Abstract
Periodically regulated cyclin-dependent kinase (Cdk) is required for DNA synthesis and mitosis. Hydroxyurea (HU) inhibits DNA synthesis by depleting dNTPs, the basic unit for DNA synthesis. HU treatment triggers the S-phase checkpoint, which arrests cells at S-phase, inhibits late origin firing and stabilizes replication forks. Using budding yeast as a model system, we found that Swe1, a negative regulator of Cdk, appears at S-phase and accumulates in HU treatment cells. Interestingly, this accumulation is not dependent on S-phase checkpoint. Δhsl1, Δhsl7, and cdc5-2 mutants, which have defects in Swe1 degradation, show HU sensitivity because of high Swe1 protein levels. We further demonstrated that their HU sensitivity is not a result of DNA damage accumulation or incomplete DNA synthesis; instead the sensitivity is due to their dramatically delayed recovery from HU-induced S-phase arrest. Strikingly, our in vivo data indicate that Swe1 inhibits the kinase activity of Clb2-Cdk1, but not that of Clb5-Cdk1. Therefore, S-phase accumulated Swe1 prevents Clb2-Cdk1–mediated mitotic activities, but has little effects on Clb5-Cdk1–associated S-phase progression.
INTRODUCTION
During each cell cycle, cells duplicate their genomic DNA and separate two sets of duplicated chromosome to two daughter cells. In all eukaryotic organisms, the key driving force of the cell cycle progression is cyclin-dependent kinase (Cdk), which is encoded by a single gene CDC28 in budding yeast Saccharomyces cerevisiae. By associating with periodically expressed cyclins, Cdc28 (Cdk1) activity is regulated during the cell cycle, peaking at S- and M-phases, as highly active Cdk1 is required for DNA replication and chromosome segregation. In budding yeast, there are total 6 B-type cyclins. Among them, CLB1–CLB4 are transcribed during G2/M-phase and promote events required for the completion of mitosis. Clb5 and Clb6 appear during S-phase, indicating their functions in DNA replication (Kuhne and Linder, 1993). However, Clb5 seems to play a more important role in DNA replication, as Δclb5 deletion mutants, but not Δclb6, results in a lengthened S-phase. It has been shown that Clb5 is responsible for both early and late origin firing, whereas Clb6 is only for early origin firing (Donaldson et al., 1998).
B-type cyclin-associated Cdk1 has been shown to phosphorylate a number of DNA replication factors, including components of the ORC, Orc1, Orc2, and Orc6, as well as Cdc6 and MCM proteins (Ubersax et al., 2003). One potential regulatory role for these phosphorylation events is to prevent reinitiation, thus restricting replication to a single round per cell cycle (Dahmann et al., 1995; Cocker et al., 1996; Piatti et al., 1996). Three other DNA replication–related proteins, Sld2/Drc1, Pol12, and Dpb2, have been shown to be the substrates of Clb-Cdk1 (Foiani et al., 1995; Masumoto et al., 2002; Kesti et al., 2004). Cdk1-dependent phosphorylation of Sld2 is the only one event known to be essential for the initiation of DNA replication in yeast. Phosphorylated Sld2 forms a complex with Dpb11 and is required for the loading of DNA polymerase α to the origins (Masumoto et al., 2002). Pol12 is the second largest subunit of Polα. Its phosphorylation is cell cycle regulated and has been shown to promote the dissociation of Polα from the chromatin (Desdouets et al., 1998). Dpb2, the regulatory subunit of budding yeast Polσ, is also a substrate of Clb-Cdk1. Its phosphorylation is cell cycle regulated and believed to promote DNA synthesis, as mutation of the Cdk1 phosphorylation sites results in a synthetic phenotype with pol2-11 mutants, which show defects in DNA synthesis (Kesti et al., 2004).
Accurate DNA replication is critical to maintain the genetic stability. During S-phase, many intrinsic and extrinsic agents can affect DNA replication. S-phase checkpoint senses interrupted DNA replication, arrests cells at S-phase, inhibits late origin firing, and stabilizes replication forks (Lopes et al., 2001). It is still unclear how cell cycle machinery responds to interrupted DNA synthesis. Evidence from fission yeast, Xenopus, and mammals indicate that Wee1, a protein kinase that phosphorylates Cdk1 and inhibits its activity, is stabilized in response to DNA damage or DNA synthesis block (O'Connell et al., 1997; Michael and Newport, 1998; Raleigh and O'Connell, 2000). Moreover, recent evidence from vertebrate cells indicates that the destruction of Wee1 is essential for the recovery from DNA damage or S-phase checkpoint arrest. Plk1 is required for the recovery process because of its role in Wee1 protein degradation (van Vugt et al., 2004; Yamada et al., 2004). In budding yeast, the Wee1 homologue, Swe1, phosphorylates tyrosine 19 of Cdc28 and inhibits Cdk1 activity. However, neither Δswe1 deletion, nor CDC28F19 mutant, which is resistant to the phosphorylation by Swe1, exhibits checkpoint defects in the presence of DNA damage or DNA synthesis block (Amon et al., 1992; Sorger and Murray, 1992). It has been thought that the phosphorylation of Cdc28 may not be a factor in DNA synthesis regulation. Therefore, the regulation of Swe1 in response to the treatment of genotoxic agents has never been fully investigated.
Here we first described that Swe1 accumulates in response to DNA synthesis block by hydroxyurea (HU), a drug that inhibits ribonucleotide reductase and depletes the dNTP pool. To our surprise, Swe1 accumulation is not dependent on S-phase checkpoint, as mec1-1 and rad53 null mutants exhibit elevated Swe1 protein levels as well as wild-type (WT) cells. Δhsl1 and Δhsl7 mutants, which have defects in Swe1 degradation (Ma et al., 1996; McMillan et al., 1999; Shulewitz et al., 1999), show Swe1-dependent HU sensitivity. Similarly, cdc5-2 mutant also shows Swe1-dependent HU sensitivity due to the failure of Swe1 protein degradation. We further demonstrated that their HU sensitivity is a result of slow recovery from S-phase arrest, but not the results of DNA damage accumulation or slowed DNA synthesis. The accumulation of Swe1 delays the Clb2-Cdk1–dependent Pol12 phosphorylation dramatically, but has little effect on the Clb5-Cdk1–dependent phosphorylation of Sld2/Drc1. Thus budding yeast cells accumulate Swe1 protein during S-phase in order to block Clb2-Cdk1 associated mitotic activities.
MATERIALS AND METHODS
Yeast Strains, Growth, and Media
The genotype and sources of the relevant yeast strains are listed in Table 1. All the strains listed are isogenic with a W303-derived Y300 strain. They were constructed using standard genetic crosses. RAD53-HA strain was made by using a PCR-based method (Longtine et al., 1998). To arrest yeast cells at G1-phase, 5 μg/ml α-factor was added into midlog cell cultures (OD600 = 0.4) and the cultures were incubated for 2.5 h. To release them into cell cycle, the cell cultures were centrifuged and washed once with H2O. Nocodazole was purchased from ICN (Costa Mesa, CA) and used at 20 μg/ml in a final concentration of 1% dimethylsulfoxide.
Table 1.
Strains used in this study
| Strains | Relevant genotype | Source |
|---|---|---|
| Y300 | Mataura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 | This study |
| 411-1-1 | Mata Δswe1::LEU2 | This study |
| 413-1-1 | Matahis- SWE1-myc-HIS2 | Lew lab |
| YYW81 | MataGAL-SWE1-LEU2 | This study |
| 426-4-3 | Mata Δhsl1::URA3 | This study |
| 427-5-2 | Matahsl7-Δ20::HIS3 | This study |
| YYW84 | Mata Δcla4::LEU2 | This study |
| 134-1-1 | Mata Δcdc55::HIS3 | This study |
| 426-2-2 | Mata Δhsl1::URA3Δ swe1::LEU2 | This study |
| 479-2-2 | Mata Δhsl1::URA3 Δcdc28::URA3 CDC28F19-TRP1 | This study |
| 433-3-2 | Mata Δhsl1::URA3 SWE1-myc-HIS2 | This study |
| YYW82-1 | MataRAD53-3HA-his5+ | This study |
| 429-4-3 | Mata Δhsl::URA3 RAD53-3HA-his5+ | This study |
| 241-1-3 | Matacdc5-2-URA3 | This study |
| 477-2-3 | Matacdc5-2-URA3 Δswe1::LEU2 | This study |
| 480-3-1 | Mata Δhsl1::URA3 cdc5-2-URA3 | This study |
| 481-7-2 | Mata Δhsl1::URA3 cdc5-2-URA3 Δswe1::LEU2 | This study |
| YYW83 | MataY300 with HSL7-GFP plasmid | This study |
| Tay179 | Matabar1 DPB2-6myc::KAN | Wittenberg lab |
| 508-3-2 | Matabar1 Δhsl1::URA3 DPB2-6myc::KAN | This study |
| YNIG12 | Matabar1 SLD2-9myc::LEU2 | Araki lab |
| 523-15-4 | Matabar1 Δhsl1::URA3 SLD2-9myc::LEU2 | This study |
| 524-6-2 | Mata Δrad53 sml1-1 SWE1-myc-HIS2 | This study |
| 413-6-4 | Mata Δcdc28::URA3 CDC28F19-TRP1 SWE1-myc-HIS2 | This study |
| 478-9-3 | Matacdc5-2::URA3 SWE1-myc-HIS2 | This study |
Cytological Techniques
Tubulin staining was performed as described previously (Wang et al., 2003). Briefly, cells were fixed by the addition of 3.7% formaldehyde to growing cultures for at least 1 h. Cells were washed in phosphate-buffered saline, and microtubules were immunostained using the rat anti-tubulin antibody YOL1/34 and a FITC-conjugated secondary anti-rat antibody. DAPI was used to visualize DNA. The spindle and nuclei structures were visualized by fluorescence microscopy (Zeiss, Thornwood, NY).
