Abstract
The Wnt signaling pathway plays a major role in development, and upon deregulation it is implicated in neoplasia. The hallmark of the canonical Wnt signal is the protection of β-catenin from ubiquitination and proteasomal degradation induced by glycogen synthase kinase (GSK)-3β inhibition. The stabilized β-catenin translocates to the nucleus where it binds to T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors, activating the expression of Wnt target genes. In the absence of Wnt signal, TCF/LEF bind to Groucho (Gro)/TLE corepressors and repress Wnt target genes. Gro/TLE bind also to Engrailed (En) transcription factors mediating En-repressive activity on En target genes. Here, we present data suggesting that En-1 serves also as a negative regulator of β-catenin transcriptional activity; however, its repressive effect is independent of Gro/TLE. Our data suggest that En-1 acts by destabilizing β-catenin via a proteasomal degradation pathway that is GSK-3β–independent. Moreover, because En-1-mediated β-catenin degradation is also Siah independent, our data imply that En-1 exerts its repressive effect by a novel mechanism negatively controlling the level of β-catenin.
INTRODUCTION
Wnts (Wingless [Wg] in Drosophila) comprise a family of secreted glycoprotein ligands that initiate signaling pathways regulating cell growth and differentiation (Akiyama, 2000; Huelsken and Birchmeier, 2001). The canonical signaling pathway initiates after binding of Wnt to the receptor/coreceptor complex comprised of the seven transmembrane Frizzled (Fz) receptor and the low-density lipoprotein receptor-related protein (LRP) 5 or LRP6 (Akiyama, 2000; Huelsken and Birchmeier, 2001). This leads to the activation of Dishevelled (Dvl), which in turn inactivates glycogen synthase kinase (GSK)-3β. In the absence of Wnt signaling, β-catenin, which associates in the “destruction complex” with Axin, adenomatous polyposis coli (APC), GSK-3β, casein kinase I (CKI), Frat and others, undergoes phosphorylation by CKI and GSK-3β, resulting in β-transducin repeat-containing protein (TrCP)–mediated ubiquitination and degradation of β-catenin by the proteasome pathway (Peifer and Polakis, 2000). Inhibition of GSK-3β activity by the activated Dvl-1 leads to the stabilization of β-catenin. In addition to the canonical Wnt-signal-mediated β-catenin stabilization, β-catenin expression is regulated posttranslationally also by other pathways, via GSK-3β–dependent or –independent mechanisms (Liu et al., 2001; Matsuzawa and Reed, 2001; Xiao et al., 2003; Nastasi et al., 2004; Sharma et al., 2004). The stabilized β-catenin translocates to the nucleus, where it associates with members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors and activates TCF/LEF transcriptional activity (Hurlstone and Clevers, 2002). In the absence of stabilized nuclear β-catenin, TCF/LEF factors serve as repressors by interacting with corepressors, such as the Drosophila Groucho (Gro) or their mammalian homologues transducin-like enhancer of split (TLE) proteins, as well as with carboxy-terminal binding protein (Hurlstone and Clevers, 2002). The direct interaction of TCF/LEF factors with the nuclear-imported β-catenin, relieves TCF/LEF from Gro/TLE-mediated repression, thus activating expression of Wnt target genes, among which are developmental regulators and other genes involved in cell cycle regulation, cell–cell interactions, and cell–matrix interactions.
Gro/TLE proteins are well-known non-DNA binding corepressors required for transcriptional repression by several types of active transcriptional repressors, including Engrailed (En) (Courey and Jia, 2001).
Engrailed proteins (Eng in Drosophila, En-1 and -2 in mammalian species) are highly conserved homeodomains, bifunctional transcriptional factors that bind to a consensus sequence TCAATTAAAT and play significant roles during development (Gibert, 2002), by acting either as repressors or activators (Serrano et al., 1997; Serrano and Maschat, 1998). Eng-repressive activity is independent of its DNA binding domain (Hanna-Rose and Hansen, 1996) and is mediated in part by its binding, via the eh1 domain (Tolkunova et al., 1998), to the C-terminal domain of Gro corepressor. In the present study, we show that En-1 represses β-catenin transcriptional activity; however, this repression is TLE-independent. We present data suggesting that En-1 acts by destabilizing β-catenin via a novel GSK-3β–independent proteasomal degradation mechanism.
MATERIALS AND METHODS
Plasmids and Reagents
To molecularly clone the human En-1 cDNA, a PC3 cDNA library in λpCEV29 (Gazit et al., 1999) was screened using primers derived from eng-1-related expressed sequence tag clones and a cDNA of ∼3 kb was isolated. The eng-1 coding region was PCR amplified using a forward 5′-GSTAAGCTTGGCCGAGCATGGAA-3′ primer and a backward 5′-GCTGAATTCCTCGCTCTCGTCTT-3′ primer, and the PCR product, cleaved with HindIII and EcoRI, was inserted into pcDNA3, in frame and upstream of a hemagglutinin (HA) epitope. For constructing En-1Δeh1-HA, the region spanning residues 103-113 was deleted by the overlapping PCR strategy: two PCR products were synthesized using pcDNA3-En-1-HA as a template, in the presence of an internal forward primer 5′-CCCAGCTGCACCGCACCGACTTCGGCTGCAAAAAGG-3′ together with SP6 primer and the complementary backward primer together with T7 primer. The PCR products were mixed, further amplified using T7 and SP6 primers, and the resultant product was cloned into HindIII/XhoI sites of pcDNA3. En-1Δeh4-HA, which lacks eh4 spanning residues 302-369, was constructed by the same strategy as En-1Δeh1-HA, except of using 5′-AAGGAGACAGGCATCAAGAAC-3′ as an internal forward primer and 5′-CGCCAGGCCGTTCTTGATGCCTGTCTCCTTCTCGTTCTTCTTCTTC-3′ as an internal backward primer. pSUPER, containing the H1 promoter for expressing short interfering RNAs (siRNAs) was kindly provided by Dr. R. Agami (The Netherlands Cancer Institute, Amsterdam, The Netherlands). pSUPER/si-En-1, which expresses siRNAs-targeting En-1 mRNA, was constructed by annealing a forward 5′-gatccccGTGACAGAGGCCAGACTGGCttcaagagaGCCAGTCTGGCCTCTGTCACtttttggaaa-3′ oligonucleotide with its complementary form 5′-agcttttccaaaaaGTGACAGAGGCCAGACTGGCtctcttgaaGCCAGTCTGGCCTCTGTCACggg-3′ and inserting the double-stranded oligonucleotide stretch into the BglII/HindIII sites of pSUPER. pSUPER/scr-En-1 expressing scrambled si-En-1 was constructed as detailed above after annealing a forward 5′-gatccccGTGACAGAGGCCAGACTGGCttcaagagaGCCAGTCTGGCCTCTGTCACtttttggaaa-3′ oligonucleotide with its complementary form 5′-agcttttccaaaaaGTGACAGAGGCCAGACTGGCtctcttgaaGCCAGTCTGGCCTCTGTCACggg-3′. Mutated En-1-HA (mEn-1-HA) containing three silent third-codon point mutations within si-En-1 target region was constructed as follows: two overlapping PCR products were