Abstract
The chromosomal passenger complex (CPC), consisting of the serine/threonine kinase Aurora B, the inner centromere protein INCENP, Survivin, and Borealin/DasraB, has essential functions at the centromere in ensuring correct chromosome alignment and segregation. Despite observations that small interfering RNA-mediated knockdown of any one member of the CPC abolishes localization of the other subunits, it remains unclear how the complex is targeted to the centromere. We have now identified a ternary subcomplex of the CPC comprising Survivin, Borealin, and the N-terminal 58 amino acids of INCENP in vitro and in vivo. This subcomplex was found to be essential and sufficient for targeting to the centromere. Notably, Aurora B kinase, the enzymatic core of the CPC, was not required for centromere localization of the subcomplex. We demonstrate that CPC targeting to the centromere does not depend on CENP-A and hMis12, two core components for kinetochore/centromere assembly, and provide evidence that the CPC may be directed to centromeric DNA directly via the Borealin subunit. Our findings thus establish a functional module within the CPC that assembles on the N terminus of INCENP and controls centromere recruitment.
INTRODUCTION
The correct localization of a kinase is generally considered as crucial for the selection and phosphorylation of a given substrate (Pines, 1999). The chromosomal passenger complex (CPC), consisting of the serine/threonine kinase Aurora B, the inner centromere protein INCENP, Survivin, and Borealin/DasraB, exhibits a highly dynamic localization throughout the cell cycle. During prometaphase and metaphase, the complex localizes to the centromere, transfers to the central spindle upon the onset of anaphase, and finally flanks the midbody during telophase and cytokinesis (Vagnarelli and Earnshaw, 2004). Reflecting this localization, the CPC has an important role in the regulation of chromosome segregation as well as in the control of cytokinesis. Specifically, Aurora B has been shown to be important for the establishment of bipolar attachment of the chromosomes to the mitotic spindle during metaphase and to be required for the formation of the central spindle during anaphase. Consequently, knockdown or inhibition of Aurora B or any of the other subunits of the CPC results in chromosome congression defects, kinetochore–microtubule attachment errors, lagging chromosomes in anaphase, and failure of cytokinesis (Vagnarelli and Earnshaw, 2004; Tanaka, 2005). Consistent with the idea that the localization of the CPC is important for target recognition by its kinase subunit, Aurora B has been demonstrated to phosphorylate proteins at the centromere, such as CENP-A and mitotic centromere-associated kinesin (MCAK) (Zeitlin et al., 2001; Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004), and at the central spindle and midbody, e.g., Mklp1, MgcRacGap, and vimentin (Goto et al., 2003; Minoshima et al., 2003; Guse et al., 2005). Yet, the molecular details of how phosphorylation by Aurora B influences its different substrates and thus regulates the different stages of the cell cycle are still not clear. Understanding how the dynamic localization of the CPC is achieved is therefore important for gaining further insight into the action of Aurora B.
Recently, it has been demonstrated that the relocalization of the complex from the centromere to the central spindle at the metaphase-to-anaphase transition is dependent on the mitotic kinesin Mklp2 (Gruneberg et al., 2004). In budding yeast, the dephosphorylation of INCENP by the Cdc14 phosphatase has been shown to be important for the transfer of INCENP from the centromere to the central spindle (Pereira and Schiebel, 2003), and this might be an additional form of regulation in human cells, too (Gruneberg et al., 2004). However, how the CPC proteins target to the centromere in the first place is unclear. A dependency analysis carried out in Xenopus extracts identified the CPC proteins as the most upstream components required for localizing essential kinetochore and spindle checkpoint proteins. Hence, no component was found that resulted in loss of Aurora B from the centromere when depleted, except the Aurora B binding partner INCENP (Vigneron et al., 2004).
While it has been well established that an interaction of Aurora B with the IN-box, a highly conserved motif within the C terminus of INCENP, is required for full activation of the kinase, a previous analysis of chicken INCENP indicated that the N terminus of the protein might be involved in targeting the CPC to the centromere (Ainsztein et al., 1998; Adams et al., 2000; Bishop and Schumacher, 2002; Bolton et al., 2002; Honda et al., 2003; Yasui et al., 2004; Sessa et al., 2005). However, the molecular basis for this observation and the involvement of the other subunits of the CPC in the localization of the complex is poorly understood. The study of CPC targeting is complicated by the fact that all the subunits of the CPC are interdependent for localization and/or protein stability. Hence, small interfering RNA (siRNA)-mediated knockdown of any component of the CPC leads to loss of the other CPC proteins from the centromere, the central spindle, and the midbody and also results in a significant decrease in the protein levels of one or more subunits of the CPC (Carvalho et al., 2003; Honda et al., 2003; Lens et al., 2003; Vader et al., 2006). This situation makes it difficult to assess individual contributions of the different subunits to the targeting of the complex.
We have now investigated the mode of centromere targeting of the CPC by biochemical in vitro analysis combined with RNA interference (RNAi)-based complementation experiments in vivo. This approach allowed us to evaluate the requirement for the different CPC members for centromere targeting and to define a CPC subcomplex necessary and sufficient for centromere localization.
MATERIALS AND METHODS
Antibodies, Antibody Production, and Aurora B Kinase Inhibition
The antibodies used in this study and their sources are as follows. Rabbit anti-INCENP and rabbit anti-Aurora B have been described previously (Honda et al., 2003); monoclonal anti-Aurora B (AIM1) (BD Biosciences PharMingen, San Diego, CA), monoclonal anti-α-tubulin (DMA1) and monoclonal anti-FLAG M2 antibody (Sigma-Aldrich, St. Louis, MO); rabbit anti-Survivin (Santa Cruz Biotechnology, Santa Cruz, CA) (for immunofluorescence) or ab469 (Abcam, Cambridge, United Kingdom) (for Western blotting); rabbit polyclonal anti-phospho-CENP-A (Ser7) and rabbit polyclonal anti-phospho-Histone H3 (Ser10) (Upstate Biotechnology, Lake Placid, NY); monoclonal anti-CENP-A antibody and monoclonal anti-Myc antibody (9E10) (Abcam); monoclonal anti-BubR1 antibody (Chemicon International, Temecula, CA); and monoclonal anti-Hec1 (GeneTex, San Antonio, TX). Polyclonal sheep anti-green fluorescent protein (GFP) antibody was a kind gift of Dr. Francis Barr (Max-Planck-Institut of Biochemistry, Martinsried, Germany). A rabbit polyclonal antibody against bacterially expressed His-Borealin was raised at Charles River Laboratories (Romans, France). Anti-Myc antibody (9E10) directly coupled to fluorescein isothiocyanate or rhodamine was purchased from Santa Cruz Biotechnology. Secondary antibodies conjugated to horseradish peroxidase, Cy2, and Cy3 were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA).
Aurora B inhibitor ZM447439 was obtained from AstraZeneca (Cancer and Infection Research, Cheshire, United Kingdom) and was used at a concentration of 10 μM.