Protein Techniques
Two milliliters of cell culture were used to prepare protein samples for time course experiments. Cells were collected in tubes with screw caps after being centrifuged and 50 μl of 20% TCA and glass beads were added. Cells were broken with a bead-beater for 2 min. Protein was precipitated by centrifugation at 3000 rpm for 2 min after glass beads were removed. Equal volume (50 μl) of 1 M Tris base and protein loading buffer were added. Dissolved protein samples were boiled for 5 min. Protein samples were resolved by 8% SDS-polyacrylamide gel electrophoresis (PAGE). Primary antibodies (anti-myc, anti-HA) were purchased from Covance (Madison, WI), and anti-Pgk1 antibody was from Molecular Probes (Eugene, OR). Anti-Pol12 was kindly provided by the Foiani lab in Italy. HRP-conjugated secondary antibody was purchased from Jackson ImmunoResearch (West Grove, PA).
Pulsed Field Gel Electrophoresis Analysis
Collected yeast cells were washed once with water and then fixed with 70% ethanol for 1 h at room temperature. To remove cell walls, cells were resuspended in LiSorb buffer and treated with zymolyase at 37°C for 1 h. Then cells were resuspended in TE buffer (10 mM Tris, 10 mM EDTA, pH 8.0). An equal volume of 1% melted agarose (after cooling down to 50°C) was added into cells. After agarose was solidified into agarose block, cells embedded in agarose blocks were subject to digestion with lysis buffer (100 mM EDTA, 10 mM Tris, 1% Sarkosyl, 100 μg/ml proteinase K, pH 8.0) overnight at 50°C. After that, agarose blocks were washed with TE buffer twice and were ready for pulsed field gel electrophoresis (PFGE) analysis. CHEF-DR II pulsed field electrophoresis systems (Bio-Rad, Richmond, CA) was used. The running time was 20 h at 6 V/cm with a 60-120-s switch time ramp (14°C).
RESULTS
Swe1 Accumulates in HU-arrested Cells
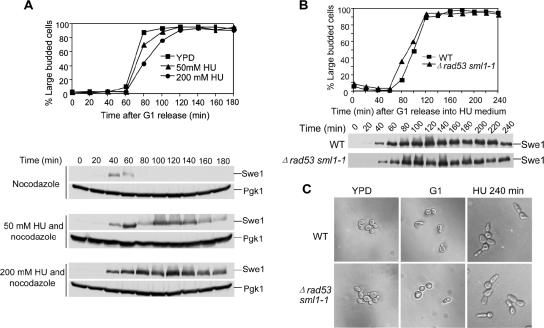
In budding yeast, Swe1 kinase phosphorylates Cdk1 (Cdc28) at tyrosine 19 and inhibits the kinase activity of Cdk1. It has been shown that overexpression of SWE1 is lethal and results in dramatically elongated cells, suggesting that Swe1 accumulation contributes to cell cycle arrest as well as hyphal growth (Booher et al., 1993). HU, an inhibitor of ribonucleotide reductase, is able to inhibit DNA synthesis and activate the S-phase checkpoint that arrests cells in S-phase. Recent evidence indicates that yeast cells exhibit Swe1-dependent hyphal growth when incubated on YPD plates containing HU (Jiang and Kang, 2003). In agreement with the previous data, we found that yeast cells also exhibited elongated cell morphology when incubated in liquid YPD medium containing 200 mM HU. Consistently, this morphology is also Swe1-dependent as Δswe1 deletion mutants did not show this phenotype (unpublished data). A reasonable explanation for these observations is that the presence of Swe1 protein in S-phase–arrested cells leads to hyphal growth. If that is the case, we expect to see high Swe1 protein levels in S-phase–arrested cells. Therefore, we examined Swe1 protein levels in the presence of HU. G1-arrested cells with myc-tagged SWE1 were released into YPD medium with different concentrations of HU at 30°C. After release, 20 μg/ml nocodazole was added into the medium to block the cells at metaphase when Swe1 protein levels are normally low. As shown in Figure 1A, Swe1 appeared at 40 and 60 min and then disappeared in the absence of HU. In the presence of HU, Swe1 appeared at the same time but kept persistent for a much longer time. The presence of either 50 or 200 mM HU resulted in Swe1 accumulation, but Swe1 protein accumulation is more dramatic in the presence of 200 mM HU. This result supports the conclusion that HU-induced hyphal growth is a result of Swe1 protein accumulation.
Figure 1.
Swe1 protein accumulates in HU-arrested S-phase cells in an S-phase checkpoint-independent manner. (A) Swe1 protein accumulates in HU-arrested S-phase cells. Strain SWE1-myc (413-1-1) was arrested in G1-phase with α-factor at 30°C and released into the media with different concentrations of HU plus 20 μg/ml nocodazole and incubated at 30°C. At the indicated times, proteins were prepared and analyzed by Western blotting with anti-myc and anti-Pgk1 antibodies. The Pgk1 protein signal was used as loading control. The budding index is shown in the top panel. (B) Swe1 protein accumulates in Δrad53 sml1-1 mutants. Strains SWE1-MYC (413-1-1) and Δrad53 sml1-1 SWE1-Myc (524-6-2) were treated as described in A. The budding index is shown in the top panel. (C) Δrad53 sml1-1 cells show hyphal growth in the presence of HU. G1-arrested SWE1-MYC (413-1-1) and Δrad53 sml1-1 SWE1-Myc (524-6-2) were released into YPD medium containing 200 mM HU and 20 μg/ml nocodazole at 30°C. Pictures were taken for cells before release and 240 min after release. The cell morphology of asynchronized 413-1-1 and 524-6-2 is also shown.
Swe1 Accumulates in an S-phase–independent Manner
Mec1 and Rad53 kinases are key components in S-phase checkpoint pathway. We have demonstrated that Swe1 accumulates in the presence of HU (Figure 1A). An obvious question is whether Swe1 accumulation depends on an S-phase checkpoint. To answer this question, we first compared Swe1 protein levels in WT and mec1-1 mutants. To our surprise, mec1-1 mutants exhibited comparable Swe1 protein levels in the presence of 200 mM of HU (Liu, unpublished data). Because the elevated Swe1 could be the trace checkpoint activity in mec1-1 point mutant, we then examined Swe1 protein levels in rad53 deletion mutants in the presence of HU. RAD53 is an essential gene, but mutation of SML1 makes rad53 viable. Therefore, SWE1-MYC and Δrad53 sml1-1 SWE1-MYC strains were used for this experiment. The cells were arrested in G1-phase and then released into YPD medium containing 200 mM HU and 20 μg/ml nocodazole and incubated at 30°C. As shown in Figure 1B, Swe1 protein appeared in both WT and Δrad53 at 40 min. Strikingly, both of them maintained high Swe1 protein levels even 240 min after G1 release, suggesting that Swe1 accumulates in an S-phase checkpoint–independent manner. Consistently, both WT and Δrad53 cells showed hyphal growth phenotype after incubation in the presence of HU (Figure 1C). Because high levels of Swe1 protein induces hyphal growth, this result further supports the conclusion that Swe1 protein accumulation in the presence of HU does not depend on the S-phase checkpoint.
