control cells with the Nrk2p-K35R-GFP transgene but without cadmium treatment (A) or wild-type cells treated with cadmium (B) or cadmium-treated Nrk2p-GFP–overproducing cells (C) and cadmium-treated Nrk2p-K35R-GFP–overproducing cells (D). (E) Cross-section of normal anterior adoral membranelle from NRK-2-dead kinase (without cadmium) showing normally occurring inclusions in these cilia. Note that inclusions are present but they are limited to only one row of cilia in the oral membranelle (F) Cross-sections of a locomotory cilium in the Nrk2p-K35R-GFP–overproducing cell. (G) Cross-section of a group of cilia at the level of ciliary tips in cells overproducing Nrk2p-GFP. Note displacements of axonemal microtubules (arrows). (H) Longitudinal section of a cilium from a Nrk2p-K35R-GFP–overproducing cell treated with cadmium. Note the lateral location of the electron-dense deposit (arrows). (I) Section of a control cell cortex treated with cadmium. (J–N) Images showing sections of the cortical regions of Nrk2p-GFP cells (J, K, M, and N) and Nrk2p-K35R-GFP cells (L) all treated with cadmium. Note electron-dense deposits around microtubules of basal bodies as well as associated microtubule bundles; these are asymmetrically localized in Nrk2p-GFP cells (K, arrows) and Nrk2p-K35R-GFP cells (L). Bar, 0.2 μm.
Figure 7.
Longitudinal sections of cilia of cells with the Nrk2p-GFP transgene with cadmium induction showing progressive shortening (A, B, D, and E). A cross-section of resorbing cilia of Nrk2p-GFP–overproducing cells is shown in C. Large arrows in A, B, and D show electron-dense deposits near the tips. Small arrows in B show IFT particle-like structures. The arrowhead in D shows an electron-dense deposit near the basal body. Bar, 0.2 μm
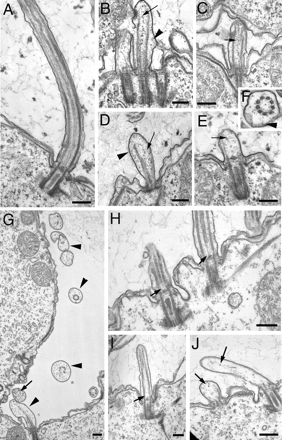
Figure 8.
(A–F) Shortening of cilia in GFP-Nrk30p–overproducing cells. (A) Control cell that carries the GFP-Nrk30p transgene but without cadmium induction. (B–E) Images showing progressive shortening of cilia in GFP-Nrk30p cells treated with cadmium. Arrowheads point at budges of ciliary membrane. Arrows show materials that seem to be breakdown products of the depolymerizing axonemes. (F) Cross-section of a shortening cilium from a GFP-Nrk30p–overproducing cells that lacks a central pair. (G–J) Shortening of cilia in GFP-Nrk17p–overproducing cells. (G) Cross-sections of several cilia (arrowheads) away from the cortical surface. Note the lack of the central pair and one or more of peripheral doublets or lack the entire axoneme cross-section. Furthermore, there is noticeable swelling of the cilium and accumulation of breakdown products. (H and I) Longitudinal sections showing gaps at the proximal end of the central pair, whereas peripheral doublets remain intact in the same area. (J) Cilia with completely depolymerized sets of axonemal structures in different stages of resorption. Bar, 0.2 μm.
In cells overproducing GFP-Nrk30p, shortening cilia had bulges of excess plasma membrane on the sides or at tips of the axonemes (Figure 8, B, D, and F, arrowheads), suggesting that the depolymerization of microtubules is not coordinated with removal of the ciliary membrane. Furthermore, the shortening axonemes generally lacked central microtubules (MTs) (Figure 8, B–F) or had fewer peripheral MTs (Figure 8, D and E). Thus, the depolymerization occurs unevenly across circumference of the axoneme and affects the central pair more strongly. In GFP-Nrk30p cilia, we did not see IFT particle-like structures, suggesting that the resorption does not involve up-regulation of IFT and could be equivalent to massive depolymerization of axonemal microtubules. Many GFP-Nrk30p axonemes had scattered materials inside the resorbing axoneme (Figure 8, B and C, arrows), suggesting breakdown products could not be removed from the axoneme using retrograde IFT.