synthesized using pcDNA3-En-1-HA as a template, in the presence of an internal forward primer 5′-CGGGTCGAGCGTGACAGGGGTCAAACTGCCGCAGGTAGAGAC-3′ oligonucleotide (the mutated nucleotides are underlined) together with SP6 primer and the complementary backward primer 5′-GTCTCTACCTGCGGCAGTTTGACCCCTGTCACGCTCGACCCG-3′ together with T7 primer. The PCR products were mixed, further amplified using T7 and SP6 primers, and the resultant product was cloned into HindIII/XhoI sites of pcDNA3. A Myc-tagged constitutively active S33Y mutant β-catenin [pCCBS/β-catenin (S33Y)-Myc], described previously (Golan et al., 2004), was kindly received from Drs. Liu Guizhong and Stuart A. Aaronson (Department of Oncological Sciences, Mount Sinai School of Medicine, New York, NY). A Myc-tagged ΔNβ-catenin lacking the 47 N-terminal residues was kindly obtained from Dr. R. Moon (University of Washington School of Medicine, Seattle, WA). A Flag-tagged β-catenin(ΔS45) in which Ser45 is deleted (Morin et al., 1997) was kindly obtained from Dr. H. Clevers (Center for Biomedical Genetics, Hubrecht, The Netherlands). HA-tagged TCF4 was described previously (Golan et al., 2004). HFz1 was described previously (Gazit et al., 1999). Wnt-3a, described previously (Gazit et al., 1999), was kindly provided by Dr. J. Kitajewski (Columbia University, New York, NY). The Wnt-responsive TCF-dependent luciferase (TCF/Luc) constructs, pGL3-OT, and its mutated version pGL3-OF, kindly provided by Dr. B. Vogelstein (John Hopkins Oncology Center, Baltimore, MD), were described previously (Golan et al., 2004). The Cyclin D1/luciferase reporter, kindly provided by Drs. R. G. Pestell and C. Albanese (Albert Einstein College of Medicine, New York, NY), harboring the β-catenin-TCF response element, was described previously (Golan et al., 2004). TLE-1 was kindly received from Dr. S. Stifani (Montreal Neurological Institute, McGill University, Montreal, Quebec, Canada). A Myc-tagged TLE-1 was constructed by PCR amplification of TLE-1 coding region and inserting it into pCCBS, downstream and in frame of a Myc tag. Siah-1 as well as dominant-negative (dn) Siah-1 and Siah-2 (Topol et al., 2003) were kindly received from Dr. Y. Yang (National Human Genome Research Institute, National Institutes of Health, Bethesda, MD). All PCR-based constructs were confirmed by sequence analysis. MG132 (Calbiochem, San Diego, CA), lithium chloride (Sigma-Aldrich, St. Louis, MO), and cycloheximide (Sigma-Aldrich) were used at concentrations as indicated.
Cell Cultures, Transfections, and Luciferase Reporter Assays
Human 293T, HCT116, and SW480 cell lines were cultured in DMEM supplemented with 10% fetal calf serum. Transfection was performed by using FuGene (Roche Diagnostics, Mannheim, Germany) or calcium phosphate precipitation method (Chen and Okayama, 1987). TCF luciferase assays were performed as described previously (Golan et al., 2004). Briefly, cells growing in six-well dishes were transfected at 60–70% confluence with 1 μg of TCF/Luc reporter, En-1, or empty vector (1 mg or as otherwise indicated in text or figure legends) and 0.1 μg of pCMV/β-galactosidase (β-Gal) plasmid (Invitrogen, Carlsbad, CA), used to evaluate the efficiency of transfection. Total amount of DNA was adjusted for equal amounts with empty vector. Forty-eight hours after transfection, the luciferase levels were measured by using the Luciferase Reporter Gene Assay kit (Roche Diagnostics) as described previously (Gazit et al., 1999). Luciferase activities were normalized for transfection efficiency by β-Gal activity measured by the β-Gal enzyme-linked immunosorbent assay kit (Roche Diagnostics). When luciferase reporter assays were conducted in SW480 cells, 0.5 μg of Renilla luciferase reporter (Promega, Madison, WI) was used to assess transfection efficiency. Luciferase levels were measured using the dual luciferase assay kit (Promega), and the firefly luciferase activity was normalized to Renilla luciferase activity. Data are presented as mean values and standard deviations for at least three independent experiments done in duplicate, compared with the level of luciferase activity obtained in the presence of empty vector presented as 1. SW480 cells, stably expressing si-En1, were prepared by cotransfection of si-En-1 or empty pSUPER and a puromycin resistant gene-expressing vector (in a 10:1 ratio) followed by selection in the presence of 1.5 mg/ml puromycin (Calbiochem).
Northern Analysis
Total RNA was extracted using the TRI Reagent isolation solution (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. RNA (20 μg) was resolved on 1.2% agarose gel containing 6% formaldehyde and transferred to nylon membrane (Hybond-N; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The membrane was prehybridized in hybridization solution (50% formamide, 5× standard saline citrate [SSC], 4× Denhardt's solution, 0.5% SDS, and 0.1 mg/ml salmon sperm DNA) for 4 h at 42°C. The hybridization was performed at 42°C using β-catenin and glyceraldelyde-3-phosphate dehydrogenase (GAPDH) probes that were generated by PCR in the presence of [32P]dCTP. Membranes were washed twice at 25°C in 2× SSC/0.1% SDS for 20 min and twice at 50°C in 0.1× SSC 0.1% SDS for 30 min and then subjected to autoradiography.
Immunoprecipitation and Western Blot Analysis
Total cell lysates were prepared by solubilization in lysing buffer (150 mM NaCl, 50 mM Tris, pH 7.5, and 0.2% NP-40). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred to nitrocellulose membranes, blocked with 5% low fat milk, and the filters were incubated with the respective antibodies. Membranes were washed in 0.001% Tween 20 in phosphate-buffered saline and incubated for 45 min with a secondary antibody. After washing in Tween/phosphate-buffered saline, membranes were subjected to enhanced chemiluminescence detection analysis (GE Healthcare), using horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). For coimmunoprecipitation analysis, cells were solubilized in lysing buffer (see above), and extracts were clarified by centrifugation at 12,000 × g for 30 min at 4°C. Four milligrams of total cell lysates was incubated with the specific antibody for 18 h at 4°C and then incubated, with rotation, with protein A beads for 2 h at 4°C. Beads were collected by centrifugation, washed three times in lysing buffer, dissolved in Laemmli buffer, and analyzed by SDS-PAGE, followed by detection with the respective antibody. Anti-β-catenin, anti-Myc, anti β-actin, and anti-HA polyclonal antibodies as well as anti-goat, -rabbit, and-mouse IgG horseradish peroxidase-conjugated secondary antibodies, were obtained from Santa Cruz Biotechnology. Rat monoclonal anti-HA antibody was obtained from Roche Diagnostics. Anti-FLAG M2 monoclonal antibody and anti-FLAG M2-agarose affinity gel were purchased from Sigma-Aldrich.