Hsl1/Hsl7 Complex Is Active in the Presence of HU
Bud neck–localized Hsl1/Hsl7 complex is responsible for bud neck recruitment and degradation of Swe1 (McMillan et al., 1999). Hsl1 is a protein kinase that phosphorylates Hsl7 and facilitates its bud neck localization (Shulewitz et al., 1999; Cid et al., 2001). Hsl7 binds to both Hsl1 and Swe1; thus, Hsl7 is thought to function as an adaptor. Recent evidence showed that the formation of the Hsl1 and Hsl7 platform at the bud neck is critical for Swe1 degradation (Asano et al., 2005). We have shown that Swe1 accumulates in the presence of HU. One possibility is that HU-induced Swe1 accumulation is attributed to Hsl1/Hsl7 inactivation. Therefore the localization of Hsl7 was examined in HU-arrested cells. WT cells with an HSL7-GFP plasmid were released into YPD medium containing 200 mM HU for 2 h, and Hsl7 localization was examined by fluorescence microscopy. As shown in Figure 2A, almost all the cells exhibited bud neck localized Hsl7 after S-phase arrest by HU. Because the bud neck localization of Hsl7 requires functional Hsl1, we conclude that Swe1 accumulation in the presence of HU is not a result of inactivation of the Hsl1/Hsl7 complex.
Figure 2.
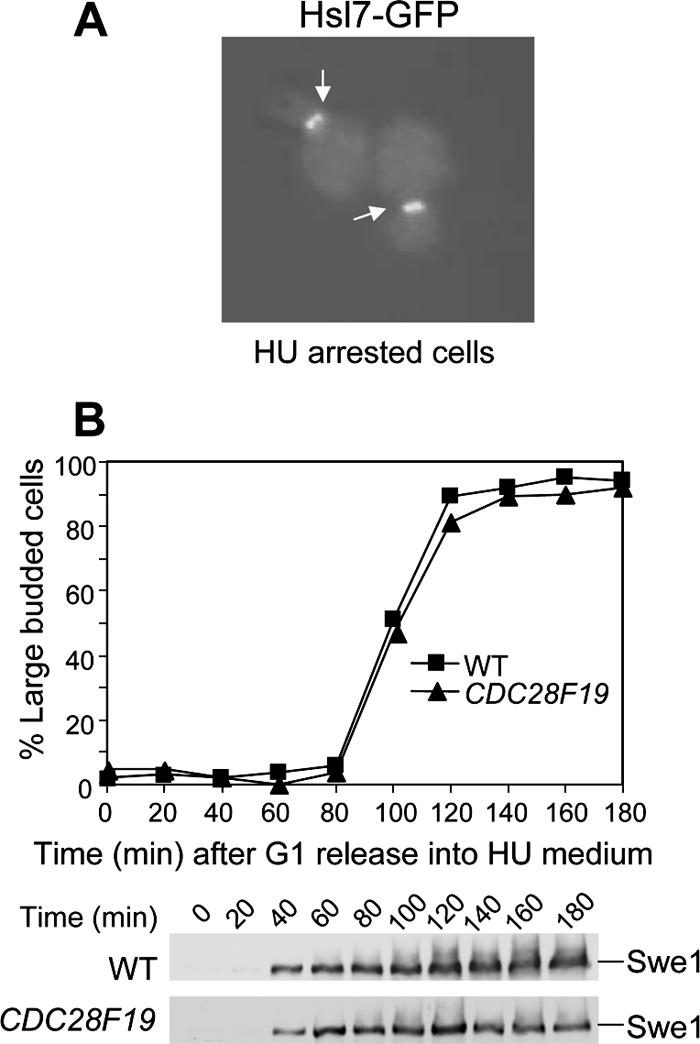
(A) Hsl7 is localized at bud neck in HU-arrested cells. WT cells with an HSL7-GFP plasmid were incubated in YPD medium containing 200 mM HU for 2 h at 30°C. Picture was taken with a fluorescence microscope. (B) Swe1 protein accumulates in CDC28F19 cells in the presence of HU. Strains SWE1-myc (413-1-1) and CDC28F19 SWE1-MYC (413-6-4) were arrested in G1-phase with α-factor and released into the media with 200 mM HU and 20 μg/ml nocodazole and incubated at 30°C. At the indicated times, proteins were prepared and analyzed by Western blotting with anti-myc antibody. The budding index is shown in the top panel.
Accumulation of Swe1 Is Not Due to Down-Regulation of Clb2-Cdk1
Swe1 kinase phosphorylates Cdc28 (Cdk1) and inactivates Cdk1. Recent research work from the Kellogg lab shows that Clb2-Cdk1 is able to phosphorylate Swe1 and promotes its degradation (Harvey et al., 2005). Because treatment of yeast cells with HU induces Cdc28 phosphorylation (Krishnan et al., 2004), it is possible that the inhibition of Clb2-Cdk1 after HU treatment results in Swe1 accumulation. Because Swe1 kinase phosphorylates Cdc28 at tyrosine 19, the CDC28F19 mutant, in which tyrosine 19 was substituted by phenylalanine, is resistant to the inhibition by Swe1 (Amon et al., 1992). If the inhibition of Cdc28 contributes to elevated Swe1 protein levels, CDC28F19 mutation should abolish HU-induced Swe1 accumulation. Thus Swe1 protein levels were compared in WT and CDC28F19 mutants. G1-arrested SWE1-MYC and CDC28F19 SWE1-MYC were released into YPD medium containing 200 mM HU and 20 μg/ml nocodazole. As shown in Figure 2B, Swe1 protein levels were persistent in both WT and CDC28F19 cells, although CDC28F19 mutant cells exhibited a little less Swe1 after a 120-min release into HU. This observation indicates that HU-induced Swe1 accumulation is not due to down-regulation of Clb2/Cdk1. Even though active Cdc28 is essential for Swe1 degradation, it is not sufficient to induce Swe1 degradation. Thus other factors essential for Swe1 degradation must be inhibited in HU-arrested yeast cells.
Mutants That Accumulate Swe1 Show HU Sensitivity
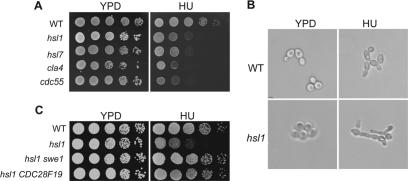
Hsl1, Hsl7, Cla4, and Cdc55 have been demonstrated to be involved in Swe1 protein degradation and deletion mutants of these genes exhibit Swe1-dependent hyphal growth. Bud-neck–localized Hsl1 and Hsl7 are able to facilitate the bud neck localization and degradation of Swe1 (McMillan et al., 1999; Shulewitz et al., 1999). Cla4 is a kinase that phosphorylates and promotes Swe1 degradation (Longtine et al., 2000). In addition, the Swe1 protein level has been shown to be elevated in the Δcdc55 mutant (Yang et al., 2000). If the elongated bud morphology in the presence of HU is a result of down-regulation of Hsl1, Hsl7, Cla4, or Cdc55, mutants of these genes will exhibit similar bud morphology in the presence or absence of HU. The hyphal growth of the four mutants was examined after incubation in the presence of 200 mM HU. Interestingly, all four mutants showed more pronounced hyphal growth in the presence of HU (Figure 3B and unpublished data), suggesting that the HU treatment and the mutation in the four genes have additive effects on the hyphal growth phenotype.
Figure 3.
(A) Mutants with defects in Swe1 degradation are sensitive to HU. Tenfold serial dilutions of saturated cell cultures (Y300, 426-4-3, 427-5-2, YYW84, 134-1-1) were spotted onto either YPD or HU (100 mM) plates. The plates were incubated at 25°C for 3 d before taking the pictures. (B) HU induces pronounced hyphal growth in Δhsl1 mutant. WT and Δhsl1 mutant cells (Y300 and 426-4-3) were grown in YPD or YPD plus 200 mM HU for 3 h at 30°C. The figure shows the bud morphology of WT and Δhsl1 mutant cells. (C) Δhsl1 mutant exhibits Swe1-dependent HU sensitivity. Strains (Y300, 426-4-3, 426-2-2, 479-2-2) were grown to saturation and then 10-fold diluted and spotted onto YPD and HU (100 mM) plates. The pictures were taken after incubation at 25°C for 3 d.