Cells overproducing GFP-Nrk17p resorbed cilia by yet another mechanism. In relatively long cilia, we observed gaps between the proximal end of the central pair microtubules and the basal body (Figure 8H). In cilia more advanced in resorption, there was fragmentation of the entire set of 9 + 2 microtubules (Figure 8, G and J). Thus, Nrk17p has a preferred site of action at the proximal end of the central pair but with time induces depolymerization of all microtubules.
Thus, despite a similar end point, the mechanisms of resorption mediated by the tested NRKs, Nrk2p, Nrk17p, and Nrk30p, are fundamentally different. These data agree with the distinct phylogenetic position of Nrk2p, Nrk17p and Nrk30p (Figure 1).
DISCUSSION
With 39 genes, Tetrahymena has 3 times the number of NRK loci present in mammals. More precisely, Tetrahymena has relatively few homologues of the most conserved subtypes of NRKs (Nima/Nek2 and Nek6) and a large number of paralogues among the subtypes of NRKs found only in unicellular eukaryotes. We found that Tetrahymena Nrk20p, one of the two close homologues of Nek2, localizes preferentially to the newly assembled BBs (our unpublished data). Thus, Nrk20p and Nrk18p, could function in the maturation of duplicated BBs, a process that therefore may be similar to the duplication of the centrosomes in which Nek2 plays an important role (Fry, 2002; Faragher and Fry, 2003).
Virtually all NRKs tested in Tetrahymena localized to cilia but were preferentially targeted to only a subset of cilia. Importantly, for Nrk1p and Nrk2p, we showed that when expressed under native promoters, their GFP fusions localized to subsets of cilia and only Nrk2p-GFP accumulated at ciliary tips (Supplemental Figure S3). Thus, specific NRK paralogues have different subcellular targeting abilities, likely controlled by the rapidly evolving nonkinase extensions. We showed here that in normal cells, the length of locomotory cilia is reduced in the anterior region. Coupled with the observation that a dominant-negative form of Nrk2p increased the steady-state length of cilia, these data suggest that Tetrahymena maintains an unequal length of cilia by varying the activities of NRKs in specific locations. The large number of NRK paralogues, each potentially having a distinct pattern of localization, could lead to a highly regionalized, combinatorial control of ciliary length. It seems, however, that some functional redundancy exists, at least among the 13 paralogues of NRK1 and NRK2, for which gene knockouts did not reveal a mutant phenotype.
Based on the ultrastructural analysis, it seems that the resorption of cilia caused by overproduction of Nrk2p occurs at the distal end of the axoneme and gradually progresses toward the BBs. The resorption is highly organized because the normal 9 + 2 structure persisted during shortening, the ciliary membrane was progressively removed and we saw both IFT particle-like structures in TEM and increased kinesin-2 in affected cilia. Thus, CNK2-type NRKs may act by increasing the rate of axoneme depolymerization and simultaneously up-regulating IFT to transport the breakdown products out of the axoneme. It has been observed that products of axonemal breakdown accumulate in mutants lacking retrograde IFT (Qin et al., 2004), indicating that retrograde IFT removes disassembled components. Recently, Pan and Snell (2005) showed that during flagellar resorption, the amount of IFT particles entering the flagellum increases, but the amount of IFT-associated assembly cargo does not, suggesting that IFT delivers empty IFT particles to be loaded with the depolymerization-derived cargo. Our data suggest that CNK2 type NRKs could phosphorylate a substrate that activates both an IFT component(s) and a factor(s) that promotes depolymerization of the distal ends of axonemal microtubules. Flagellar resorption and autotomy in Chlamydomonas requires phosphorylation of the Aurora kinase CALK (Pan et al., 2004). It is therefore possible that CNK2 type NRKs and CALK function in the same pathway.
Nrk2p seems to participate in the regulation of ciliary length. This hypothesis is based on the observation that a kinase-inactive form of Nrk2p caused ciliary elongation. Our results agree fully with recent observations by Bradley and Quarmby (2005), who found that ectopic expression of the related Cnk2p shortened flagella, whereas an RNA interference-based depletion made flagella longer. It is intriguing that Nrk1p and Nrk2p preferentially localized to assembling cilia. It is possible that the process of assembly of cilia of proper length requires a proper balance between activities that deliver and remove structural components.