Immunofluorescence Staining
Forty-eight hours after transfection, cells were fixed and permeabilized with methanol/acetone. Cells were treated for 1 h with anti-β-catenin monoclonal antibodies or anti-HA antibodies (Santa Cruz Biotechnology) and then washed and exposed for 1 h to fluorescein isothiocyanate (FITC)-conjugated anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA) or rhodamine red anti-rabbit antibody (Jackson ImmunoResearch Laboratories), respectively. Slides were analyzed by confocal microscopy.
Subcellular Fractionation
Forty-eight hours after transfection cells were lysed in a buffer containing 1% Triton X-100, 50 mM Tris-HCl, pH 7.5, 1 mM EGTA, 5 mM EDTA, 150 mM NaCl, 10% glycerol, 1 mM MgCl2, 0.5 mM phenylmethylsulfonyl fluoride, and Complete EDTA-free protease. After two successive centrifugations at 330 × g for 20 min at 4°C and 15,000 rpm for 15 min at 4°C, the anucleated supernatant was centrifuged at 100,000 × g at 4°C for 1 h. The Triton-soluble supernatant was collected for measuring the level of cytosolic β-catenin by Western analysis. The residual pellet, representing the Triton-insoluble fraction, was resuspended in a buffer containing 1%SDS, 50 mM Tris-HCl, 1 mM EGTA, and 5 mM EDTA and subjected to Western blot analysis for measuring the level of membrane-bound β-catenin.
RESULTS
The Native En-1 Represses β-Catenin Transcriptional Activity by Decreasing the Level of β-Catenin
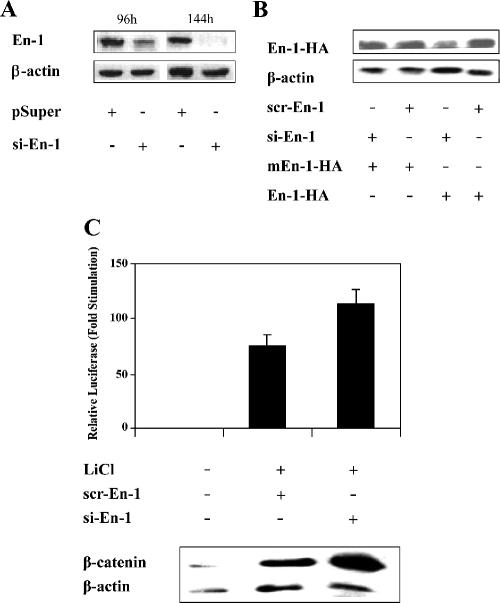
Because Eng directly interacts with Gro, and Gro/TLE serve as corepressors for the Wnt canonical pathway, we investigated whether En-1 might affect the canonical Wnt signal. The RNA interference (RNAi) approach (Elbashir et al., 2001) was used to silence the expression of the native En-1 and assess the magnitude of the Wnt signal in its absence. To this end, we constructed pSUPER expressing short interfering En-1-specific RNAs (si-En-1) and showed that the level of expression of the native En-1 was reduced in the presence of si-En-1 (Figure 1A). Moreover, si-En-1 also reduced the level of ectopically expressed En-1-HA (Figure 1B), compared with scr-En-1 expressing scrambled si-En-1, which did not affect the level of En-1-HA. To further confirm the specificity of si-En-1 silencing activity, we showed that in contrast to wild-type En-1, si-En-1 did not reduce the level of mEn-1-HA, which contains three silent mutations in the region targeted by si-En-1 and accordingly was refractory to si-En-1 silencing activity (Figure 1B). To determine the effect of En-1 on β-catenin transcriptional activity, 293T cells were treated with LiCl, a known inhibitor of GSK-3β activity (Klein and Melton, 1996). Data showed that LiCl-induced β-catenin transcriptional activity was elevated in the presence of si-En-1, compared with scr-En-1 (Figure 1C). Interestingly, immunoblot analysis of cell lysates revealed an elevation of the steady-state level of endogenous β-catenin in the presence of si-En-1, compared with scr-En-1 (Figure 1C). These data strongly suggested that En-1 negatively regulates β-catenin transcriptional activity by decreasing the level of β-catenin.
Figure 1.
Silencing of En-1 expression stimulates β-catenin transcriptional activity. (A) Si-En-1 decreases the expression of the native En-1. 293T cells were cotransfected with si-En-1 or empty pSUPER (4 μg). At the indicated times after transfection, 100 μg of cell lysates were analyzed by Western blotting using anti-En-1 or anti-β-actin antibody. (B) si-En-1 decreases the level of expression of ectopically expressed En-1. 293T cells were cotransfected with si-En-1 or scr-En-1 (2.9 μg) and En-1-HA or mEn-1-HA (0.01 μg). Seventy-two hours after transfection, 100 μg of cell lysates was analyzed by Western blotting using anti-HA or anti-β-actin antibody. (C) LiCl-activated β-catenin transcriptional activity is stimulated in the presence of si-En-1. 293T cells were cotransfected with si-En-1, scr-En-1, or empty vector (2.5 μg), TCF/Luc reporter (1 μg) and β-Gal (0.1 μg). Twenty-four hours after transfection, cells were treated with 30 mM LiCl and after 48 h, cell lysates were measured for the levels of luciferase and β-Gal activities. Data are presented as mean values and standard deviations for at least three independent experiments done in duplicate, compared with the level of luciferase activity obtained in the absence of LiCl treatment presented as 1 (top). Cell lysates (100 μg) were subjected to Western analysis using anti-β-catenin or anti-β-actin antibody (bottom).