Accumulation of Swe1 protein is toxic to yeast cells. If HU treatment leads to additional accumulation of Swe1 in Δhsl1, Δhsl7, Δcla4, and Δcdc55 mutants, these mutants may exhibit HU sensitivity. Indeed, we found that all four mutants showed HU sensitivity because they failed to form colonies on YPD plates containing 100 mM HU (Figure 3A). Because the four mutants exhibit defects in Swe1 protein degradation, it is likely that their HU sensitivity is a result of Swe1 accumulation. If this is the case, deletion of SWE1 should suppress their HU sensitivity. Therefore Δhsl1 Δswe1, Δhsl7 Δswe1, Δcdc55 Δswe1, and Δcla4 Δswe1 double mutants were constructed, and their HU sensitivity was examined on YPD plates containing 100 mM HU. Surprisingly, deletion of SWE1 only suppressed the HU sensitivity of Δhsl1 and Δhsl7 mutants; Δcdc55 Δswe1 and Δcla4 Δswe1 exhibited HU sensitivity similar to that of the single mutants (Figure 3C and unpublished data). However, the hyphal growth phenotype of the four mutants was all suppressed by SWE1 deletion, indicating that other Swe1-independent defects in Δcdc55 and Δcla4 mutants also contribute to HU sensitivity. Because Swe1 inhibits Cdc28 activity by phosphorylating a highly conserved tyrosine residue (Y19) at the N-terminus (Booher et al., 1993), we also tested whether the HU sensitivity of the Δhsl1 mutant depends on the phosphorylation of Cdc28 tyrosine 19. For this purpose, we examined the HU sensitivity of Δhsl1 CDC28F19, in which tyrosine 19 was substituted by phenylalanine. We found that CDC28F19 also suppressed the HU sensitivity of Δhsl1, suggesting that the inactivation of Cdk1 activity by elevated Swe1 protein leads to the HU sensitivity of the Δhsl1 mutant (Figure 3C).
cdc5-2 Mutant Shows Swe1-dependent HU Sensitivity
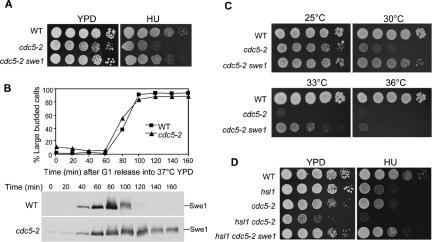
Cdc5, together with Cdk1 and Cla4, is involved in Swe1 phosphorylation and degradation (Sakchaisri et al., 2004). Because we have shown that Δhsl1 mutants exhibit HU sensitivity because of the failure of Swe1 degradation, cdc5 mutants defective in Swe1 degradation should also exhibit HU sensitivity. Thus, the HU sensitivity of two cdc5 mutant alleles was examined at room temperature, and cdc5-2 but not cdc5-1 exhibited HU sensitivity (Figure 4A and unpublished data). We reasoned that the HU sensitivity of cdc5-2 mutants should be a result of Swe1 accumulation, and this idea was tested by analyzing the HU sensitivity of cdc5-2 Δswe1 double mutants. As shown in Figure 4A, the Δswe1 null mutation completely suppressed the HU sensitivity of cdc5-2 mutants. The differential HU sensitivity of cdc5-1 and cdc5-2 mutants could reflect their distinct defects in Swe1 degradation. It is likely that the cdc5-2 mutant, but not cdc5-1, exhibits compromised Swe1 degradation.
Figure 4.
cdc5-2 mutant is defective in Swe1 degradation and shows Swe1-dependent HU sensitivity. (A) cdc5-2 shows Swe1-dependent HU sensitivity. Saturated cell cultures of WT (Y300), cdc5-2 (241-1-3), and cdc5-2 Δswe1 (477-2-3) were serial diluted 10-fold and then spotted onto either YPD or HU (100 mM) plates. The plates were incubated at 25°C for 3 d. (B) Swe1 protein accumulates in cdc5-2 mutants. SWE1-myc (413-1-1) and cdc5-2 SWE1-myc (478-9-3) strains were arrested in G1-phase at 25°C and released into YPD containing 20 μg/ml nocodazole at 37°C. Protein samples were prepared and analyzed by immunoblotting with anti-myc antibody. The budding index and Swe1 protein levels are shown. (C) Deletion of SWE1 partially suppresses the temperature sensitivity of cdc5-2. Tenfold serial dilutions of saturated WT (Y300), cdc5-2 (241-1-3), and cdc5-2 Δswe1 (477-2-3) cells were spotted onto YPD plates and incubated at various temperatures for 2 d. (D) cdc5-2 Δhsl1 shows synthetic phenotype. Tenfold serial dilutions of saturated WT (Y300), cdc5-2 (241-1-3), Δhsl1 (426-4-3), cdc5-2 Δhsl1 (480-3-1), and cdc5-2 Δhsl1 Δswe1 (481-7-2) cell cultures were spotted onto YPD and HU (100 mM) plates and incubated at 25°C for 3 d.
To clarify the defects of Swe1 protein degradation in cdc5-2 mutants, Swe1 protein levels were examined in cdc5-2 mutants. G1-phase–synchronized WT and cdc5-2 mutant cells were released into YPD medium containing nocodazole and incubated at 37°C. Protein samples were prepared and Swe1 protein levels were examined after Western blot analysis. Swe1 protein appeared at ∼40 min and disappeared after 100 min in WT cells. In cdc5-2 mutants, Swe1 protein existed even after release for 160 min. Moreover, even G1-arrested cdc5-2 mutants exhibited low levels of Swe1 protein, indicating the defects of Swe1 degradation (Figure 4B).
Swe1 degradation is one of the essential functions of Cdc5 kinase (Asano et al., 2005). Because the cdc5-2 mutant accumulates Swe1 protein, the temperature-sensitive phenotype of cdc5-2 mutants could result from defects in Swe1 degradation. If this is true, deletion of SWE1 would suppress or partially suppress the temperature sensitivity of cdc5-2 mutant. Thus the growth of cdc5-2 and cdc5-2 Δswe1 mutants was examined at various temperatures. As expected, cdc5-2 Δswe1 double mutants grew much better than cdc5-2 single mutants at 30 or 33°C, the semipermissive temperature for cdc5-2. However, cdc5-2 Δswe1 failed to grow at 36°C, indicating that the cdc5-2 allele has other defects in addition to Swe1 protein degradation (Figure 4C). Indeed, we have previously demonstrated that cdc5-2 mutants exhibited defects in Bfa1 phosphorylation, which is required for mitotic exit (Hu et al., 2001).
Both Hsl1 and Cdc5 are required for Swe1 protein degradation, and Δhsl1 and cdc5-2 mutants exhibit HU sensitivity. Hsl1 and Cdc5 may function in a single pathway to control Swe1 degradation, or they regulate Swe1 protein levels in parallel pathways. To distinguish these possibilities, the Δhsl1 cdc5-2 double mutant was constructed, and its growth and HU sensitivity were examined. The double mutant displayed poor growth phenotype even when incubated at 25°C, and it was more HU sensitive than each single mutant (Figure 4D). Moreover, the sickness and the HU sensitivity of Δhsl1 cdc5-2 double mutants were suppressed by deleting SWE1 gene. Because Δhsl1 cdc5-2 double mutants exhibit a more severe phenotype than the single mutants, it is likely that Hsl1 and Cdc5 act in different pathways to control Swe1 protein degradation.
Swe1 Accumulation in Δhsl1 Mutant Causes a Slow Recovery from S-phase Arrest
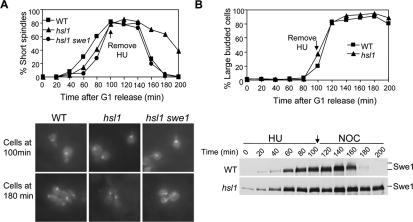
Next we asked why Δhsl1 and Δhsl7 mutants show Swe1-dependent HU sensitivity. One possibility is that Δhsl1 and Δhsl7 mutants fail to arrest the cell cycle in the presence of HU. To test this, we examined the spindle morphology of WT and Δhsl1 mutant cells in the presence of 200 mM HU. G1-synchronized WT and Δhsl1 mutant cells were released into YPD medium containing 200 mM HU and incubated at 30°C. The spindle staining of the collected cells revealed that both WT and Δhsl1 mutant cells exhibited short spindle structure even after incubation for 3 h (unpublished data). The typical S-phase spindle structures in Δhsl1 mutant cells in the presence of HU suggests the intact S-phase checkpoint function (Alcasabas et al., 2001). Moreover, no obvious loss of viability was observed for Δhsl1 mutant cells after incubation in the presence of 200 mM HU for 6 h.
As our result indicates no checkpoint defects in Δhsl1 mutants, we then tested the possibility that persistent Swe1 protein levels prevent cell cycle progression after the removal of HU. Recent data indicate that the degradation of Wee1 protein, the Swe1 homologue in higher eukaryotes, is required to resume cell cycle progression after removal of genotoxic agents (van Vugt et al., 2004; Yamada et al., 2004). It is possible that budding yeast shares the same regulation. To test this possibility, G1-arrested WT, Δhsl1, and Δhsl1 Δswe1 mutant cells were released into YPD medium containing 200 mM HU. After incubation in the presence of HU for 100 min, HU was washed off and the cells were resuspended in YPD medium. α-factor was then added into the medium to block the second round of the cell cycle. Cells were collected and fixed for spindle staining and budding index. In the presence of HU, WT, Δhsl1, and Δhsl1 Δswe1 were arrested in S-phase with short spindle structures (Figure 5A). Sixty minutes after HU removal, the short spindle structure disappeared in majority of WT cells, suggesting that cells entered anaphase. However, it took Δhsl1 mutant cells much longer to enter anaphase after HU removal. One hundred minutes after HU removal, more than 40% of Δhsl1 mutant cells still exhibited short spindle structure. Strikingly, Δswe1 deletion completely suppressed the delay of anaphase entry in Δhsl1 mutants after HU removal (Figure 5A), indicating that Swe1-dependent delay of cell cycle resumption after HU removal contributes to the HU sensitivity of Δhsl1 and Δhsl7 mutants.