Directed Yeast Two-Hybrid Assays
Borealin, INCENP, INCENP1-530, INCENP531-789, INCENP790-919, and INCENP59-919 were cloned into the bait vector pFBT9, a version of pGBT9 (Clontech, Mountain View, CA) modified to encode kanamycin resistance (Haas et al., 2005) and the prey vector pACT2 (Clontech), respectively. The corresponding Survivin and Aurora B constructs were a kind gift of Dr. Francis Barr. Cotransformations of bait and prey vector into yeast were tested for their ability to grow on selective medium (quadruple drop-out [QDO]), compared with nonselective medium (−LW).
Cloning, Expression, and Purification of Recombinant Proteins
The cDNAs for Aurora B, Survivin, and INCENP have been described previously (Honda et al., 2003). The image clone for human Borealin IMAGp958PO68Q was purchased from the Deutsches Ressourcenzentrum für Genomforschung (rzpd). The gene was amplified by PCR and cloned into pcDNA3.1-3xMyc. For gene-fusion with the hexa-histidine tag (His-tag), Survivin and Borealin were cloned into pQE vectors (QIAGEN, Hilden, Germany). For fusion to the maltose binding protein (MBP), Borealin and INCENP1-58 were cloned into the pMAL vector (New England Biolabs, Beverly, MA). Glutathione S-transferase (GST) was expressed from the pGEX vector (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and GST-His-Survivin was a gift of Dr. Francis Barr. His-Plk1 purified from insect cells was a gift of Christoph Baumann. MBP-Borealin was expressed by induction with 1 mM isopropyl β-d-thiogalactoside (IPTG) for 4 h at 30°C in Escherichia coli BL21 cells. All other recombinant proteins were expressed by induction with 0.1 mM IPTG at 18°C overnight. His-, GST-, and MBP-fusion proteins were purified according to standard protocols.
In Vitro Binding Assays
Five micrograms of purified His-Borealin, GST-His-Survivin, or 10 μg of purified GST was bound to Ni-NTA agarose (QIAGEN) or glutathione-Sepharose beads (GE Healthcare), respectively, in binding buffer [20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM dithiothreitol (DTT), and 0.1% (vol/vol) Triton X-100] for 2 h at 4°C. After incubation, the beads were washed two times in binding buffer. Binding partners were added at 10 μg/ml (MBP-INCENP1-58 and MBP-Borealin) or 20 μg/ml (MBP), respectively, in binding buffer. Zn2+ was added to a final concentration of 20 mM where indicated. Samples were incubated on a rotating wheel at 4°C for 2 h. The beads were washed three times with binding buffer, boiled in 2× SDS sample buffer, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
For DNA-binding assays, 5 μg of purified protein (Histone H3, His-Survivin, His-Borealin, MBP-Borealin, MBP-INCENP1-58, GST-Cdc20, and His-Plk1) or 10 μg of purified protein (MBP) was added into binding buffer [50 mM Tris-Cl, pH 8.0, 4 mM MgCl2, 1 mM DTT, 150 mM NaCl, and 0.1% (vol/vol) Triton X-100] and bound to calf-thymus double-stranded DNA-cellulose (Sigma-Aldrich) for 2 h at 4°C. Zn2+ was added at 20 mM where indicated. DNA-cellulose was washed three times with binding buffer, boiled in 2× SDS sample buffer, and analyzed by SDS-PAGE.
Coimmunoprecipitation
For coimmunoprecipitation experiments with different GFP-INCENP constructs, HeLa S3 cells in 15-cm dishes were transfected for 36 h and synchronized by aphidicolin release followed by mitotic shake-off to enrich for mitotic cells. Cell pellets were lysed in 500 μl of lysis buffer [50 mM Tris, pH 7.4, 400 mM NaCl, 40 mM β-glycerol phosphate, 10 mM NaF, 0.5% (vol/vol) IGEPAL, 0.1% deoxycholate, 30 μg/ml RNase, 80 U/ml micrococcal nuclease (Sigma-Aldrich), 2 mM Prefabloc, protease inhibitor cocktail tablets (Roche Diagnostics, Mannheim, Germany), 100 μM ATP, 100 μM MgCl2, 100 nM okadaic acid, and 0.3 mM Na-vanadate] for 30 min at 4°C on ice. GFP-tagged proteins were precipitated from the cleared lysate with sheep anti-GFP antibodies. To test for dimerization, HeLa S3 cells were transfected with Flag- and Myc-constructs and processed in a similar way except that the immunoprecipitations were performed with mouse anti-FLAG and rabbit anti-Myc antibodies, respectively.
RNAi and Rescue Experiments
RNAi was performed as described previously (Elbashir et al., 2001) using Oligofectamine (Invitrogen, Carlsbad, CA). To test for siRNA-mediated knockdown of chromosomal passenger proteins, HeLa S3 cells were grown on coverslips in six-well plates and incubated with the respective RNAi oligonucleotide for 36 h. For rescue experiments, RNAi and plasmid transfection using FuGENE (Roche Diagnostics) were performed in parallel. Cells were incubated for 36 h and fixed in 3% paraformaldehyde. Thirty mitotic, nontransfected cells on each coverslip were analyzed for the presence of chromosomal passenger proteins by staining with the appropriate antibodies. Slides were discarded if more than two of these 30 cells showed chromosomal passenger staining, indicating an RNAi efficiency lower than 94%. On coverslips meeting this criterion, between eight and 23 transfected mitotic cells could be analyzed on a single coverslip for staining of the chromosomal passenger proteins or phospho-CENP-A. Each rescue experiment was done in triplicate. To biochemically analyze the potential of GFP-INCENP construct to rescue Aurora B levels in a background free of endogenous INCENP, HeLa S3 cells were treated with siRNA oligonucleotides and transfected with corresponding constructs as described above. Puromycin was added at 2 μg/ml to enrich for transfected cells 24 h before lysate preparation.
The 3′ untranslated region (UTR) regions of the chromosomal passenger protein transcripts were targeted as follows: INCENP, 5′-GGCTTGGCCAGGTGTATATdTdT-3′; Aurora B, 5′-GGAAAGAAGGGATCCCTAAdTdT-3′; Borealin, 5′-AGGTAGAGCTGTCTGTTCAdTdT-3′; Aurora C, 5′-GCTGAATCATTTCATACCAdTdT-3′; and Survivin, Survivin HP validated siRNA 1027400 (catalog no. SI02652958, QIAGEN). CENP-A was targeted with 5′-CTCGTGGTGTGGACTTCAAdTdT-3′ and humanMis12 with 5′-GGACATTTTGATAACCTTTdTdT-3′ (Goshima et al., 2003). All siRNA oligonucleotides were purchased from QIAGEN. The Lamin-A control has been described previously (Elbashir et al., 2001). For the rescue experiments, INCENP wild-type and deletion constructs were cloned into the pEGFP-C2 vector (Clontech) and a modified version of pcDNA4/TO (Invitrogen) encoding the enhanced green fluorescent protein (EGFP)-tag and puromycin resistance (a gift from Dr. Francis Barr). Borealin, Survivin, and INCENPfull-length were cloned into modified versions of pcDNA3.1 (Invitrogen), encoding a 3xMyc or Flag-tag, respectively.
Immunoblotting and Indirect Immunofluorescence
To analyze RNAi efficiency, HeLa S3 cells in six-well dishes were solubilized in hot 2× Laemmli buffer, boiled for 5 min, and resolved by SDS-PAGE followed by Western blotting.