En-1–repressive Activity Is Exerted Downstream of β-Catenin Destruction Complex
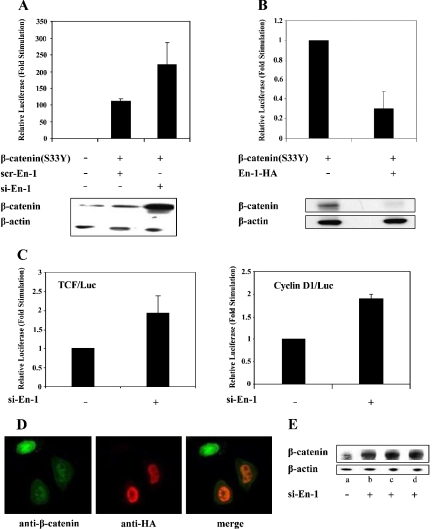
Because En-1 activity was displayed in the presence of LiCl, a GSK-3β inhibitor, it was suggested that En-1 effect on β-catenin expression is not mediated via affecting GSK-3β activity. It was therefore reasonable to assume that En-1 would also inhibit the transcriptional activity of a mutant β-catenin that is refractory to GSK-3β–phosphorylating activity. Hence, we used the constitutively active β-catenin(S33Y) mutant that is insensitive to GSK-3β–mediated phosphorylation and proteasomal degradation (Kolligs et al., 1999). Data showed that β-catenin(S33Y) transcriptional activity was elevated in the presence of si-En-1 compared with scr-En-1 (Figure 2A). Moreover, consistent with our previous observations, silencing of the native En-1 expression resulted in an increase in the steady-state level of β-catenin(S33Y) (Figure 2A).
Figure 2.
En-1 represses β-catenin(S33Y) transcriptional activity, concomitantly with decreasing β-catenin level. (A) Silencing of the native En-1 augments β-catenin(S33Y)-transcriptional activity. 293T cells were cotransfected with β-catenin(S33Y) or empty vector (0.1 μg), si-En-1 or scr-En-1 (2.5 μg each), TCF/Luc reporter (1 μg), and β-Gal (0.1 μg). Luciferase and β-Gal levels were measured as described in the legend to Figure 1. Cell lysates (100 μg) were subjected to Western analysis using anti-β-catenin or β-actin antibody. (B) Ectopically expressed En-1 represses β-catenin(S33Y)-transcriptional activity. 293T cells were cotransfected with β-catenin(S33Y) or empty vector (0.1 μg), En-1-HA or empty vector (1 μg), Cyclin D1/Luc (1 μg), and β-Gal (0.1 μg). Luciferase and β-Gal levels were measured as described in Figure 1. Cell lysates (30 μg) were subjected to Western analysis using anti-β-catenin or anti-β-actin antibody. (C) En-1 represses the constitutively active β-catenin in SW480 cell line. SW480 cells were transiently transfected with si-En-1 or empty vector (2.5 μg), TCF/Luc or CyclinD1/Luc reporter (1 μg), and Renilla luciferase vector (0.5 μg). Forty-eight hours after transfection, the luciferase activity was measured, and the luciferase levels were normalized according to Renilla luciferase activity. (D) Immunohistochemistry assays showing that En-1 decreases the level of native β-catenin. SW480 cells were transiently transfected with En-1-HA, and 48 h after transfection cells were subjected to immunofluorescence analysis with anti-β-catenin and anti-HA antibody, followed by FITC-conjugated anti-mouse antibody and rhodamine anti-rabbit antibody, respectively. Green, β-catenin expression; red, En-1-HA expression. (E) Silencing of the endogenous En-1 augments the level of native β-catenin in SW480 cells. SW480 cells were stably transfected with si-En-1 (clones b–d) or empty pSUPER (a), and the level of native β-catenin was quantitated by Western analysis using anti-β-catenin or anti-β-actin antibody.
Because our experiments, investigating the effect of En-1 on β-catenin transcriptional activity, used a TCF-dependent luciferase reporter harboring three copies of a TCF-binding motif (Korinek et al., 1997), we next investigated the effect of En-1 on a native TCF-responsive promoter. To this end, we used a luciferase reporter in which the expression of luciferase is governed by the promoter of Cyclin D1, a Wnt target gene containing a TCF-binding domain within its transcriptional unit (Shtutman et al., 1999). 293T cells were transfected with β-catenin(S33Y), En-1-HA, and CyclinD1/Luc, and the level of luciferase was assessed. Data showed that ectopically expressed En-1 repressed β-catenin(S33Y)–mediated stimulation of CyclinD1 promoter transcriptional activity (Figure 2B). Consistent with our previous results, this repression was concomitantly associated with a reduction in the level of β-catenin(S33Y) (Figure 2B).
Next, we investigated whether En-1 would affect a constitutively active Wnt signal in a human colon cancer (CRC) derived SW480 cell line harboring a mutated APC (Munemitsu et al., 1995). SW480 cells were transiently transfected with si-En-1 or empty vector and either TCF/Luc or CyclinD1/Luc reporter, and the level of luciferase was measured. Data showed that silencing of the native En-1 stimulated the constitutively active β-catenin in SW480 cells (Figure 2C). Moreover, immunohistochemistry assays showed that when SW480 cells were transiently transfected with En-1-HA, 20–50% of En-1-HA transfectants expressed dramatically lower levels of endogenous β-catenin, compared with neighboring untransfected cells (Figure 2D). Consistent with these results, SW480 cells that were stably transfected with si-En-1 expressed higher levels of β-catenin (Figure 2E, b–d), compared with empty-vector SW480 transfectants (Figure 2E, a). Together, our data imply that En-1 acts downstream of β-catenin destruction complex.
En-1–repressive Activity Is TLE Independent
Gro/TLE serve as Wnt corepresssors via direct interaction with TCF/LEF (Cavallo et al., 1998; Roose et al., 1998). Because the Drosophila Eng binds to Gro via the Eng eh1 domain (Tolkunova et al., 1998), we investigated whether En-1 mutant lacking eh1, which lost its ability to interact with TLE-1 (Figure 3A), would retain its repressive effect on β-catenin transcriptional activity. 293T cells were transfected with β-catenin(S33Y), Cyclin D1/Luc, and either En-1Δeh1-HA or empty vector, and the level of luciferase was measured. Data showed that β-catenin transcriptional activity was repressed also by En-1Δeh1, and this repression was associated with a decrease in the level of β-catenin (Figure 3B). These data suggested that En-1–repressive activity on β-catenin is TLE independent.
Figure 3.