Figure 5.
Compromised Swe1 degradation in Δhsl1 causes slow recovery from S-phase arrest. (A) Δhsl1 mutant shows a delayed mitotic entry after HU treatment. G1-arrested WT, Δhsl1, and Δhsl1 Δswe1 mutant cells (Y300, 426-4-3, and 426-2-3) were released into YPD medium containing 200 mM HU at 30°C for 100 min. HU was then washed off by centrifugation and released into YPD medium containing α-factor in order to block the second round of cell cycle. Cells were collected every 20 min and fixed with 3.7% formaldehyde for spindle staining. The percentage of short spindles is shown in the top panel. The bottom panel exhibits the spindle structure of the cells with indicated genotype at 100 and 180 min. (B) Δhsl1 mutant shows delayed Swe1 degradation. G1-arrested WT (413-1-1) and Δhsl1 mutant (433-3-2) cells with myc-tagged SWE1 were released into YPD medium containing 200 mM HU and incubated at 30°C for 100 min. Then HU was washed off by centrifugation and the cells were released into YPD medium containing 20 μg/ml nocodazole. Protein samples were prepared at 20 min intervals and analyzed by immunoblotting with anti-myc antibody. The budding index and Swe1 protein levels are shown in the top and bottom panel respectively.
To further examine if the delayed entry into mitosis in Δhsl1 mutants correlates with Swe1 accumulation, Swe1 protein levels were examined in WT and Δhsl1 mutants using a protocol similar to the one mentioned above. Briefly, G1-synchronized cells were released into YPD medium containing 200 mM HU for 100 min. After HU was washed off, the cells were released into YPD medium containing 20 μg/ml nocodazole to block the cell cycle at metaphase when Swe1 protein levels are low. In WT cells, Swe1 protein disappeared 60 min after HU removal, consistent with the timing of anaphase entry. However, Swe1 protein levels were persistent in Δhsl1 mutant cells even at the 200-min time point when HU was removed for 100 min (Figure 5B). Thus Δhsl1 mutant exhibits dramatically delayed disappearance of Swe1 after HU treatment compared with WT cells. This observation suggests that the accumulation of Swe1 protein in the Δhsl1 mutant is responsible for slower recovery from HU-induced S-phase arrest.
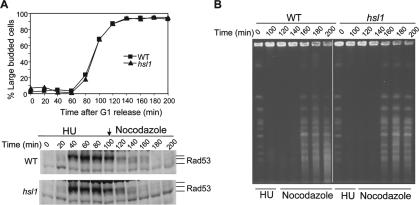
The Slow Recovery in the Δhsl1 Mutant Is Not a Result of Accumulated DNA Damage
It has been shown that deletion of CLB5 is lethal in the absence of RAD53 due to the accumulated DNA damage in Δclb5 mutants (Gibson et al., 2004). This observation raises a possibility that the slow recovery in Δhsl1 mutants could be a result of accumulation of damaged DNA due to the inhibition of Clb5-Cdk1 by Swe1. If that is the case, damaged DNA in Δhsl1 mutants should activate the DNA damage checkpoint pathway and delay recovery from S-phase arrest. Thus the existence of damaged DNA was examined in Δhsl1 mutants by analyzing the phosphorylation of Rad53, a key player in both S-phase and DNA damage checkpoint pathways. Rad53 is phosphorylated by Mec1 kinase in response to DNA damage or incomplete DNA synthesis (Sanchez et al., 1996). The phosphorylation of Rad53 has been widely used as a marker for checkpoint activation. RAD53-HA and Δhsl1 RAD53-HA strains were synchronized at G1-phase and then released into YPD medium containing 200 mM HU. One hundred minutes later, HU was washed off and the cells were resuspended in YPD medium containing nocodazole as described earlier. Protein samples were prepared to examine the phosphorylation of Rad53. As shown in Figure 5A, both WT and Δhsl1 mutant cells exhibited phosphorylated Rad53-HA in the presence of HU, as judged by a protein band shift. After the removal of HU, the phosphorylated Rad53 protein disappeared in ∼60 min in WT cells. Both WT and Δhsl1 mutant cells exhibited similar kinetics for the disappearance of phosphorylated Rad53 (Figure 6A). Thus the slow recovery phenotype in Δhsl1 mutants is not due to the presence of DNA damage.
Figure 6.
The delayed recovery in Δhsl1 mutant after HU challenge is not due to DNA damage accumulation or slowed DNA synthesis. (A) The slow recovery in Δhsl1 mutant is not caused by the accumulation of DNA damage. WT (YYW82-1) and Δhsl1 mutant (429-4-3) cells with HA-tagged Rad53 were synchronized at G1-phase and then released into 30°C YPD medium containing 200 mM HU for 100 min. Then HU was washed off and the cells were resuspended in YPD medium containing 20 μg/ml nocodazole. Cells were collected every 20 min for protein preparation. The budding index and Rad53 protein levels are shown at the top and bottom panels, respectively. (B) Δhsl1 mutant and WT cells complete DNA synthesis with the same kinetics after HU challenge. G1-synchronized WT (Y300) and Δhsl1 (426-4-3) were treated as described in Figure 5A. Cells were collected every 20 min for chromosome preparation and the chromosome DNA was separated by PFGE.
The Slow Recovery in Δhsl1 Mutants Is Not a Result of Delayed DNA Synthesis
The dramatically delayed recovery in Δhsl1 mutants after exposure to HU could be a result of slowed DNA synthesis due to the high levels of Swe1. If so, we expect to observe delayed completion of DNA synthesis in Δhsl1 mutants after they are challenged by HU. Completion of chromosome replication allows chromosomes to be resolved by PFGE. Before completion of DNA replication, chromosomes contain structures such as replication forks and bubbles that prevent migration of chromosomal DNA into the gel (Desany et al., 1998; Schollaert et al., 2004). Thus we used PFGE to follow the completion of DNA synthesis in WT and Δhsl1 mutant cells after HU treatment.
Cells synchronized in G1 were released into the cell cycle in medium containing 200 mM HU. After 100 min, the cells were washed and resuspended in medium containing nocodazole as described earlier. The chromosomes from cells collected during and after HU treatment were separated by PFGE. As shown in Figure 6B, WT cells recovered from transient exposure to 200 mM HU and completed DNA replication, as evidenced by the appearance of the chromosomes signal after 160 min. Surprisingly Δhsl1 mutant cells completed DNA replication at the same time (160 min) under these conditions (Figure 6B). Therefore, the delayed recovery in Δhsl1 mutants is not a result of slow DNA synthesis. In another word, Swe1 accumulation does not affect DNA synthesis. Indeed, we have found that overexpression of Swe1 results in the cell cycle arrest and hyphal growth, but the S-phase progression was not affected by high dosage of Swe1 based on the FACS analysis (unpublished data).
Clb2-Cdk1 Might Be Down-regulated in Δhsl1 and Δhsl7 Mutants
If the failure of the growth of Δhsl1 and Δhsl7 on HU plates is the result of inhibition of Clb-Cdk1, overexpression of B-type cyclins may suppress the HU sensitivity of Δhsl1 and Δhsl7 mutants. Thus a PGAL-CLB5, PGAL-CLB2, and a control vector were transformed into Δhsl7 mutants, and the growth of the transformants was examined on URA dropout plates containing galactose and 100 mM HU. Overexpression of either CLB5 or CLB2 suppressed the HU sensitivity of Δhsl7 mutants, but overexpression of CLB2 exhibited a more dramatic suppression phenotype (Figure 7A). This result indicates that Clb2-Cdk1 might be the physiological target of Swe1 and overproduction of Clb2 reverses the inhibition of Clb2-Cdk1 by Swe1.
Figure 7.