Nrk30p and Nrk17p caused massive depolymerization of the axoneme, apparently without up-regulation of IFT (based on lack of IFT particle-like structures and swelling of cilia presumably due to the presence of breakdown components that could not be removed by retrograde IFT). Although the depolymerization mediated by GFP-Nrk30p also started distally, the central pair disassembled more rapidly, and there was an asymmetry in the rate of depolymerization among individual doublet microtubules. First, we need to consider that Nrk30p activity inhibits IFT, which in turn could lead to shortening of cilia and accumulation of breakdown products caused by lack of retrograde IFT. However, depolymerization of cilia at steady state is a slow process in Tetrahymena, because it takes ∼80 h to completely exchange tubulins in already assembled Tetrahymena cilia (Thazhath et al., 2004). Thus, inhibition of IFT seems to be an insufficient explanation of the phenotype caused by excessive NRK30p. Nrk30p is more likely to act by accelerating the rate of depolymerization, for example, by phosphorylating a factor that destabilizes the ends of axonemal microtubules. It is tempting to speculate that one of the (direct or indirect) targets of Nrk30p activity is an EB1-like end-stabilizing protein (Pedersen et al., 2003) or an MCAK microtubule end depolymerizer (Hunter et al., 2003; Walczak, 2003).
Nrk17p, an NRK related to Fa2p NRK of Chlamydomonas, also caused rapid resorption of cilia. Fa2p is restricted to the transitional region between the BB and the axoneme (Mahjoub et al., 2004), and FA2 mutants are defective in flagellar autotomy, a process that is triggered by microtubule severing in the transitional region (Mahjoub et al., 2002). We show here that Nrk17p is mainly localized with the BBs and the most proximal part of the cilium. Remarkably, we observed preferential depolymerization of the central pair microtubules at their proximal end. Therefore, NRKs of the FA2 type could be stimulating a microtubule-severing activity at the proximal ends of ciliary microtubules. Proteins related to Nrk17p could be responsible for regulating severing of microtubules during autotomy as has been shown for Fa2p (Mahjoub et al., 2002). Because Fa2p mutations also reduce the rate of flagellar disassembly at steady state, it is likely that Fa2p type kinases promote microtubule turnover specifically in the proximal region of the axoneme. Based on the mutant phenotype, Fa2p seems to affect the doublet microtubules, whereas Nrk17p preferentially affects the central pair. It is possible that the one or more of the remaining three paralogues of Nrk17p in Tetrahymena act preferentially on the outer doublets.
It is tempting to speculate that Nrk17p and Nrk30p both function in turnover of axonemal components at the proximal and distal end of the cilium, respectively. At the native level of expression, Nrk17p and Nrk30p could be acting preferentially at the ends of the central pair microtubules. The complete resorption of cilia observed in our study requires both kinases to act on the doublet microtubules as well and could be the result of excessive level of these kinases.
The responses to resorption signals were not uniform across the Tetrahymena cell. For example, Nrk2p did not resorb a small subset of posterior and dorsal cilia and Nrk17p showed a strong preference for resorption of posterior cilia. With increased time, a wave of resorption induced by overexpressed Nrk17p progressed to the more anterior region (Figure 5). This wave is reminiscent of a naturally occurring wave of resorption that traverses the preoral region of Paramecium before conjugation (Vivier and Andre, 1961). Cilia are also selectively lost from the preoral region of mating Tetrahymena (Wolfe and Grimes, 1979; Wolfe, 1985; Kiersnowska and Kaczanowski, 1993). It is therefore tempting to speculate that ciliates use a combination of NRK paralogues to regulate the size of cilia in defined cortical locations.
The key question now is what are the phosphorylation substrates of NRKs. Based on the distinct consequences of overexpression and phylogenetic position, we speculate that Nrk2p, Nrk17p, and Nrk30p phosphorylate different substrates. The Tetrahymena strains with inducible expression of NRKs presented in this article will provide powerful tools for identification of NRK phosphorylation targets.