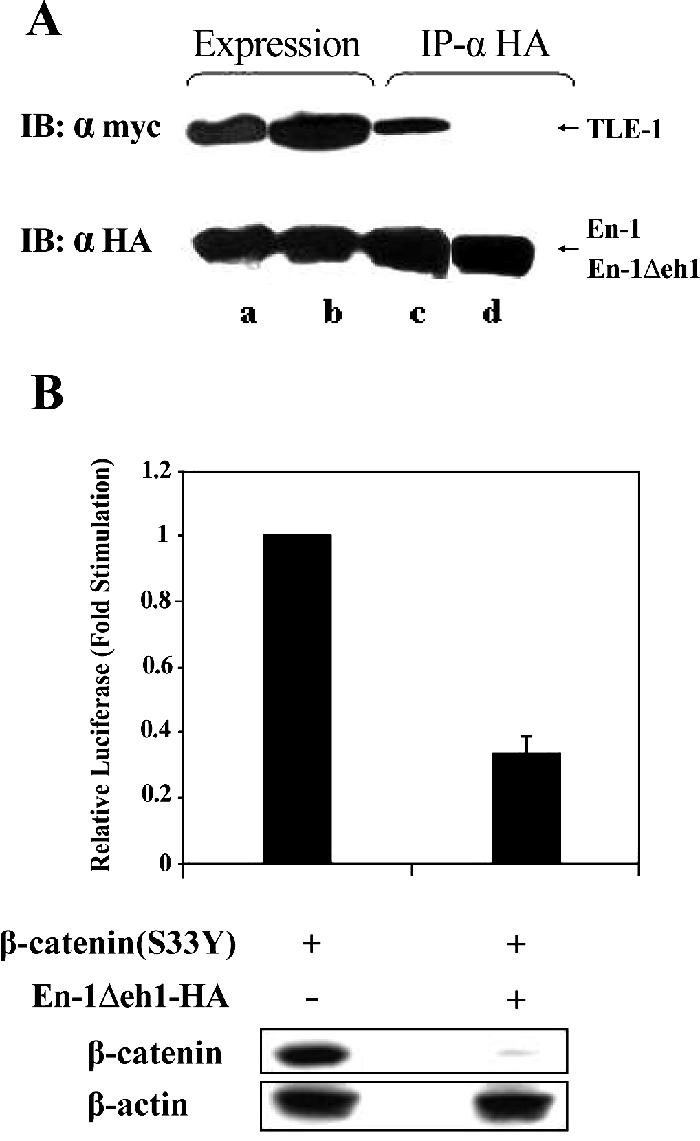
En-1 repressive activity is TLE-independent. (A) En-1Δeh1-HA lacks the ability to bind to TLE-1. 293T cells were cotransfected with TLE-1-Myc (2 μg) and either En-1-HA (a and c) or En-1Δeh1-HA (b and d) (2 μg). Lysates were immunoprecipitated using anti-HA antibody, and Western blots were treated with anti-Myc or anti-HA antibody before (a and b) or after (c and d) immunoprecipitation. (B) En-1Δeh1-HA represses β-catenin-transcriptional activity. 293T cells cotransfected with β-catenin(S33Y) (0.1 μg), En-1Δeh1-HA or empty vector (1 μg), Cyclin D1/Luc (1 μg), and β-Gal (0.1 μg), and the luciferase and β-Gal levels were measured as described in the legend to Figure 1 (top). Cell lysates (30 μg) were subjected to Western analysis using anti-β-catenin or anti-β-actin antibody (bottom).
En-1 Affects β-Catenin Stability
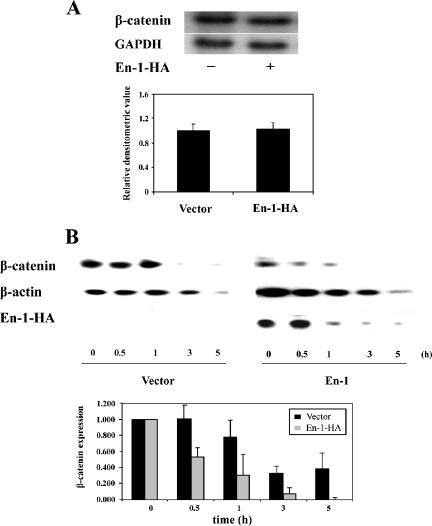
Because our data suggested that En-1 diminished the level of β-catenin, Northern analysis was performed to investigate whether En-1 affects β-catenin expression at the transcriptional level. Data showed the same level of β-catenin transcripts in the presence or absence of En-1 (Figure 4A). To investigate whether En-1 affects β-catenin stability, we followed up the degradation rate of β-catenin after cycloheximide addition. 293T cells were cotransfected with β-catenin(S33Y) and either En-1-HA or empty vector. Five hours after transfection, 80 μg/ml cycloheximide was added, and the level of β-catenin was measured at the indicated times. Data showed that already 5 h after transfection, before the addition of cycloheximide, the level of β-catenin was significantly lower in the presence of En-1 (Figure 4B). Moreover, although the apparent half-life of β-catenin in the presence of empty vector was ∼120 min, β-catenin apparent half-life in the presence of En-1 was ∼40 min. This reduction in β-catenin stability was detected even though En-1 itself was gradually degraded after cycloheximide addition (Figure 4B, top). Our data, showing that the degradation rate of β-catenin was accelerated in the presence of En-1, implied that En-1 affects the level of β-catenin via a posttranslational mode.
Figure 4.
En-1 affects the level of β-catenin by a posttranslational mode. (A) En-1 does not affect the level of β-catenin transcription. 293T cells were cotransfected with β-catenin(S33Y) and either En-1 or empty vector. Forty-eight hours after transfection, Northern analysis was performed by hybridizing the blots to either β-catenin or GAPDH probe. A representative gel is shown (top). Densitometric analysis of six independent experiments is shown (bottom). Each bar denotes the mean ± SD, compared with the level of GAPDH expression. (B) En-1 accelerates the rate of β-catenin(S33Y) degradation after cycloheximide addition. 293T cells were cotransfected with β-catenin(S33Y) and either En-1-HA or empty vector. Five hours after transfection, 80 μg/ml cycloheximide was added, and at the indicated hours postcycloheximide addition, lysates were subjected to Western analysis using anti-β-catenin, anti-HA, or anti-β-actin antibody. Representative gel is shown (top). The mean of the densitometric analysis of three independent experiments is shown (bottom). Each bar denotes the level of β-catenin divided by the level of β-actin at each time point, compared with the level of β-catenin at time zero, taken as 1.
En-1 Destabilizes β-Catenin via a Proteasomal Degradation Pathway
The hallmark of the canonical Wnt pathway is blocking the GSK-3β–dependent β-catenin proteasomal degradation (Aberle et al., 1997; Orford et al., 1997). However, because En-1–repressive effect on β-catenin was GSK-3β independent, we further investigated whether En-1–mediated β-catenin destabilization proceeds by the proteasomal pathway and as such would be relieved in the presence of the proteasome inhibitor MG132. 293T cells transfected with β-catenin(S33Y) and either En-1 or empty vector were treated with 25 μM MG132 for 4 h. Data showed that treating 293T cell transfectants with MG132, severely affected En-1 capacity to destabilize β-catenin (Figure 5). These data argue that En-1 affects the level of β-catenin via a GSK-3β–independent proteasomal degradation pathway.
Figure 5.