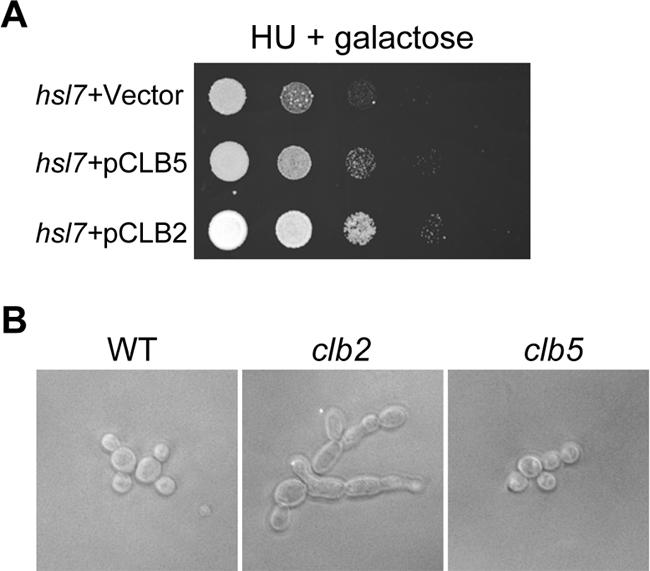
(A) Overexpression of CLB2 suppresses the HU sensitivity of Δhsl7 mutants. Ten-time serial dilutions of saturated cell cultures with indicated genotype were spotted onto a URA3 dropout plate containing galactose and 100 mM HU. The picture was taken after incubated at 25°C for 3 d. (B) Δclb2 deletion mutants exhibit obvious hyphal growth in the presence of HU. WT, Δclb2, and Δclb5 cells were incubated in YPD medium containing 200 mM HU for 3 h at 25°C before taking the picture.
One function of B-type cyclins in yeast is to suppress G1 cyclin–related activities. It is the G1 cyclins that induce polarized growth. When B-type cyclins appear later in the cell cycle, they induce a switch from polarized to isotropic bud growth (Lew and Reed, 1993; Kellogg and Murray, 1995). Thus we speculate that Swe1-dependent hyphal growth in the presence of HU should be a result of inhibition of Clb-Cdk1. If that is the case, deletion of B-type cyclins should promote hyphal growth in the presence of HU. Therefore we examined the cell morphology of all the B-type cyclin deletion mutants (from ATCC, Manassas, VA) in the presence of 200 mM HU. Among all the B-type cyclin deletion mutants (Δclb1, Δclb2, Δclb3, Δclb4, Δclb5, and Δclb6), only the Δclb2 mutant exhibited dramatic hyphal growth (Figure 7B). This observation suggests that the inhibition of Clb2-Cdk1, but not other B-type cyclin-associated Cdk1, contributes to the hyphal growth phenotype in the presence of HU.
Clb5-Cdk and Clb2-Cdk1 Exhibit Differential Sensitivity to Swe1
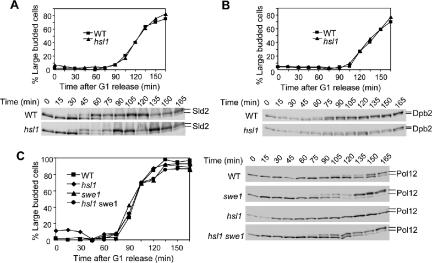
It is well established that Swe1 phosphorylates Cdk1 and inhibits its kinase activity, but our observation indicates that accumulated Swe1 does not affect DNA synthesis (Figure 6). Moreover, overexpression of CLB2 suppresses the HU sensitivity of Δhsl7 better than CLB5. Clb5 and Clb6 are S-phase cyclins and Clb5-Cdk1 plays a major role in DNA synthesis. One obvious question is whether S-phase cyclin-conjugated Cdk1 is inhibited by accumulated Swe1 during S-phase. Recently, the Morgan laboratory compared the specificity of the S-phase cyclin Clb5 and the M-phase cyclin Clb2 in the phosphorylation of 150 Cdk1 substrates. Some of these proteins were phosphorylated more efficiently by Clb5-Cdk1 than Clb2-Cdk1. Sld2 is one of the Clb5-specific targets and it is involved in early S-phase events (Loog and Morgan, 2005). Sld2/Drc1 is required for DNA replication and plays a role in checkpoint signaling of stalled replication forks (Wang and Elledge, 1999). Phosphorylation of Sld2 by Clb5-Cdk1 promotes its association with Dpb11 and the subsequent loading of Cdc45 and DNA polymerase (Masumoto et al., 2002). So we first examined if the phosphorylation of Sld2 is delayed in Δhsl1 mutants.
To examine the effect of Swe1 on Sld2 phosphorylation, G1-arrested WT and Δhsl1 mutant cells with 9myc-tagged SLD2 were released into YPD medium containing 50 mM HU and 20 μg/ml nocodazole at 25°C. Protein samples were prepared every 15 min, and Sld2 protein phosphorylation was examined after Western blot analysis. At 30 min after G1 release, both WT and Δhsl1 mutant cells exhibited a slow migrating band of Sld2, indicating that Sld2 is phosphorylated by Clb5-Cdk1 at this time. Clearly, there is no any delay in Sld2 phosphorylation in Δhsl1 mutants (Figure 8A). Because we have shown that Swe1 accumulates in the presence of 50 mM HU (Figure 1), it is very likely that Clb5-Cdk1 is not inhibited by the presence of Swe1.
Figure 8.
Phosphorylation of Cdk1 substrates shows differential sensitivity to high levels of Swe1 protein. (A) Sld2 exhibits similar phosphorylation kinetics in WT and Δhsl1 mutants in the presence of 50 mM HU. WT and Δhsl1 mutant cells with myc-tagged SLD2 were arrested in G1-phase and then released into 25°C YPD medium containing 50 mM HU and 20 μg/ml nocodazole. Protein samples were prepared every 15 min. Anti-myc antibody was used to detect Sld2 protein. The top panel shows the budding index and the phosphorylation of Sld2 protein is indicated at the bottom panel. (B) Dpb2 shows delayed phosphorylation kinetics in Δhsl1 mutants in the presence of 50 mM HU. Strain TAY179 (DPB2-myc) and 508-3-2 (Δhsl1 DPB2-myc) were synchronized in G1 and treated as described in Figure 8A. The top panel shows the budding index and the bottom panel shows the phosphorylation of Dpb2 based on band shift. (C) Elevated Swe1 blocks Pol12 phosphorylation. WT, Δswe1, Δhsl1, and Δhsl1 Δswe1 mutant cells were synchronized in G1-phase and then treated as described in A. The phosphorylation of Pol12 was determined by Western blot analysis with anti-Pol12 antibody.
In addition to Sld2, Dpb2, and Pol12 have been demonstrated to be Cdk1 substrates and they are also involved in DNA replication (Desdouets et al., 1998; Masumoto et al., 2002; Kesti et al., 2004). Like Sld2, Dpb2 is phosphorylated more efficiently by Clb5-Cdk1 than Clb2-Cdk1, whereas Pol12 is not a Clb5-Cdk1–specific target (Loog and Morgan, 2005). Because Clb2-Cdk1 possesses higher intrinsic kinase activity than Clb5-Cdk1 (Loog and Morgan, 2005), we speculate that Pol12 is more efficiently phosphorylated by Clb2-Cdk1. To determine if the phosphorylation of Pol12 and Dpb2 is inhibited by Swe1 accumulation, WT cells, Δhsl1, DBP2-myc, and Δhsl1 DPB2-myc were synchronized at G1-phase and then released into YPD medium containing 50 mM HU and 20 μg/ml nocodazole as described in Figure 8A. For WT cells, the phosphorylated Dpb2 appeared at 45 min, and the majority of Dpb2 exhibited as hyperphosphorylated forms at 105 min after G1 release (Figure 8B). In Δhsl1 mutants, phosphorylated Dpb2 did not appear until 60 min after G1 release and the majority of Dpb2 became phosphorylated 150 min after G1 release. Moreover, at 75 min, WT cells exhibited equal amount of hypo- and hyperphosphorylated Dpb2 protein, but it took 105 min for Δhsl1 mutants to show a similar ratio (Figure 8B). It is clear that the phosphorylation of Dpb2 by Cdk1 is delayed, but it could be phosphorylated eventually.
The kinetics of Pol12 phosphorylation was also examined in WT and Δhsl1 mutants with anti-Pol12 antibody (Piatti et al., 1996). In WT cells, the phosphorylated Pol12 appeared at 105 min after G1 release, much later than Sld2 and Dpb2 (Figure 8C). However, obvious phosphorylation of Pol12 was not observed even at the last time point in Δhsl1 mutants in the presence of 50 mM HU (180 min; Figure 8C), indicating that HU treatment blocks Pol12 phosphorylation completely. We then asked if the delayed Pol12 phosphorylation depends on Swe1 accumulation. For this purpose, Δswe1 and Δswe1 Δhsl1 mutant cells were synchronized at G1-phase and then released into YPD medium containing 50 mM HU and 20 μg/ml nocodazole at 25°C. As shown in Figure 8C, the phosphorylated Pol12 appeared at 75 min in Δswe1 mutant cells, 30 min earlier than WT cells. Moreover, we did not observed any difference between Δswe1 and Δswe1 Δhsl1 mutants regarding to the kinetics of Pol12 phosphorylation (Figure 8C). These observations indicate that both HU treatment and Δhsl1 deletion contribute to delayed Pol12 phosphorylation, and this delay is a result of Swe1 accumulation.