Indirect immunofluorescence was performed as described previously (Gruneberg et al., 2004). Anti-Survivin antibody (Santa Cruz Biotechnology) was used at a dilution of 1:200 and anti-Borealin serum at a dilution of 1:3000. All other antibodies were diluted to 1:1000.
RESULTS
The N-Terminal 58 Amino Acids of Human INCENP Are Sufficient for Centromere Targeting
Analysis of how the CPC targets to the centromere has so far concentrated on INCENP, the binding partner of Aurora B (Adams et al., 2001a). For chicken INCENP, it has been demonstrated that amino acids 1-68 are sufficient to direct the protein to centromeres (Ainsztein et al., 1998). In budding yeast, the removal of cyclin-dependent kinase (Cdk)1 phosphorylation sites from INCENP seems to be important for the transfer of INCENP from the centromere to the central spindle, raising the possibility that Cdk1 phosphorylation might play a role in the centromere targeting of INCENP (Pereira and Schiebel, 2003). Sequence analysis of the first 68 amino acids of human INCENP indicated the presence of a conserved Cdk1-consensus site at position T59 and an in vitro kinase assay showed that T59 is the only Cdk phosphorylation site within INCENP1-68 (our unpublished data). To investigate the targeting of human INCENP to the centromere and the potential role of Cdk1 in this process, constructs were prepared comprising either residues 1-68, residues 1-58 (lacking the potential Cdk1-site) and residues 59-919, all tagged with an N-terminal GFP-tag. In HeLa S3 cells both INCENP1-58 and INCENP1-68 localized to the centromere from prophase to metaphase (Figure 1A; our unpublished data). Conversely, the construct lacking residues 1-58 (INCENP59-919) was dispersed in the cytoplasm and failed to target to the centromere (Figure 1B). Neither INCENP1-58 nor INCENP59-919 transferred to the central spindle or the midbody, and both were cytoplasmic in anaphase and telophase cells (Figure 1, A and B) or associated with the chromatin (our unpublished data). These data indicate that the N-terminal 58 amino acids of the INCENP protein must carry information that determines centromere targeting and that Cdk phosphorylation of INCENP is not required for this process.
Figure 1.
The N terminus of human INCENP (GFP-INCENP1-58) targets to the centromere. HeLa S3 cells were transfected with INCENP1-58 or INCENP59-919 N-terminally fused to GFP. (A) GFP-INCENP1-58 targets to the centromere in prometaphase and metaphase (independently of its expression level) but remains cytoplasmic in anaphase and telophase. (B) A GFP-INCENP construct lacking the first 58 amino acids (GFP-INCENP59-919) does not localize to any defined cellular structure during mitosis. Bar, 10 μm.
Borealin and Survivin Bind to the N Terminus of INCENP In Vitro
To determine whether any of the other chromosomal passenger proteins bind to the N terminus of INCENP and may contribute to the centromere targeting a combination of directed yeast two-hybrid analysis and in vitro binding assays with recombinant proteins was used.
In two-hybrid assays, Aurora B interacted with full-length INCENP (IN1-919) and the C terminus of INCENP (IN790-919) containing the IN-box domain (Figure 2A, I, top and two bottom rows) but not with the coiled-coil domain of INCENP (IN531-789) or its N terminus (IN1-530) (Figure 2A, I, rows 2 and 3). (Note that for the two-hybrid assays, an N-terminal construct of INCENP comprising residues 1-530 had to be used because shorter N-terminal fragments were self-activating.) An interaction between full-length INCENP and Borealin was found (Figure 2A, II, top row), in line with a previous report (Gassmann et al., 2004). This interaction was mapped to the N terminus of INCENP (INCENP1-530; Figure 2A, II, second row from top). Interestingly, depletion of the first 58 amino acids of INCENP abolished this interaction (Figure 2A, II, bottom). No interaction between INCENP and Survivin was observed in the two-hybrid assay (Figure 2A, III). Borealin, in contrast, associated with Survivin (Figure 2A, V). Neither Survivin nor Borealin showed an interaction with Aurora B using this approach (Figure 2A, IV).
Figure 2.
INCENP1-58, Borealin, and Survivin form a complex. (A) pFBT9-Aurora B (I and IV), pFBT9-Borealin (II and V), and pFBT9-Survivin (III) were tested against different fragments of INCENP (I–III) and full-length Borealin or Survivin (IV and V) in pACT2 by two-hybrid analysis. INCENP fragments were created based on a coiled-coil prediction tool (Berger et al., 1995). INCENP1-58 could not be used in this assay because it was self-activating. −LW indicates plates lacking leucine and tryptophane, whereas QDO indicates plates lacking leucine, tryptophane, histidine and adenine. (B) MBP (lanes 1 and 2) or MBP-INCENP1-58 (lanes 3–5) was incubated with His-Borealin immobilized on Ni-NTA agarose (lanes 2 and 4) or Ni-NTA agarose alone (lane 5). Input and protein bound to the beads were analyzed by SDS-PAGE. (C) MBP (lanes 1, 4, 8, and 9), MBP-INCENP1-58 (lanes 2, 5, 6, 10, and 11), and MBP-Borealin (lanes 3, 7, and 12) were incubated with GST (lanes 4–7) or GST-His-Survivin immobilized on glutathione-Sepharose (lanes 8–12). Zn2+ (20 mM) was added to the reactions where indicated (lanes 6, 9, and 11).
To corroborate the yeast two-hybrid observations and explore whether Borealin and Survivin could bind directly to the N-terminal 58 amino acids of INCENP, in vitro binding assays were used. In agreement with the two-hybrid data, an interaction between His-tagged Borealin (His-Borealin) and maltose binding protein-tagged INCENP1-58 (MBP-IN1-58) was observed (Figure 2B, lane 4). Furthermore, both MBP-Borealin and MBP-INCENP1-58 bound to immobilized GST-His-Survivin (Figure 2C, lanes 11 and 12). The interaction between MBP-INCENP1-58 and GST-His-Survivin was only observed in the presence of excess Zn2+ (Figure 2C, compare lanes 10 and 11), whereas the interaction between MBP-Borealin and GST-His-Survivin was not dependent on this (lane 12). This observation is consistent with structural data on the three-dimensional folding and dimerization state of Survivin, which requires Zn2+ (Chantalat et al., 2000; Muchmore et al., 2000; Verdecia et al., 2000). We suppose that within the yeast nucleus the specific folding of Survivin required to interact with INCENP cannot be generated (see yeast two-hybrid data in Figure 2A, column III). In contrast, the interaction between Borealin and Survivin was not affected by Zn2+, as noted previously (Gassmann et al., 2004). These data confirm and extend previously reported interactions between the CPC proteins (Adams et al., 2000; Bishop and Schumacher, 2002; Honda et al., 2003; Gassmann et al., 2004; Sessa et al., 2005). While Aurora B associates with the C terminus of INCENP via the IN-box, Survivin and Borealin interact with the N-terminal 58 amino acids of INCENP and bind to each other.