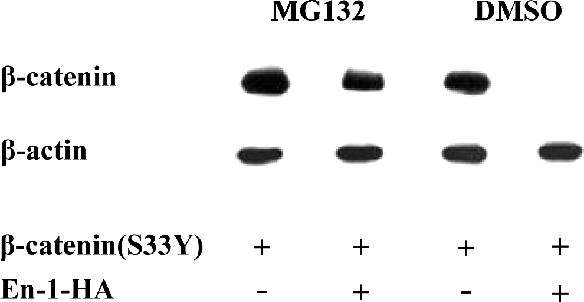
En-1 mediated β-catenin destabilization effect is blocked in the presence of the proteasome inhibitor MG132. 293T cells transfected with β-catenin(S33Y) (0.1 μg) and either En-1-HA or empty vector (1 μg) were treated with 25 μM MG132 or dimethyl sulfoxide. Four hours later, 50 μg of cell lysates were analyzed by Western blot analysis using anti-β-catenin or anti-β-actin antibody.
En-1–mediated Destabilization Is Siah Independent
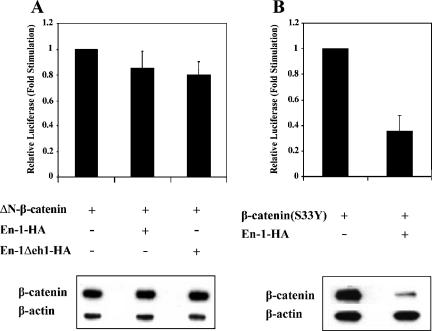
Because the N-terminal 50-amino acid–spanning domain of β-catenin is targeted by the GSK-3β–regulated β-catenin degradation pathway, we next determined whether the N terminus of β-catenin was dispensable for En-1–repressive effect. Data showed that the transcriptional activity of ΔN-β-catenin, lacking the N-terminal domain, was not repressed by either En-1 or En-1-Δeh1 (Figure 6A, top), in contrast to β-catenin(S33Y) (Figure 6B, top). Consistent with this result, ΔN-β-catenin was refractory to En-1 or En-1-Δeh1–mediated β-catenin destabilization (Figure 6A, bottom), in contrast to β-catenin(S33Y) (Figure 6B, bottom). These data suggested that the N terminus of β-catenin is indispensable for En-1–destabilizing effect. Because β-catenin was shown to be degraded also via the Siah-1/p53 pathway, which also targets the N terminus of β-catenin (Liu et al., 2001; Matsuzawa and Reed, 2001), we next investigated whether En-1–mediated β-catenin proteosomal degradation is Siah dependent and would thus be blocked by abolishing the Siah pathway using dnSiah mutants. 293T cells were cotransfected with Myc-tagged β-catenin(S33Y) mutant, En-1-HA, and either dnSiah-1, dnSiah-2, or empty vector. Data showed that both dnSiah-1 (Figure 7A) and -2 (our unpublished data) neither blocked En-1–repressive effect on β-catenin transcriptional activity, nor did they abolish En-1–mediated β-catenin destabilization. This was in contrast to the Siah1-mediated decrease in the level of β-catenin, which was abolished by dnSiah1 (Figure 7B). These data imply that En-1–destabilizing activity on β-catenin occurs via a distinct proteasomal pathway that is Siah independent. This conclusion is substantiated by our data showing En-1–repressive activity in SW480 cells (Figure 2D), which do not express the wild-type APC and are refractory to the Siah pathway (Liu et al., 2001).
Figure 6.
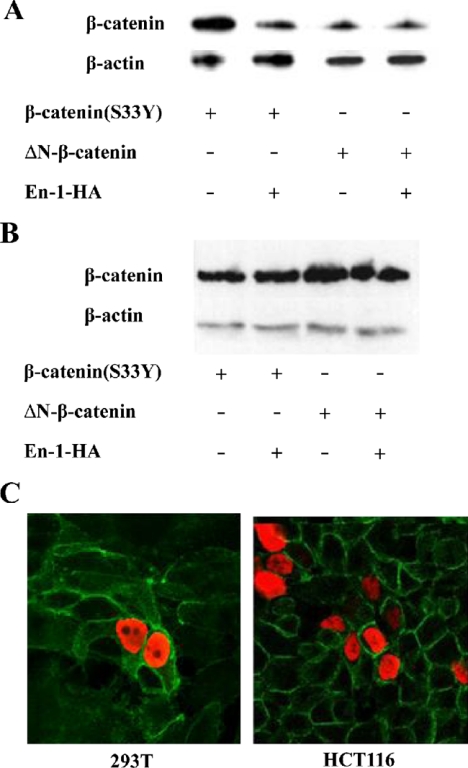
The N terminus of β-catenin is indispensable for En-1–destabilizing effect. (A) 293T cells were cotransfected with ΔN-β-catenin (0.1 μg), En-1-HA, En-1Δeh1-HA, or empty vector (1 μg), Cyclin D1/Luc (1 μg), and β-Gal (0.1 μg). (B) 293T cells were cotransfected with β-catenin(S33Y) (0.1 μg), En-1-HA or empty vector (1 μg), Cyclin D1/Luc (1 μg), and β-Gal (0.1 μg). Luciferase and β-Gal levels were measured as described in the legend to Figure 1 (top). Cell lysates (30 μg) were subjected to Western analysis using anti-β-catenin or anti-β-actin antibody (bottom).
Figure 7.