It is obvious that accumulated Swe1 protein delays the phosphorylation of some Cdk1 substrates, but different Cdk1 substrates exhibit a distinct response to the accumulation of Swe1. We noticed that the timing of the phosphorylation of Sld2, Dpb2, and Pol12 during the cell cycle was different. In the presence of 50 mM HU, WT cells showed phosphorylated Sld2, Dpb2, and Pol12 at 30, 45, and 120 min, respectively (Figure 8). It seems that proteins that are phosphorylated later in the cell cycle are more sensitive to Swe1 accumulation. Moreover, Sld2 and Dpb2 are two Clb5-Cdk1–specific substrates; thus the accumulation of Swe1 only inhibits Clb2-Cdk1, but not Clb5-Cdk1.
DISCUSSION
Here we described the regulation and function of Swe1 during S-phase progression in the budding yeast S. cerevisiae. Because Swe1 phosphorylates Cdk1 and inhibits its kinase activity, it could be a target of checkpoints used to inhibit Clb-Cdk1–associated activities, including DNA replication and chromosome segregation. We have shown that budding yeast cells accumulate Swe1 during S-phase, and the treatment of yeast cells with DNA synthesis inhibitor, HU, leads to Swe1 accumulation. Surprisingly, Swe1 accumulation is independent of the S-phase checkpoint. Δhsl1 and Δhsl7 mutants, which are unable to destroy Swe1, exhibited HU sensitivity because of the failure of the resumption of cell cycle after HU treatment. We further demonstrated that the delayed recovery after HU treatment in Δhsl1 and Δhsl7 mutants is not a result of slowed DNA synthesis or DNA damage accumulation. Interestingly, the kinase activity of Clb2-Cdk1, but not Clb5-Cdk1, is inhibited in the presence of high levels of Swe1 protein. Therefore, the presence of Swe1 during S-phase inhibits Clb2-Cdk1–associated mitotic activities. On the other hand, the destruction of Swe1 after DNA synthesis is required to initiate mitosis.
In the presence of HU, activated S-phase checkpoint arrests cells at S-phase, inhibits late origin firing, and stabilizes replication forks (Osborn et al., 2002). We demonstrated that yeast cells treated with HU maintain high levels of Swe1. To our surprise, mec1-1 and Δrad53 mutants also exhibit persistent levels of Swe1 protein in the presence of HU, arguing against the possibility that the activated S-phase checkpoint leads to Swe1 accumulation. In contrary to previous data, in which HU-induced hyphal growth is suppressed by the S-phase checkpoint (Jiang and Kang, 2003), we found that Δrad53 deletion mutants exhibit obvious hyphal growth when incubated in liquid medium containing HU (Figure 1C), further supporting the notion that Swe1 accumulates in the absence of the S-phase checkpoint. The difference could be due to the different incubation conditions. We examined the morphology of cells grown in liquid YPD medium containing 200 mM HU, whereas the previous work was done with YPD plates containing 100 mM HU.
The degradation of Swe1 requires protein kinases Cdk1, Cdc5, and Cla4. Hsl1 and Hsl7 complex provides a platform at the bud neck for Swe1 degradation (Asano et al., 2005). It has been shown that the Swe1 protein level is elevated in Δcdc55 mutants, and this is consistent with the observation that Δcdc55 mutants exhibit Swe1-dependent hyphal growth (Yang et al., 2000). We found that Δhsl1, Δhsl7 mutants, but not Δcla4 and Δcdc55, showed Swe1-dependent HU sensitivity. Thus other defects than Swe1 degradation in Δcla4 and Δcdc55 mutants may also contribute to their HU sensitivity. Indeed, Cla4 is required for mitotic exit, and Cdc55 plays a role in mitotic exit and sister chromatid separation (Wang and Ng, 2006). Moreover, recent research already showed that the inhibition of PP2A impairs DNA repair and cells are hypersensitive to DNA damage (Chowdhury et al., 2005), suggesting that HU sensitivity of Δcdc55 may be due to defects in DNA repair. It will be interesting to understand the mechanism of Swe1-independent HU sensitivity of Δcla4 and Δcdc55 mutants.
Different from Δhsl1 and Δhsl7, cdc5-2 mutant exhibited Swe1-dependent HU sensitivity without any obvious hyphal growth phenotype (unpublished data). This observation indicates that Swe1-induced hyphal growth in Δhsl1 and Δhsl7 mutants is not the cause of their HU sensitivity. Instead, the elevated Swe1 protein levels in Δhsl1 or Δhsl7 mutants result in both HU sensitivity and hyphal growth. It is possible that the hyphal growth phenotype is suppressed in cdc5-2 mutants. For example, the defect of cdc5-2 in anaphase entry and mitotic exit may lead to higher Clb2 levels that inhibit hyphal growth (Alexandru et al., 2001; Hu et al., 2001).
It is not clear how HU-induced S-phase arrest leads to Swe1 accumulation. One possibility is that SWE1 is transcribed during S-phase, and HU arrests cells at S-phase, which contributes to consistent SWE1 transcription. Alternatively, the presence of HU may induce Swe1 accumulation through inhibiting Swe1 degradation machinery. HU-arrested S-phase cells exhibit bud-neck localized Hsl7 protein, indicating that the Hsl7 pathway is functional in the presence of HU. Moreover, the inactivation of Cdk1 may not be the reason for Swe1 accumulation, because the CDC28F19 mutant, which is resistant to the inhibitory phosphorylation of Swe1, also exhibits comparable Swe1 protein levels in the presence of HU. The inhibition of Cdc5 kinase activity in HU-treated cells could contribute to Swe1 stabilization, based on the following observations. First, cdc5-2 shows Swe1-dependent HU sensitivity, and Swe1 protein degradation is compromised in cdc5-2 mutants. Moreover, Cdc5 kinase activity is largely inhibited in the presence of HU because there is little phosphorylation of its substrate Bfa1 (Hu et al., 2001). The fact that treatment of Cdc5 protein with phosphatase abolishes its kinase activity indicates the presence of a protein kinase responsible for the phosphorylation and activation of Cdc5 in budding yeast (Cheng et al., 1998). Recent data from fission yeast indicate that a protein kinase phosphorylates Plo1, the yeast Cdc5 homologue, at Ser 402 and this phosphorylation is required for recovery after stress (Petersen and Hagan, 2005). Thus, to identify the protein kinase that phosphorylates Cdc5 will be the key to understand the regulation Swe1 protein levels in response to DNA synthesis block.
We have demonstrated that Swe1 degradation is required for the recovery from HU-induced S-phase arrest. Similarly, destruction of Wee1 has been shown to be required to resume the cell cycle in vertebrate cells after challenge by DNA damage or DNA synthesis interruption. Plk1, the Cdc5 homologue in mammals, is required for the recovery after DNA damage (van Vugt and Medema, 2004). In Xenopus, it has been demonstrated that Wee1 exhibits Hsl7-dependent accumulation after DNA synthesis inhibition by HU (Yamada et al., 2004). It remains to be tested if degradation of Wee1 is required for the recovery after DNA synthesis block in mammals. It is likely that all eukaryotic cells share a similar mechanism to recover from the cell cycle arrest after stress. This process might be achieved by activating Cdc5 kinase in budding yeast or its homologues in other eukaryotic organisms.
What is the biological significance of Swe1 accumulation in response to interrupted DNA synthesis? One possibility is that the presence of Swe1 in the presence of HU inhibits DNA synthesis. For example, Clb5-Cdk1 might be inhibited so that the later origin firing is blocked. But our observations argue against this model. First, the phosphorylation of Sld2/Drc1, one of the Clb5-Cdk1 specific substrate, exhibits similar kinetics in WT and Δhsl1 mutants, even though there is much more Swe1 protein in Δhsl1 mutants. Second, after exposed with HU, Δhsl1 mutants complete DNA synthesis without any obvious delay compared with WT cells, indicating that the inhibition of late origin firing after S-phase checkpoint activation in the presence of HU is not dependent on Swe1. Thus, the delayed DNA synthesis does not contribute to the slow recovery in Δhsl1 mutants. We also clarified that the slow recovery phenotype in Δhsl1 mutants is not a result of DNA damage accumulation.