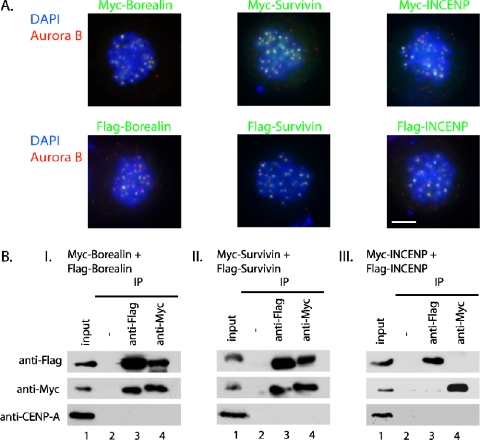
Borealin and Survivin Form Higher Order Structures at the Centromere
The finding that both Borealin and Survivin interact with the first 58 amino acids of INCENP as well as with each other suggests the existence of a heterotrimeric complex assembled on the N terminus of INCENP. Because oligomerization of Survivin as well as Borealin had been observed in vitro (Chantalat et al., 2000; Muchmore et al., 2000; Verdecia et al., 2000; Gassmann et al., 2004), we next asked whether this also occurred in vivo at the centromere. To this end, HeLa S3 cells were cotransfected with Flag- and Myc-tagged constructs of either Survivin, Borealin, or INCENP. The cells were then arrested with nocodazole to enrich for transfected mitotic cells with centromere-localized tagged CPC subunits (Figure 3A). Coimmunoprecipitation of Flag- and Myc-tagged constructs was assessed by Western blotting. Oligomerization was observed for Borealin and Survivin but not INCENP (Figure 3B, compare lanes 3 and 4 in I–III). Based on these experiments, we propose that Survivin and Borealin form higher order structures at the centromere that assemble on the N terminus of INCENP.
Figure 3.
Borealin and Survivin form oligomeres at the centromere. (A) HeLa S3 cells were transiently transfected with the indicated constructs, arrested in prometaphase by treatment with nocodazole for 16 h, and then processed for indirect immunofluorescence analysis using anti-Flag or anti-Myc antibodies (green), respectively, and anti-Aurora B antibody (red). Bar, 10 μm. (B) Myc- or Flag-tagged CPC constructs were cooverexpressed in HeLa S3 cells followed by Myc and Flag pull-downs and Western blotting with anti-Flag or anti-Myc antibodies. Both Borealin and Survivin form oligomeres in this assay (compare lanes 3 and 4 of I, II, and III).
The N Terminus of INCENP Forms a Complex with Survivin and Borealin In Vivo
To verify that the N-terminal domain of INCENP can also interact with Survivin and Borealin in vivo, different GFP-tagged INCENP-constructs were transfected into HeLa S3 cells and immunoprecipitations were performed on mitotic lysates using anti-GFP antibodies. Full-length GFP-INCENP coprecipitated Aurora B, Borealin, and Survivin (Figure 4, lane 1). INCENP-fragments containing the C-terminal IN-box motif of INCENP pulled down Aurora B but not Survivin or Borealin (Figure 4, lanes 5 and 7). In contrast, N-terminal fragments of INCENP containing residues 1-58 precipitated Borealin and Survivin but not Aurora B (lanes 3 and 4), whereas the coiled-coil domain of INCENP lacking both the N and the C terminus did not associate with any of the other CPC components (lane 6). None of the constructs coprecipitated the centromeric protein CENP-A (lanes 1–7, second row from bottom) or the kinetochore protein Hec1 (lanes 1–7, bottom row). Together, these results support data on the interaction between the C terminus of INCENP containing the IN-box motif and Aurora B (Bishop and Schumacher, 2002; Bolton et al., 2002; Honda et al., 2003; Yasui et al., 2004; Sessa et al., 2005) and show that a ternary complex between Borealin, Survivin, and the first 58 amino acids of INCENP (INCENP1-58) exists in vivo.
Figure 4.
INCENP1-58, Borealin, and Survivin form a ternary complex in vivo. (A) Different fragments of INCENP fused to GFP were transfected into HeLa S3 cells and precipitated from mitotic lysates with sheep anti-GFP antibodies. Coprecipitating passenger proteins or the control centromere/kinetochore proteins CENP-A and Hec1 were visualized by Western blotting. Asterisks indicate GFP constructs. The anti-GFP blot was cut into two halves to remove the strong signal of the immunoglobulin heavy chain.
Ectopic GFP-INCENPfull-length Can Replace Endogenous, siRNA-knocked-down INCENP and Restore CPC Function
To examine the physiological relevance of these findings for the targeting of the CPC to the centromere in vivo, an RNAi rescue assay was established. INCENP or Aurora B were effectively knocked down by a 36-h treatment with siRNA oligonucleotides targeting the 3′ UTR of the corresponding transcripts. In line with previous results (Honda et al., 2003; Gassmann et al., 2004; Vader et al., 2006), the knockdown of Aurora B or INCENP resulted in the loss of all other CPC components from the centromere, the central spindle and the midbody (Figure 5A). Furthermore, Western blotting showed that knockdown of INCENP resulted in the simultaneous knockdown of Aurora B and vice versa and strong reduction of Survivin levels, whereas the protein levels of Borealin were less affected (Figure 5B, lanes 2 and 3). Also, knockdown of Borealin did not significantly reduce Aurora B and INCENP levels, consistent with the idea of an Aurora B/INCENP complex devoid of Borealin and Survivin (Gassmann et al., 2004).
Figure 5.
siRNA suppression and simultaneous rescue of INCENP by ectopic expression. (A) HeLa S3 cells were treated with siRNA oligonucleotides targeting the 3′ UTR of INCENP, Aurora B, Borealin, or Survivin transcripts, fixed, and then costained with antibodies against INCENP and Aurora B (INCENP- and Aurora B-RNAi, top) or Borealin and Survivin (Borealin- and Survivin-RNAi, bottom). Mitotic cells are indicated by arrows. Bar, 10 μm. (B) Cell extracts from cells depleted for the different passenger proteins by treatment with corresponding siRNA oligonucleotides for 36 h were analyzed by Western blotting. The blots were reprobed with antibodies against CENP-A to demonstrate equal loading (bottom row). (C) HeLa S3 cells treated with the indicated 3′ UTR siRNA oligonucleotides targeting chromosomal passenger proteins for 36 h were stained with antibodies against Aurora B. One hundred cells were assessed in each case for the presence of Aurora B centromere staining. (D) HeLa S3 cells were treated with 3′ UTR siRNA oligonucleotides targeting INCENP and simultaneously transfected with GFP-INCENPfull-length. Transfected cells were analyzed for the presence of the other chromosomal passenger proteins or phospho-CENP-A staining as a readout for Aurora B activity. All images shown are representatives of three independent experiments. Bar, 10 μm.