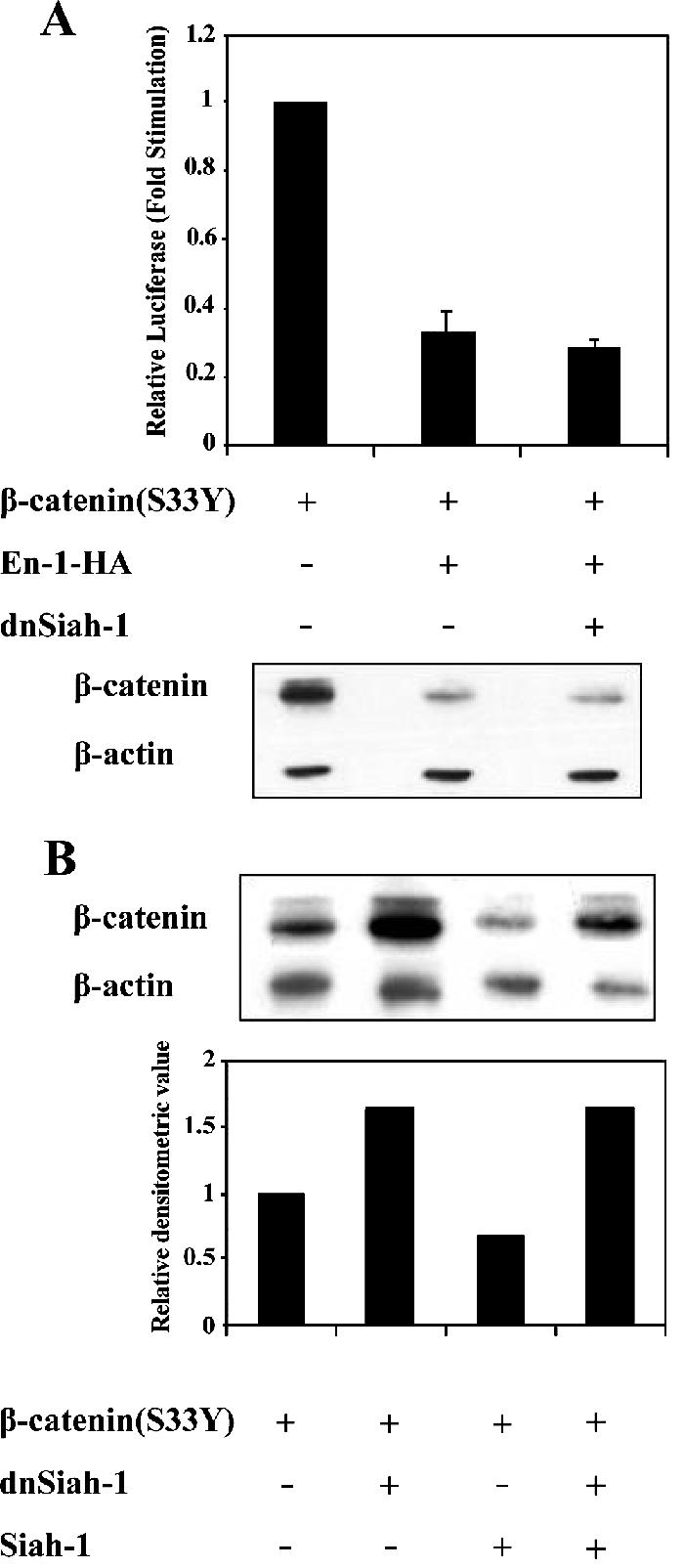
En-1–mediated β-catenin destabilization is Siah-1 independent. (A) The presence of dnSiah-1 does not affect En-1–repressive activity. 293T cells were transfected with β-catenin(S33Y) (0.1 μg), En-1-HA or empty vector (1 μg), dnSiah-1(1 μg) or empty vector, Cyclin D1/Luc (1 μg), and β-Gal (0.1 μg), and the luciferase and β-Gal levels were measured as described in the legend to Figure 1 (top). Cell lysates (30 μg) were subjected to Western analysis using anti-β-catenin or anti-β-actin antibody (bottom). (B) The presence of dnSiah-1 blocks Siah-1–mediated β-catenin degradation. 293T cells were transfected with β-catenin(S33Y) (0.1 μg), Siah-1 (0.1 μg) or empty vector, and dnSiah-1(1 μg) or empty vector. Cell lysates (30 μg) were subjected to Western analysis using anti-β-catenin or β-actin antibody (top). Densitometric analysis of the levels of β-catenin compared with β-actin levels is shown (bottom).
En-1 Does Not Affect the Stability of Membrane-bound β-Catenin
Because membrane-bound β-catenin degradation was recently shown to be specifically regulated, we sought to determine whether En-1 would affect also the stability of membrane bound β-catenin. 293T cells were cotransfected with β-actenin(S33Y) or ΔNβ-catenin, and either En-1-HA or empty vector, and the level of cytosolic as well as membrane bound β-catenin was determined. Data showed that En-1 reduced the level of cytosolic β-catenin(S33Y) but not cytosolic ΔNβ-catenin (Figure 8A). In contrast, En-1 did not affect the level of membrane-bound forms of β-catenin in either β-catenin(S33Y) or ΔNβ-catenin transfectants (Figure 8B). These results were further confirmed by immunohistochemistry assays, which showed that the level of membrane bound endogenous β-catenin was not affected by En-1 overexpression either in naive 293T cells or in HCT116 cells (Figure 8C).
Figure 8.
En-1 does not affect the stability of membrane-bound β-catenin. 293T cells were cotransfected with β-catenin(S33Y) or ΔNβ-catenin, and either En-1-HA or empty vector. The level of β-catenin was measured in cytosolic (A) or membrane (B) fraction by Western analysis, using anti-β-catenin or β-actin antibody. (C) 293T or HCT116 cells were transiently transfected with En-1-HA, and 48 h after transfection cells were subjected to immunofluorescence analysis with anti-β-catenin and anti-HA antibodies, followed by FITC-conjugated anti-mouse antibody and rhodamine-conjugated anti-rabbit antibody, respectively. Green, β-catenin expression; red, En-1-HA expression.
En-1–repressive Effect on β-Catenin Is Not Exerted via En-1 Target Genes
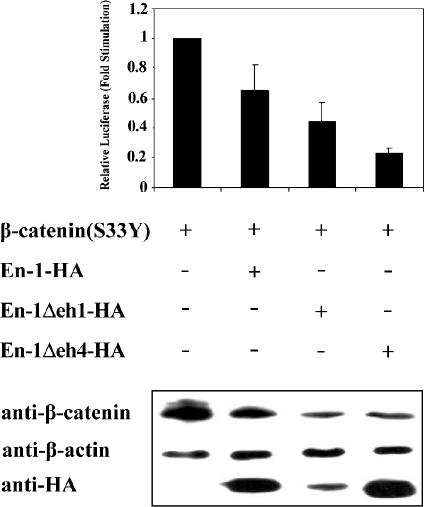
To explore whether En-1–destabilizing effect on β-catenin is transmitted via En-1 target genes, we deleted eh4, representing En-1 homeobox domain (Logan et al., 1992), and investigated whether En-1–repressive effect on β-catenin would be abrogated. 293T cells were cotransfected with β-catenin(S33Y) and En-1-HA, En-1Δeh1-HA, En-1Δeh4-HA, or empty vector, and the level of β-catenin transcriptional activity was measured. Data showed that the lack of eh4 domain did not interfere with En-1–repressive activity. These results were confirmed both by luciferase assay using CyclinD1/Luc reporter (Figure 9, top) and by Western analysis (Figure 9, bottom), showing that both Cyclin D1 promoter activity and the level of β-catenin, respectively, were reduced by overexpressing En-1Δeh4. These data suggested that eh4 homeobox domain of En-1 is dispensable for En-1 destabilizing effect on β-catenin.
Figure 9.
En-1–repressive effect on Wnt signal is not exerted via En-1 target genes. 293T cells were cotransfected with β-catenin(S33Y) (0.1 μg), and En-1-HA, En-1Δeh1-HA, En-1Δeh4-HA, or empty vector (1 μg), Cyclin D1/Luc (1 μg), and β-Gal (0.1 μg). Luciferase and β-Gal levels were measured as described in Figure 1. Cell lysates (30 μg) were subjected to Western analysis using anti-β-catenin, anti-HA, or anti-β-actin antibody.