Supplementary Material
ACKNOWLEDGMENTS
Preliminary sequence data for T. thermophila were obtained from The Institute for Genomic Research at http://www.tigr.org. We thank the staff of the Laboratory for Electron Microscopy at the Nencki Institute (Warsaw, Poland) for excellent technical assistance. We thank Lynne Quarmby (Simon Fraser University, British Columbia, Canada), for sharing Cnk2p data before publication. We are grateful to Joseph Frankel (University of Iowa, Iowa City, IA) for numerous discussions. We thank the following researchers for providing essential reagents: Aaron Turkewitz (University of Chicago, Chicago, IL) for NRK2 EST plasmid, Marie-Helene Bré (Université Paris-Sud, Orsay, France) for TAP952 antibodies, Martin Gorovsky (University of Rochester, Rochester, NY) for SG antibodies, Klaus Weber (Max Planck Institute, Goettingen, Germany), for ID5 antibodies, Jeffrey Salisbury (Mayo Clinic, Rochester, MN) for 20H5 centrin antibodies, and Joseph Frankel for 12G10 antibodies. The 12G10 antibodies are available from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa. This work was supported by National Science Foundation award MBC-0235826 (to J. G.), the statute grant to the Nencki Institute from Committee of Scientific Research (Poland) (to M.J.-D.), and funding from the Razavi-Newman Center for Bioinformatics (to G. M.). D. W. was supported by a fellowship from the Kosciuszko Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-05-0450) on April 12, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Barsel S. E., Wexler D. E., Lefebvre P. A. Genetic analysis of long-flagella mutants of Chlamydomonas reinhardtii. Genetics. 1988;118:637–648. doi: 10.1093/genetics/118.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman S. A., Wilson N. F., Haas N. A., Lefebvre P. A. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 2003;13:1145–1149. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- Bradley B. A., Quarmby L. M. A NIMA-related kinase, Cnk2p, regulates both flagellar length and cell size in Chlamydomonas. J. Cell Sci. 2005;118:3317–3326. doi: 10.1242/jcs.02455. [DOI] [PubMed] [Google Scholar]
- Bradley B. A., Wagner J. J., Quarmby L. M. Identification and sequence analysis of six new members of the NIMA-related kinase family in Chlamydomonas. J. Eukaryot. Microbiol. 2004;51:66–72. doi: 10.1111/j.1550-7408.2004.tb00164.x. [DOI] [PubMed] [Google Scholar]
- Brown J. M., Fine N. A., Pandiyan G., Thazhath R., Gaertig J. Hypoxia regulates assembly of cilia in suppressors of Tetrahymena lacking an intraflagellar transport subunit gene. Mol. Biol. Cell. 2003;14:3192–3207. doi: 10.1091/mbc.E03-03-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M., Marsala C., Kosoy R., Gaertig J. Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol. Biol. Cell. 1999;10:3081–3096. doi: 10.1091/mbc.10.10.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J. Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- Christensen S. T., Guerra C. F., Awan A., Wheatley D. N., Satir P. Insulin receptor-like proteins in Tetrahymena thermophila ciliary membranes. Curr. Biol. 2003;13:R50–R52. doi: 10.1016/s0960-9822(02)01425-2. [DOI] [PubMed] [Google Scholar]
- Coyne B., Rosenbaum J. L. Flagellar elongation and shortening in Chlamydomonas. II. Re-utilization of flagellar proteins. J. Cell Biol. 1970;47:771–781. doi: 10.1083/jcb.47.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faragher A. J., Fry A. M. Nek2A kinase stimulates centrosome dysjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 1997;46:101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]
- Fry A. M. The Nek2 protein kinase: a novel regulator of centrosome structure. Oncogene. 2002;21:6184–6194. doi: 10.1038/sj.onc.1205711. [DOI] [PubMed] [Google Scholar]
- Fry A. M., Meraldi P., Nigg E. A. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A. M., Schultz S. J., Bartek J., Nigg E. A. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J. Biol. Chem. 1995;270:12899–12905. doi: 10.1074/jbc.270.21.12899. [DOI] [PubMed] [Google Scholar]