To express GFP-tagged INCENP constructs in the absence of the endogenous protein, cells were transfected simultaneously with INCENP siRNA oligonucleotides and the respective rescue constructs (see Materials and Methods for precise protocol). The effectiveness of INCENP knockdown in these experiments was assessed by staining with antibodies against Aurora B and was found to be close to 100% (Figure 5C). The transfection efficiency of the rescue constructs was typically between 10 and 20%. To validate the assay, control rescue experiments using the combination of INCENP-siRNA treatment and transfection with full-length GFP-tagged INCENP were performed. In cells treated with siRNA duplexes targeting endogenous INCENP, the transfected GFP-INCENPfull-length construct efficiently localized to the centromere and restored Borealin, Survivin, and Aurora B staining (Figure 5D). In addition, phospho-CENP-A staining, a marker for Aurora B kinase activity (Zeitlin et al., 2001), was lost completely in INCENP knocked-down cells but restored upon expression of GFP-INCENPfull-length (Figure 5D, bottom, and Supplemental Figure S1). We note that phospho-CENP-A staining was used as a read-out because it was found to be more sensitive to siRNA-mediated knockdown of chromosomal passenger proteins than staining with anti-phospho-histone H3 antibody (Supplemental Figure S1).
Ectopic GFP-INCENP1-58 Can Target Survivin and Borealin but Not Aurora B to the Centromere in the Absence of Endogenous INCENP
Having established the RNAi-rescue assay, this approach was used to assess the ability of GFP-tagged INCENP1-58 to target to the centromere in cells with knocked-down endogenous INCENP. GFP-INCENP1-58 efficiently localized to the centromere in this scenario. The transfected cells were also positive for Survivin and Borealin but, consistent with the coprecipitation data (Figure 4), not for Aurora B nor phospho-CENP-A (Figure 6A). Biochemical analysis of cells treated as described above but enriched for transfected cells by puromycin selection showed that in contrast to GFP-INCENPfull-length and GFP-INCENP59-919 (both containing the Aurora B-binding IN-box), GFP-INCENP1-58 could not rescue the protein levels of Aurora B (Figure 6B, compare lane 5 with lanes 3 and 4) in cells lacking endogenous INCENP. Immunofluorescence analysis showed that RNAi-rescue with GFP-INCENP59-919 resulted in mislocalized, diffuse INCENP staining in mitotic cells (Figure 6C). Interestingly, the rescue of Aurora B protein levels in the absence of correct localization was not sufficient to restore Aurora B function at the centromere/kinetochore as indicated by the absence of phospho-CENP-A staining in these cells (Figure 6B, lane 4, and C). This stresses the importance of the first 58 amino acids of INCENP not only for correct Aurora B localization but also for function of the kinase at the centromere.
Figure 6.
GFP-INCENP1-58 targets to the centromere in the absence of endogenous INCENP and restores Survivin and Borealin but not Aurora B staining. (A) HeLa S3 cells were treated with 3′ UTR siRNA oligonucleotides targeting INCENP and simultaneously transfected with GFP-INCENP1-58. Transfected cells were analyzed for the presence of the other chromosomal passenger proteins or phospho-CENP-A staining as a readout for Aurora B activity. (B) HeLa S3 cells were transfected with the indicated constructs in pcDNA4/TO-EGFP encoding puromycin resistance and treated with the indicated siRNA oligonucleotides for 36 h. Twenty-four hours before harvesting, the cells were treated with 2 μg/ml puromycin. Asterisks indicate GFP constructs. (C) INCENP siRNA-treated HeLa S3 cells transfected with INCENP59-919 were treated as in A and costained for pCENP-A. (D) Recruitment of BubR1 to the kinetochore as a readout for normal chromosomal passenger function was visualized by immunofluorescence in INCENP knockdown cells transfected with either GFP-INCENPfull-length (top) or GFP-INCENP1-58 (bottom). (E) The percentage of transfected cells with properly aligned metaphase plates (left), prometaphase-like chromosome configurations (middle), or with metaphases with uncongressed chromosomes (right) was quantitated in cells treated with 3′ UTR siRNA oligonucleotides against INCENP and transfected with either GFP-INCENPfull-length (gray columns) or GFP-INCENP1-58 (black columns). (F and G) HeLa S3 cells were treated as in A but in addition to INCENP, Survivin (F) or Borealin (G) was simultaneously knocked down. All immunofluorescence images are representatives of three independent experiments.
Together, these results indicate that a complex consisting of INCENP1-58, Survivin, and Borealin is sufficient for centromere targeting, and, remarkably, it does not require the presence of the Aurora B protein. Although Western blotting showed that in INCENP siRNA-treated cells, Aurora B was co-knocked down to undetectable levels (Figure 5B, lane 2), the possibility remained that the above-mentioned observations were due to minimal residual Aurora B in INCENP siRNA-treated cells, which might be sufficient to mediate centromere targeting. To exclude this, INCENP and Aurora B were simultaneously knocked down and GFP-tagged INCENP1-58 was expressed. The additional knockdown of Aurora B did not change the result that INCENP1-58, Borealin, and Survivin could target to the centromere (Supplemental Figure S2, top row).
One protein that could potentially replace Aurora B function is the related Aurora C kinase. Aurora C was shown to localize like a chromosomal passenger protein, but it is expressed at lower levels than Aurora B and might be functionally redundant with Aurora B (Li et al., 2004; Sasai et al., 2004). However, simultaneous knockdown of INCENP and Aurora C or INCENP, Aurora C, and Aurora B followed by rescue with GFP-INCENP1-58 resulted in centromere localization of the ternary INCENP1-58–Survivin–Borealin complex, excluding a compensatory role for Aurora C (Supplemental Figure S2, middle and bottom rows).
Aurora B Kinase Activity Is Not Required for the Maintenance of the CPC at the Centromere
The above-mentioned findings show that at least for the initial targeting of the CPC to the centromere Aurora B kinase activity is not required. However, prolonged high-level expression of kinase-dead Aurora B leads to loss of Aurora B itself and the other chromosomal passengers from the centromere and spreading throughout the chromatin (Ditchfield et al., 2003; Honda et al., 2003). This suggests either a requirement for Aurora B kinase activity for the maintenance of the CPC at the centromere, or, alternatively, that overexpression of the kinase-dead Aurora B exerts a dominant-negative effect on the localization of the CPC. Because the prolonged direct inhibition of Aurora B kinase activity by a chemical inhibitor does not affect the association of Aurora B itself or the other chromosomal passengers with the centromere (Ditchfield et al., 2003) (Supplemental Figure S3), the latter explanation seems more likely.
Ectopic GFP-INCENP1-58 Cannot Functionally Rescue INCENP siRNA Knockdown
Together with the results from the RNAi rescue experiments, these data demonstrate that Aurora B kinase activity is neither required for the initial recruitment of the chromosomal passenger proteins to the centromere nor for the maintenance of the proteins at the centromere but that a complex of INCENP1-58, Survivin, and Borealin is essential for centromere targeting. However, consistent with the well documented requirement for Aurora B kinase for chromosome segregation (Schumacher et al., 1998; Adams et al., 2001b; Giet and Glover, 2001; Ditchfield et al., 2003; Hauf et al., 2003), a functional rescue, as judged by the ability of the transfected cells to recruit the spindle checkpoint component BubR1 (Ditchfield et al., 2003; Hauf et al., 2003) or to form a properly aligned metaphase plate, was not observed in INCENP siRNA-treated cells that had been transfected with GFP-INCENP1-58 (Figure 6, D and E).