We compared the phosphorylation kinetics of three Cdk1 substrates, Sld2/Drc1, Dpb2 and Pol12, in response to Swe1 accumulation. Surprisingly, the three Cdk1 substrates showed differential sensitivity to high Swe1 protein levels. As shown in Figure 8, the phosphorylation of Sld2/Drc1 does not show any sensitivity to Swe1 accumulation. In contrast, Pol12 exhibits the most dramatic sensitivity to Swe1 accumulation. As Sld2/Drc1 is a specific substrate of Clb5-Cdk1, whereas Pol12 is a Clb2-Cdk1 substrate, we therefore conclude that Clb2-Cdk1 is more sensitive to Swe1 than Clb5-Cdk1. In agreement with this, we found that deletion of CLB2, but not other B-type cyclins, results in dramatic hyphal growth in the presence of HU (Figure 7), indicating that Swe1 induced hyphal growth is due to the inhibition of Clb2-Cdk1. Recent published data from Aparicio's lab confirm this speculation. The in vitro kinase activity of Clb2-Cdk1 was inhibited dramatically in the presence Swe1, but Clb5-Cdk1 was unaffected (Hu and Aparicio, 2005). Thus, this conclusion can well explain the observation that high levels of Swe1 during S-phase do not affect Clb5-Cdk1–dependent S-phase progression.
Our results suggest that Swe1 prevents Clb2-associated mitotic activities during S-phase. If that is the case, deletion of SWE1 should exhibit premature mitotic activities in S-phase–blocked cells. We did observed earlier Pol12 phosphorylation in swe1 deletion mutants, indicating the premature activation of Clb2-Cdk1. However, the Δswe1 mutant does not show any obvious HU sensitivity. It is likely that the periodic expression of mitotic cyclins limits their activity during S-phase and makes Swe1 dispensable for S-phase regulation. Therefore, both Swe1 and lower transcription levels of CLB2 gene in S-phase may prevent any mitotic activities stimulated by Clb2-Cdk1. It is also possible that other unidentified pathways keep Clb2-Cdk1 inactive in S-phase–blocked cells.
In summary, we have identified a new type regulation in response to DNA synthesis block in budding yeast. Swe1 accumulates when DNA synthesis is interrupted, and degradation of Swe1 is required for the resumption of cell cycle after the removal of DNA replication stress. This accumulation may be a result of the inhibition of Cdc5 kinase activity. Accumulated Swe1 prevent Clb2-Cdk1 activation, but has no effect on S-phase cyclin-associated Cdk1 activity. This regulation is conserved from yeast to human cells, and studies on the regulation and function of Swe1 in response to interrupted DNA synthesis will help us understand how eukaryotic cells maintain the genome integrity when genotoxic agents are present.
ACKNOWLEDGMENTS
We thank Drs. Elledge, Lew, Thorner, Wittenberg, Araki, Ganjun, and Foiani for research materials and Dr. Gunjan for critical reading of the manuscript. We also thank Tuen-Yung Ng for constructing RAD53-HA strain. This work was supported by James and Esther King Biomedical Research Program (04NIR13) from Florida State Department of Health and an American Heart Association Scientist Development grant to Y. W.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1093/mbc.E05-11-1093) on March 29, 2006.
REFERENCES
- Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., Bousset K., Furuya K., Diffley J. F., Carr A. M., Elledge S. J. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Alexandru G., Uhlmann F., Mechtler K., Poupart M. A., Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Amon A., Surana U., Muroff I., Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- Asano S., Park J. E., Sakchaisri K., Yu L. R., Song S., Supavilai P., Veenstra T. D., Lee K. S. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 2005;24:2194–2204. doi: 10.1038/sj.emboj.7600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R. N., Deshaies R. J., Kirschner M. W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Hunke L., Hardy C. F. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol. Cell. Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Keogh M. C., Ishii H., Peterson C. L., Buratowski S., Lieberman J. Gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Cid U. J., Shulewitz M. J., McDonald K. L., Thorner J. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol. Biol. Cell. 2001;12:1645–1669. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker J. H., Piatti S., Santocanale C., Nasmyth K., Diffley J. F. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Diffley J. F., Nasmyth K. A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouets C., Santocanale C., Drury L. S., Perkins G., Foiani M., Plevani P., Diffley J. F. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase alpha. EMBO J. 1998;17:4139–4146. doi: 10.1093/emboj/17.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A. D., Raghuraman M. K., Friedman K. L., Cross F. R., Brewer B. J., Fangman W. L. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Foiani M., Liberi G., Lucchini G., Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol. Cell. Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Aparicio J. G., Hu F., Aparicio O. M. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:10208–10222. doi: 10.1128/MCB.24.23.10208-10222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S. L., Charlet A., Haas W., Gygi S. P., Kellogg D. R. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–420. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Hu F., Aparicio O. M. Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2005;102:8910–8915. doi: 10.1073/pnas.0406987102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S. J. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Jiang Y. W., Kang C. M. Induction of S. cerevisiae filamentous differentiation by slowed DNA synthesis involves Mec1, Rad53 and Swe1 checkpoint proteins. Mol. Biol. Cell. 2003;14:5116–5124. doi: 10.1091/mbc.E03-06-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D. R., Murray A. W. NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. J. Cell Biol. 1995;130:675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T., McDonald W. H., Yates J. R., 3rd, Wittenberg C. Cell cycle-dependent phosphorylation of the DNA polymerase epsilon subunit, Dpb2, by the Cdc28 cyclin-dependent protein kinase. J. Biol. Chem. 2004;279:14245–14255. doi: 10.1074/jbc.M313289200. [DOI] [PubMed] [Google Scholar]
- Krishnan V., Nirantar S., Crasta K., Cheng A. Y., Surana U. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell. 2004;16:687–700. doi: 10.1016/j.molcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Kuhne C., Linder P. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 1993;12:3437–3447. doi: 10.1002/j.1460-2075.1993.tb06018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Theesfeld C. L., McMillan J. N., Weaver E., Pringle J. R., Lew D. J. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog M., Morgan D. O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C. S., Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Ma X. J., Lu Q., Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Muramatsu S., Kamimura Y., Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- McMillan J. N., Longtine M. S., Sia R. A., Theesfeld C. L., Bardes E. S., Pringle J. R., Lew D. J. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W. M., Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- O'Connell M. J., Raleigh J. M., Verkade H. M., Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J., Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- Petersen J., Hagan I. M. Polo kinase links the stress pathway to cell cycle control and tip growth in fission yeast. Nature. 2005;435:507–512. doi: 10.1038/nature03590. [DOI] [PubMed] [Google Scholar]
- Piatti S., Bohm T., Cocker J. H., Diffley J. F., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Raleigh J. M., O'Connell M. J. The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. J. Cell Sci. 2000;113(Pt 10):1727–1736. doi: 10.1242/jcs.113.10.1727. [DOI] [PubMed] [Google Scholar]
- Sakchaisri K., Asano S., Yu L. R., Shulewitz M. J., Park C. J., Park J. E., Cho Y. W., Veenstra T. D., Thorner J., Lee K. S. Coupling morphogenesis to mitotic entry. Proc. Natl. Acad. Sci. USA. 2004;101:4124–4129. doi: 10.1073/pnas.0400641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Desany B. A., Jones W. J., Liu Q., Wang B., Elledge S. J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Schollaert K. L., Poisson J. M., Searle J. S., Schwanekamp J. A., Tomlinson C. R., Sanchez Y. A role for Saccharomyces cerevisiae Chk1p in the response to replication blocks. Mol. Biol. Cell. 2004;15:4051–4063. doi: 10.1091/mbc.E03-11-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulewitz M. J., Inouye C. J., Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Murray A. W. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature. 1992;355:365–368. doi: 10.1038/355365a0. [DOI] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- van Vugt M. A., Bras A., Medema R. H. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- van Vugt M. A., Medema R. H. Checkpoint adaptation and recovery: back with Polo after the break. Cell Cycle. 2004;3:1383–1386. doi: 10.4161/cc.3.11.1248. [DOI] [PubMed] [Google Scholar]
- Wang H., Elledge S. J. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ng T. Y. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:80–89. doi: 10.1091/mbc.E04-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shirogane T., Liu D., Harper J. W., Elledge S. J. Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell. 2003;112:697–709. doi: 10.1016/s0092-8674(03)00121-1. [DOI] [PubMed] [Google Scholar]
- Yamada A., Duffy B., Perry J. A., Kornbluth S. DNA replication checkpoint control of Wee1 stability by vertebrate Hsl7. J. Cell Biol. 2004;167:841–849. doi: 10.1083/jcb.200406048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Jiang W., Gentry M., Hallberg R. L. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol. Cell. Biol. 2000;20:8143–8156. doi: 10.1128/mcb.20.21.8143-8156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]