- Gaertig J., Gao Y., Tishgarten T., Clark T. G., Dickerson H. W. Surface display of a parasite antigen in the ciliate Tetrahymena thermophila. Nat. Biotech. 1999;17:462–465. doi: 10.1038/8638. [DOI] [PubMed] [Google Scholar]
- Galtier N., Gouy M., Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Hunter A. W., Caplow M., Coy D. L., Hancock W. O., Diez S., Wordeman L., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerka-Dziadosz M., Strzyzewska-Jowko I., Wojsa-Lugowska U., Krawczynska W., Krzywicka A. The dynamics of filamentous structures in the apical band, oral crescent, fission line and the postoral meridional filament in Tetrahymena thermophila revealed by the monoclonal antibody 12G9. Protist. 2001;152:53–67. doi: 10.1078/1434-4610-00043. [DOI] [PubMed] [Google Scholar]
- Kiersnowska M., Kaczanowski A. Inhibition of oral morphogenesis during conjugation of Tetrahymena thermophila and its resumption after cell separation. Eur. J. Protistol. 1993;29:359–369. doi: 10.1016/S0932-4739(11)80398-6. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Beech P. L., Rosenbaum J. L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin R. A., Lee T. H., Fang L. S. Effects of various agents on flagellar activity, flagellar autotomy and cell viability in four species of Chlamydomonas (Chlorophyta: Volvocales) Symp. Soc. Exp. Biol. 1982;35:421–437. [PubMed] [Google Scholar]
- Liu S., Lu W., Obara T., Kuida S., Lehoczky J., Dewar K., Drummond I. A., Beier D. R. A defect in a novel Nek-family kinase causes cystic kidney disease in the mouse and in zebrafish. Development. 2002;129:5839–5846. doi: 10.1242/dev.00173. [DOI] [PubMed] [Google Scholar]
- Mahjoub M. R., Montpetit B., Zhao L., Finst R. J., Goh B., Kim A. C., Quarmby L. M. The FA2 gene of Chlamydomonas encodes a NIMA family kinase with roles in cell cycle progression and microtubule severing during deflagellation. J. Cell Sci. 2002;115:1759–1768. doi: 10.1242/jcs.115.8.1759. [DOI] [PubMed] [Google Scholar]
- Mahjoub M. R., Qasim Rasi M., Quarmby L. M. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol. Biol. Cell. 2004;15:5172–5186. doi: 10.1091/mbc.E04-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. Size control in dynamic organelles. Trends Cell Biol. 2002;12:414–419. doi: 10.1016/s0962-8924(02)02341-3. [DOI] [PubMed] [Google Scholar]
- Marshall W. F., Qin H., Rodrigo Brenni M., Rosenbaum J. L. Flagellar length control system: testing a simple model based on intraflagellar transport and turnover. Mol. Biol. Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F., Rosenbaum J. L. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen E. M. Transformation in Tetrahymena thermophila. Development of an inducible phenotype. Dev. Biol. 1978;66:17–31. doi: 10.1016/0012-1606(78)90270-1. [DOI] [PubMed] [Google Scholar]
- Nelsen E. M., DeBault L. E. Transformation in Tetrahymena pyriformis: description of an inducible phenotype. J. Protozool. 1978;25:113–119. doi: 10.1111/j.1550-7408.1978.tb03880.x. [DOI] [PubMed] [Google Scholar]
- Nelsen E. M., Frankel J., Martel E. Development of the ciliature of Tetrahymena thermophila. I. Temporal coordination with oral development. Dev. Biol. 1981;88:27–38. doi: 10.1016/0012-1606(81)90216-5. [DOI] [PubMed] [Google Scholar]
- Osmani S. A., Engle D. B., Doonan J. H., Morris N. R. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell. 1988;52:241–251. doi: 10.1016/0092-8674(88)90513-2. [DOI] [PubMed] [Google Scholar]
- Pan J., Snell W. J. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev. Cell. 2005;9:431–438. doi: 10.1016/j.devcel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Pan J., Wang Q., Snell W. J. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev. Cell. 2004;6:445–451. doi: 10.1016/s1534-5807(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Parker J. D., Quarmby L. M. Chlamydomonas fla mutants reveal a link between deflagellation and intraflagellar transport. BMC Cell Biol. 2003;4:11. doi: 10.1186/1471-2121-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L. B., Geimer S., Sloboda R. D., Rosenbaum J. L. The microtubule plus end tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr. Biol. 2003;13:1969–1974. doi: 10.1016/j.cub.2003.10.058. [DOI] [PubMed] [Google Scholar]