The above-mentioned finding that GFP-INCENP1-58 could target to the centromere in the absence of Aurora B raised the possibility that this part of INCENP could localize autonomously. To analyze whether INCENP1-58 might be able to target to the centromere independently of Survivin and Borealin, simultaneous knockdowns of INCENP and Survivin or INCENP and Borealin, respectively, followed by rescue transfections with GFP-INCENP1-58 were performed. In the absence of Survivin (Figure 6F) or Borealin (Figure 6G), GFP-INCENP1-58 was unable to target to the centromere. This provides clear evidence that INCENP1-58 can only target to the centromere in association with its binding partners Survivin and Borealin.
CPC Loading onto the Centromere Is Independent of CENP-A and hMis12
Two major branches of centromere/kinetochore assembly depending on the core centromere proteins CENP-A and hMis12, respectively, have been identified (for review, see Chan et al., 2005). The above-mentioned results raise the question of how the CPC targets to the centromere. The histone H3 variant CENP-A that defines the centromere (Earnshaw and Rothfield, 1985; Howman et al., 2000; Yoda et al., 2000) has been implicated in this process. A recent study reported that human CENP-A phosphorylated by Aurora A at serine 7 is required to restrict Aurora B to the centromere (Kunitoku et al., 2003). In chicken DT-40 cells and C. elegans embryos, however, CENP-A seems to be dispensable for the localization of INCENP (Oegema et al., 2001; Regnier et al., 2005). To assess the effect of the absence of CENP-A on CPC localization in human cells, CENP-A was knocked down using corresponding siRNA oligonucleotides. CENP-A levels were found to be efficiently reduced to undetectable levels, as judged by Western blotting and immunofluorescence analysis, but the localization of the CPC proteins to the centromere was normal (Figure 7, A and B). According to these results, it seems unlikely that CENP-A plays a critical role in the recruitment of the CPC to the centromere.
Figure 7.
Centromere targeting of the CPC is independent of CENP-A and hMis12. (A) Lamin A (control) and CENP-A were knocked down by siRNA treatment. HeLa S3 cell lysates were probed by Western blotting using the indicated antibodies. (B) CENP-A or hMis12 siRNA-treated HeLa S3 cells were analyzed by immunofluorescence for the presence of CENP-A, Borealin, or Aurora B. Bar, 10 μm.
Besides CENP-A, the human homologue of the fission yeast Mis12 protein, hMis12, has been reported to constitute a second CENP-A–independent pathway for centromere/kinetochore assembly. In agreement with previous data using the published siRNA duplexes (Goshima et al., 2003), we found hMis12 siRNA treatment to result in defective chromosome alignment in metaphase, lagging chromosomes in anaphase and the formation of micronuclei. However, knockdown of hMis12 did not alter localization of the chromosomal passenger proteins (Figure 7B). Thus, centromere localization of the CPC does not depend on the two described branches of centromere/kinetochore assembly. As described for CENP-A and hMis12, we tested whether other components of the centromere/kinetochore might play a role in centromere recruitment of the CPC. Among 10 different centromere/kinetochore proteins tested, none was found to be required for localizing the CPC to the centromere (our unpublished data).
Borealin Binds to Double-stranded DNA In Vitro, but It Requires Survivin and INCENP1-58 for In Vivo Localization
Because no centromere/kinetochore component so far analyzed was found to influence CPC recruitment to the centromere the possibility that one or more of the CPC subunits might be able to bind to DNA directly was considered. To test this hypothesis, purified histone H3, recombinant MBP-Borealin, His-Borealin, MBP-INCENP1-58, MBP, GST-Cdc20, and His-Plk1 were incubated with native double-stranded calf-thymus DNA-cellulose. Only His-Borealin and MBP-Borealin as well as the positive control protein histone H3 bound to the DNA (Figure 8A, lanes 7, 9, and 2, respectively). The centromere/kinetochore proteins Cdc20 and Plk1, both of which exhibit a similar basic pI as Borealin, did not bind to the DNA cellulose arguing against a nonspecific charge-based association of Borealin with the cellulose. Furthermore, increased salt concentrations reduced binding of His-Borealin to the DNA (Figure 8B). Importantly, when His-Borealin, His-Survivin, and MBP-INCENP1-58 were mixed and incubated with DNA-cellulose, His-Borealin could recruit the two other proteins to the DNA (Figure 8C, lane 4). These data suggest that Borealin might be the subunit within the CPC that can directly bind to DNA and thus localize the CPC to the centromere.
Figure 8.
Borealin binds to double-stranded DNA in vitro and recruits Survivin and INCENP1-58 to DNA. (A) Histone H3, His-Survivin, His-Borealin, MBP-Borealin, MBP-INCENP1-58, MBP alone, GST-Cdc20, and His-Plk1 were tested for DNA-binding activity by incubation with calf-thymus DNA-cellulose. Input (lanes 1, 3, 6, 8, 10, 12, 14, and 16) and proteins bound to the cellulose (lanes 2, 4, 5, 7, 9, 11, 13, 15, and 17) were analyzed by SDS-PAGE. Only histone H3 (lane 2), His-Borealin (lane 7), and MBP-Borealin (lane 9) bound to the DNA cellulose. (B) His-Borealin was incubated with DNA-cellulose as in A in the presence of increasing NaCl concentrations. (C) His-Borealin, His-Survivin, and MBP-INCENP1-58 were mixed at equimolar concentrations and incubated with calf-thymus DNA-cellulose as described above. In the presence of His-Borealin, His-Survivin and MBP-INCENP1-58 were found in the DNA-binding fraction (lane 4).
Next, it was tested whether Borealin could target independently to the centromere in vivo. Endogenous Borealin was knocked down with 3′ UTR–siRNA duplexes (see Figure 5B, lane 4, for Western blot) and replaced by transfected Myc-tagged Borealin. Myc-Borealin localized correctly to the centromere in the absence of endogenous Borealin and restored targeting of the other passenger proteins to the centromere as well as phospho-CENP-A staining (Figure 9A). In contrast, centromere targeting of Myc-Borealin was abolished when Borealin was co-knocked down with either INCENP (Figure 9B) or Survivin (Figure 9C). Hence, even though Borealin is able to bind to DNA in vitro, in vivo the presence of INCENP and Survivin is required for targeting to the centromere. Similarly, Myc-Survivin was also unable to localize to the centromere in the absence of one of its binding partners in Survivin siRNA-treated cells (compare Figure 9, D with E and F). In summary, these data demonstrate the requirement for a complex of Borealin, Survivin, and INCENP1-58 for targeting to the centromere, and they have implications for the recruitment and regulation of kinetochore proteins (Figure 10).
Figure 9.
In vivo targeting of Borealin and Survivin to the centromere requires the presence of their binding partners. (A) HeLa S3 cells were treated with 3′ UTR siRNA oligonucleotides targeting Borealin and simultaneously transfected with Myc-Borealin (see Figure 5C for RNAi efficiency). Transfected cells were analyzed for the presence of the other chromosomal passenger proteins. (B and C) Cells were treated as in A except that INCENP (B) or Survivin (C) were knocked down in parallel with Borealin. (D) HeLa S3 cells were treated with siRNA oligonucleotides targeting the 3′ UTR of the Survivin transcript (see Figure 5C for RNAi efficiency) and simultaneously transfected with Myc-Survivin. Transfected cells were analyzed for the presence of INCENP, Borealin, and phospho-CENP-A by indirect immunofluorescence. (E and F) HeLa S3 cells were treated as described for D but in addition to Survivin, either INCENP (E) or Borealin (F) was knocked down. All immunofluorescence images are representatives of three independent experiments. Bar, 10 μm.