- Piperno G., Mead K., Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1(FLA10) to reach the distal part of flagella in Chlamydomonas. J. Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Burnette D. T., Bae Y. K., Forscher P., Barr M. M., Rosenbaum J. L. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Qin H., Diener D. R., Geimer S., Cole D. G., Rosenbaum J. L. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby L. M. Cellular deflagellation. Int. Rev. Cytol. 2004;233:47–91. doi: 10.1016/S0074-7696(04)33002-0. [DOI] [PubMed] [Google Scholar]
- Quarmby L. M., Mahjoub M. R. Caught Nek-ing: cilia and centrioles. J. Cell Sci. 2005;118:5161–5169. doi: 10.1242/jcs.02681. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Jensen C. G., Jensen L. C. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J. Ultrastruct. Res. 1979;68:173–185. doi: 10.1016/s0022-5320(79)90152-7. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Moulder J. E., Ringo D. L. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schliwa M., van Blerkom J. Structural interaction of cytoskeletal components. J. Cell Biol. 1981;90:222–235. doi: 10.1083/jcb.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J. M. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J., Gorovsky M. A. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor D., Wedaman K. P., Orozco J. T., Dwyer N. D., Bargmann C. I., Rose L. S., Scholey J. M. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell. Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Dentler W. Flagellar protein dynamics in Chlamydomonas. J. Biol. Chem. 2001;276:29754–29763. doi: 10.1074/jbc.M103184200. [DOI] [PubMed] [Google Scholar]
- Stephens R. E. Synthesis and turnover of embryonic sea urchin ciliary proteins during selective inhibition of tubulin synthesis and assembly. Mol. Biol. Cell. 1997;8:2187–2198. doi: 10.1091/mbc.8.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazhath R., Jerka-Dziadosz M., Duan J., Wloga D., Gorovsky M. A., Frankel J., Gaertig J. Cell context-specific effects of the beta-tubulin glycylation domain on assembly and size of microtubular organelles. Mol. Biol. Cell. 2004;15:4136–4147. doi: 10.1091/mbc.E04-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazhath R., Liu C., Gaertig J. Polyglycylation domain of beta-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 2002;4:256–259. doi: 10.1038/ncb764. [DOI] [PubMed] [Google Scholar]
- Upadhya P., Birkenmeier E. H., Birkenmeier C. S., Barker J. E. Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc. Natl. Acad. Sci. USA. 2000;97:217–221. doi: 10.1073/pnas.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E., Andre J. Donnes structurales et ultrastructurales nouvelles sur la conjugasion de Paramecium caudatum. J. Protozool. 1961;8:416. [Google Scholar]
- Walczak C. E. The Kin I kinesins are microtubule end-stimulated ATPases. Mol. Cell. 2003;11:286–288. doi: 10.1016/s1097-2765(03)00067-4. [DOI] [PubMed] [Google Scholar]
- Wang S., Nakashima S., Sakai H., Numata O., Fujiu K., Nozawa Y. Molecular cloning and cell-cycle-dependent expression of a novel NIMA (never-in-mitosis in Aspergillus nidulans)-related protein kinase (TpNrk) in Tetrahymena cells. Biochem. J. 1998;334:197–203. doi: 10.1042/bj3340197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. E. Preparation of cytoskeletal fractions from Tetrahymena thermophila. Methods Cell Biol. 2000;62:441–447. doi: 10.1016/s0091-679x(08)61548-4. [DOI] [PubMed] [Google Scholar]
- Wolfe J. Cytoskeletal reorganization and plasma membrane fusion in conjugating Tetrahymena. J. Cell Sci. 1985;73:69–85. doi: 10.1242/jcs.73.1.69. [DOI] [PubMed] [Google Scholar]
- Wolfe J., Grimes G. W. Tip transformation in Tetrahymena: morphogenetic response to interactions between mating types. J. Protozool. 1979;26:82–89. [Google Scholar]
- Yin M. J., Shao L., Voehringer D., Smeal T., Jallal B. The serine/threonine kinase Nek6 is required for cell cycle progression through mitosis. J. Biol. Chem. 2003;278:52454–52460. doi: 10.1074/jbc.M308080200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.