Figure 10.
Model for centromere targeting of the CPC. (A) Our data suggest that Survivin and Borealin, both forming higher molecular order structures at the centromere, bind to the first 58 amino acids of INCENP. Aurora B, in contrast, associates with the C-terminal IN-box of INCENP. We propose that Borealin targets the CPC to the centromere by binding to DNA directly (dashed arrow) but can only do so when present within a functional subcomplex. Aurora B kinase is not involved in this process. (B) The centromere recruitment of the CPC is independent of the CENP-A and the hMis12 assembly pathways. Both pathways are required for the targeting of CENP-I and CENP-H (not depicted); in addition, CENP-A recruits the Hec1/Nuf2-complex. The CPC recruits MCAK to the inner centromere and checkpoint proteins to the kinetochore.
DISCUSSION
Progress toward understanding how the CPC targets to the centromere has so far been hampered by the interdependency of the different subunits for protein stability and localization, making it difficult to assess their individual contributions to the process of targeting. The RNAi rescue approach used in this study revealed the presence of a functional subcomplex of the CPC that bound to the centromere independently of the kinase Aurora B. Based on biochemical studies and both in vitro and in vivo analyses, we show that this subcomplex comprises the N-terminal 58 amino acids of INCENP and at least dimers of both Survivin and Borealin. Our data also suggest that, within this module, Borealin might target the complex to the centromere by directly binding to DNA.
Aurora B-independent Recruitment of Survivin, Borealin, and INCENP1-58 to the Centromere
How can the finding of Aurora B-independent targeting of a CPC subcomplex be reconciled with siRNA-mediated knockdown of Aurora B leading to loss of the other CPC components from the centromere? We suggest that the requirement for Aurora B is indirect, and results from the instability of full-length INCENP in the absence of Aurora B. That GFP-INCENP1-58 is stable in the absence of Aurora B suggests that the region that confers instability to INCENP must lie within the central coiled-coil domain or the C terminus of the protein, the region that interacts with the Aurora B kinase (Adams et al., 2000; Bishop and Schumacher, 2002; Bolton et al., 2002; Honda et al., 2003; Yasui et al., 2004; Sessa et al., 2005). Furthermore, the RNAi-rescue results with GFP-INCENP1-58 show that expression of the first 58 amino acids of INCENP rescues the protein levels of Survivin and Borealin in the transfected cells. Similarly, we found Aurora B levels to be rescued by an INCENP construct that comprises the IN-box but does not localize to the centromere. These findings argue that the interaction with INCENP stabilizes its binding partners of the CPC.
Interestingly, in C. elegans, in contrast to mammalian cells, localization of CSC-1 (a remote Borealin homologue), BIR-1 (C. elegans Survivin), and ICP-1 (C. elegans INCENP) is not dependent on Aurora B (AIR-2), whereas, conversely, AIR-2 depends on all three for localization (Speliotes et al., 2000; Romano et al., 2003). This difference might be explained by the different sizes of C. elegans and mammalian ICP-1/INCENP. C. elegans ICP-1 (∼70 kDa) is much smaller than mammalian INCENP (∼120 kDa) and its stability might therefore be independent of AIR-2/Aurora B.
Targeting the Survivin–Borealin–INCENP1-58 Subcomplex to the Centromere via Borealin-mediated DNA Binding
The chromosomal passenger proteins bind to the inner centromere during prometaphase and metaphase, but the precise mechanism(s) by which the CPC proteins localize to the inner centromere have not been elucidated previously. Importantly, no kinetochore/centromere protein has yet been found whose knockdown results in the loss of the CPC from the centromere. Our present data raise the intriguing possibility that INCENP targeting is ultimately determined by a DNA binding activity associated with Borealin but that in vivo this activity is only displayed in the context of a functional ternary complex of Borealin with INCENP1-58 and Survivin (see Figures 8 and 9 and model in Figure 10A). This assumption predicts that fragments and/or deletion mutants of INCENP1-58, Borealin, or Survivin that do not interact with their binding partners of the ternary complex cannot target to the centromere. In agreement with this, it has been reported that Borealin fragments that do not bind INCENP as well as an N-terminal fragment of Survivin lacking the Borealin-binding domain were unable to localize to the centromere (Gassmann et al., 2004; Lens et al., 2006). Interestingly, in the latter study, a C-terminal Survivin fragment containing the Borealin-binding domain still failed to target to the centromere. In the light of our findings, a possible explanation for the latter data is that in this case the Survivin-INCENP or the Survivin-Survivin self-interaction is lost. Further supporting the requirement for the holo-CPC complex, attempts of restoring the CPC by supplementing Aurora B depleted Xenopus egg extract with INCENP, Aurora B, and Survivin failed (Vigneron et al., 2004). This result is likely due to the absence of Borealin in the corresponding experiment and indicates that localization and stability of the CPC requires the Borealin protein.
On the basis of biochemical experiments, it has been proposed that in addition to the holo-CPC, Aurora B and INCENP can form a separate, independent complex (Gassmann et al., 2004). Our data would suggest that such a complex should not be able to target to the centromere but rather be cytoplasmic. This idea is consistent with the observation that GFP-Aurora B exhibits a dynamic behavior at the centromere, exchanging rapidly with a cytoplasmic pool (Murata-Hori et al., 2002).
Other factors beside the CPC composition may also contribute to the centromere localization of the CPC, such as specific modification states of (peri)centromeric histones (Sullivan and Karpen, 2004) and chromatin structure. Also, it has very recently been reported that the ubiquitination state of Survivin affects the dynamic localization of chromosomal passengers to the centromere in mitosis (Vong et al., 2005). Recruitment of the CPC to the centromere might therefore include a larger number of regulatory steps than previously assumed. Cooperation of these mechanisms may be necessary to ensure that the CPC targets specifically to the centromeres rather than the entire chromosome.
Assembly of the Centromere/Kinetochore
The CENP-A and hMis12 proteins have been identified as two important founder proteins for the formation of an intact centromere (Chan et al., 2005). Both proteins contribute to the kinetochore recruitment of CENP-I and CENP-H, but they are independent of each other for localization (Goshima et al., 2003). In addition CENP-A has been shown to recruit CENP-C and the conserved Hec1/Nuf2 complex to the kinetochore (Chan et al., 2005; Regnier et al., 2005). A recent study suggested that phosphorylated CENP-A is important for specifying the centromere targeting of the CPC (Kunitoku et al., 2003). However, consistent with results from other organisms and our conclusion that Borealin may directly bind to DNA and thereby direct the CPC to the centromere, siRNA knockdown of CENP-A or hMis12 did not influence centromere localization of the CPC (Oegema et al., 2001; Vigneron et al., 2004; Regnier et al., 2005) (see Figures 7 and 8 and model in Figure 10A). As we demonstrate (Figure 6A and Supplemental Figure S2), the centromere targeting of the CPC per se is independent of Aurora B (although Aurora B is required for INCENP stability). However, Aurora B kinase activity is clearly required at the centromere for the recruitment of other proteins, notably the MCAK (Andrews et al., 2004; Lan et al., 2004), and downstream checkpoint components, including BubR1, CENP-E, and Mad2 (Ditchfield et al., 2003; Hauf et al., 2003; Vigneron et al., 2004) (Figures 6D and 10B). Thus, the CPC may itself represent another important branch of centromere/kinetochore assembly.
In summary, we conclude that vertebrate INCENP comprises two functional modules. First, the C-terminal IN-box, which has previously been shown to function as an effector module in the binding and activation of Aurora B (Adams et al., 2000; Bishop and Schumacher, 2002; Honda et al., 2003; Yasui et al., 2004; Sessa et al., 2005); and second, a functional module required for centromere targeting associated with the N terminus of the INCENP protein (Figure 10A). In the future, it will be important to elucidate the precise stoichiometry of the targeting subcomplex identified in this study and to analyze potential Aurora B-independent functions of this ternary module.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Francis A. Barr and Robert Kopajtich, Dr. Herman Sillje, Dr. Andreas Uldschmid, Christoph Baumann, and Dr. Sabine Elowe for reagents. We are grateful to Dr. Francis A. Barr and Dr. Sabine Elowe for comments on the manuscript, and we thank Christoph Baumann for helpful discussions. We also thank Dr. Claire Crafter and AstraZeneca for providing the Aurora B inhibitor ZM447439. We acknowledge financial support from the “Max-Planck-Gesellschaft,” the “Fonds der Chemischen Industrie,” and the Deutsche Forschungsgemeinschaft (SFB646). U.R.K. is supported by a Ph.D. fellowship by the Boehringer Ingelheim Fonds (B.I.F.).
Abbreviations used:
- CPC
chromosomal passenger complex
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-12-1133) on March 29, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams R. R., Carmena M., Earnshaw W. C. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001a;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Adams R. R., Maiato H., Earnshaw W. C., Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 2001b;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. R., Wheatley S. P., Gouldsworthy A. M., Kandels-Lewis S. E., Carmena M., Smythe C., Gerloff D. L., Earnshaw W. C. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Ainsztein A. M., Kandels-Lewis S. E., Mackay A. M., Earnshaw W. C. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J. R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Berger B., Wilson D. B., Wolf E., Tonchev T., Milla M., Kim P. S. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. D., Schumacher J. M. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M. A., Lan W., Powers S. E., McCleland M. L., Kuang J., Stukenberg P. T. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A., Carmena M., Sambade C., Earnshaw W. C., Wheatley S. P. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 2003;116:2987–2998. doi: 10.1242/jcs.00612. [DOI] [PubMed] [Google Scholar]
- Chan G. K., Liu S. T., Yen T. J. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Chantalat L., Skoufias D. A., Kleman J. P., Jung B., Dideberg O., Margolis R. L. Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual alpha-helical extensions. Mol. Cell. 2000;6:183–189. [PubMed] [Google Scholar]
- Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S. S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A. J., Ruchaud S., Hudson D. F., Honda R., Nigg E. A., Gerloff D. L., Earnshaw W. C. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Glover D. M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Yasui Y., Kawajiri A., Nigg E. A., Terada Y., Tatsuka M., Nagata K., Inagaki M. Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J. Biol. Chem. 2003;278:8526–8530. doi: 10.1074/jbc.M210892200. [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Honda R., Nigg E. A., Barr F. A. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Mishima M., Glotzer M. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 2005;15:778–786. doi: 10.1016/j.cub.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Haas A. K., Fuchs E., Kopajtich R., Barr F. A. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., Peters J. M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Korner R., Nigg E. A. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman E. V., Fowler K. J., Newson A. J., Redward S., MacDonald A. C., Kalitsis P., Choo K. H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitoku N., Sasayama T., Marumoto T., Zhang D., Honda S., Kobayashi O., Hatakeyama K., Ushio Y., Saya H., Hirota T. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell. 2003;5:853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lens S. M., Rodriguez J. A., Vader G., Span S. W., Giaccone G., Medema R. H. Uncoupling the central spindle-associated function of the chromosomal passenger complex from its role at centromeres. Mol Biol. Cell. 2006;17:1897–1909. doi: 10.1091/mbc.E05-08-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens S. M., Wolthuis R. M., Klompmaker R., Kauw J., Agami R., Brummelkamp T., Kops G., Medema R. H. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003;22:2934–2947. doi: 10.1093/emboj/cdg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Sakashita G., Matsuzaki H., Sugimoto K., Kimura K., Hanaoka F., Taniguchi H., Furukawa K., Urano T. Direct association with inner centromere protein (INCENP) activates the novel chromosomal passenger protein, Aurora-C. J. Biol. Chem. 2004;279:47201–47211. doi: 10.1074/jbc.M403029200. [DOI] [PubMed] [Google Scholar]
- Minoshima Y., et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell. 2003;4:549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Muchmore S. W., Chen J., Jakob C., Zakula D., Matayoshi E. D., Wu W., Zhang H., Li F., Ng S. C., Altieri D. C. Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell. 2000;6:173–182. [PubMed] [Google Scholar]
- Murata-Hori M., Tatsuka M., Wang Y. L. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell. 2002;13:1099–1108. doi: 10.1091/mbc.01-09-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T. J. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Pines J. Four-dimensional control of the cell cycle. Nat. Cell Biol. 1999;1:E73–E79. doi: 10.1038/11041. [DOI] [PubMed] [Google Scholar]
- Regnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A., Guse A., Krascenicova I., Schnabel H., Schnabel R., Glotzer M. CSC-1, a subunit of the Aurora B kinase complex that binds to the survivin-like protein BIR-1 and the INCENP-like protein ICP-1. J. Cell Biol. 2003;161:229–236. doi: 10.1083/jcb.200207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai K., et al. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil. Cytoskeleton. 2004;59:249–263. doi: 10.1002/cm.20039. [DOI] [PubMed] [Google Scholar]
- Schumacher J. M., Golden A., Donovan P. J. AIR-2, an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Speliotes E. K., Uren A., Vaux D., Horvitz H. R. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Sullivan B. A., Karpen G. H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. U. Chromosome bi-orientation on the mitotic spindle. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2005;360:581–589. doi: 10.1098/rstb.2004.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., Kauw J. J., Medema R. H., Lens S. M. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2006;7:85–92. doi: 10.1038/sj.embor.7400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Earnshaw W. C. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 2004;113:211–222. doi: 10.1007/s00412-004-0307-3. [DOI] [PubMed] [Google Scholar]
- Verdecia M. A., Huang H., Dutil E., Kaiser D. A., Hunter T., Noel J. P. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 2000;7:602–608. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- Vigneron S., Prieto S., Bernis C., Labbe J. C., Castro A., Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong Q. P., Cao K., Li H. Y., Iglesias P. A., Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- Yasui Y., Urano T., Kawajiri A., Nagata K., Tatsuka M., Saya H., Furukawa K., Takahashi T., Izawa I., Inagaki M. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 2004;279:12997–13003. doi: 10.1074/jbc.M311128200. [DOI] [PubMed] [Google Scholar]
- Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S. G., Shelby R. D., Sullivan K. F. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.