DISCUSSION
Interactions between eng and wg signaling pathways have been extensively described previously (Bejsovec and Martinez Arias, 1991; Heemskerk et al., 1991; DiNardo et al., 1988; Martinez-Arias, 1998; Araki and Nakamura, 1999). A regulation of Fz receptor expression by Eng has been shown (Lecourtois et al., 2001), and recently Eng was shown to act as a direct repressor of Fz2 promoter (Solano et al., 2003). Because the spreading of the Wg signal is crucial for establishing the pattern of differentiated cell types within tissues and organs, the level of Fz2 expression that was shown to be repressed by Eng, was suggested to be significant for differentiation (Solano et al., 2003). The present study suggests that En-1 serves as a negative regulator of β-catenin. We showed that knocking down the endogenous En-1 by the RNAi approach increased β-catenin transcriptional activity activated by LiCl treatment or by overexpressing a mutant β-catenin. Moreover, this stimulation was associated with an increase in the level of β-catenin. Consistent with these results, En-1-HA overexpression resulted in a decrease in β-catenin transcriptional activity, concomitantly with a decrease in the steady-state level of ectopically expressed β-catenin in 293T cells as well as the level of the native β-catenin in LiCl-treated cells. Moreover, ectopically expressed En-1 also decreased the level of β-catenin stabilized by Wnt–3a/Hfz1 interaction (our unpublished data). Consistent with these results, our data showed that En-1 decreased the amount of TCF/β-catenin coimmunoprecipitates as well as the amount of TCF/β-catenin complexes bound to TCF target promoters when analyzed by gel electrophoretic mobility shift assay (our unpublished data).
Because En-1 did not affect the level of β-catenin transcripts, but it accelerated the degradation rate of β-catenin, our data strongly imply that En-1 represses β-catenin transcriptional activity by targeting the destruction of β-catenin. Moreover, our data suggested that En-1–promoted β-catenin degradation is proteasome dependent.
Interestingly, immunohistochemistry assays showed that when En-1 was transiently transfected into SW480 cells, 20–50% of En-1-HA–expressing cells displayed a dramatic decrease in the level of nuclear β-catenin. Because a fraction of En-1-HA–expressing SW480 cells were refractory to En-1 effect on the level of β-catenin, further studies are in progress to establish whether β-catenin sensitivity to En-1–destabilizing effect is cell cycle (Olmeda et al., 2003) or cell density (Brabletz et al., 2001; Davies et al., 2004) dependent.
The hallmark of the canonical Wnt pathway is the inhibition of GSK-3β/APC-dependent β-catenin ubiquitin-mediated proteasomal degradation. Our data showed that En-1 repressed LiCl-stimulated β-catenin transcriptional activity. En-1 destabilized also β-catenin(S33Y), which is insensitive to GSK-3β phosphorylation. Moreover, En-1 destabilized also β-cateninΔS45 in which Ser45 was deleted (our unpublished data). Because CK-1α–mediated phosphorylation of Ser45 was shown to prime β-catenin for further phosphorylation by GSK-3β (Polakis, 2002), our data suggest that β-catenin phosphorylation at Ser33, Ser37, and Thr41 are dispensable for En-1 effect on β-catenin. Hence, our data strongly imply that En-1–promoted β-catenin destabilization is GSK-3β independent. Although we showed that En-1–mediated β-catenin degradation is proteasome-dependent, further studies are required to establish whether En-1 effect on β-catenin is ubiquitin dependent.
In addition to GSK3β-dependent β-catenin turnover, recent observations suggested that β-catenin is down-regulated also by other pathways. One of these pathways, also APC mediated, is a p53-inducible proteasomal pathway transmitted via Siah-1 (Liu et al., 2001; Matsuzawa and Reed, 2001). However, our data showed that neither dnSiah-1 nor dnSiah-2 abolished En-1–repressive effect. Moreover, En-1 represses β-catenin transcriptional activity also in SW480 cells, which are impaired in Siah-mediated degradation due to loss of function mutations of both APC and p53 (Liu et al., 2001). These data thus imply that En-1–mediated β-catenin degradation is Siah and p53 independent. In addition to the two APC-dependent pathways, β-catenin down-regulation is executed also via a newly identified retinoid X receptor (RXR)-mediated APC-independent proteasomal degradation pathway that does not target the N terminus of β-catenin (Xiao et al., 2003). Because the N terminus of β-catenin was indispensable for En-1 activity, it is suggested that En-1 effect is not transmitted via the RXR-mediated degradation pathway. Additionally, our data showed that En-1 did not affect the stability of membrane-bound β-catenin, suggesting that En-1–destabilizing effect on β-catenin is not exerted via Ozz-mediated ubiquitination (Nastasi et al., 2004). Together, our results suggest that En-1 controls β-catenin turnover via a novel pathway that is independent of APC but still targets the N terminus of β-catenin. Further studies are in progress to investigate whether En-1–repressive activity is mediated via the peroxisome proliferator-activated receptor γ, recently shown to destabilize β-catenin by an APC- and Siah-independent pathway that targets the N terminus of β-catenin (Sharma et al., 2004). Notably, our data showed that En-1–repressive activity on β-catenin is not exerted via En-1 target genes.
Previously, we have shown that Wnt2 was overexpressed in CRC samples (Vider et al., 1996). Moreover, we found that En-1 was concomitantly overexpressed with Wnt2 in breast tumor cell lines as well in CRC samples (unpublished data). These data are in accordance with previous studies showing that the Drosophila Eng is a Wg target gene (Ohkuma et al., 1990; Bejsovec and Arias, 1991; Heemskerk et al., 1991) and that the expression of Wnt-1 was indirectly activated by En-1 (Heemskerk et al., 1991). Our data, showing that En-1 serves as a negative regulator of β-catenin transcriptional activity, suggest that En-1 is part of a negative feedback loop that is up-regulated in response to Wnt signaling. In this respect, En-1 resembles other negative regulators of the Wnt pathway, such as β-TrCP (Spiegelman et al., 2000), coductin/axin2 (Leung et al., 2002; Lustig et al., 2002), naked cuticle (Nkd) (Rousset et al., 2001), Dickkopf-1 (Gonzalez-Sancho et al., 2005) as well as the dominant forms of TCF-1 (Roose and Clevers, 1999), all of which are activated by Wnt signaling and thus serve, via a negative feedback loop, to down-regulate the activated Wnt pathway.
ACKNOWLEDGMENTS
We are grateful to Drs. Liu Guizhong and Stuart A. Aaronson as well as to Drs. R. Agami, W. Birchmeier, H. Clevers, J. Kitajewski, R. Moon, R. G. Pestell, C. Albanese, S. Stifani, B. Vogelstein, and Y. Yang for kindly providing various reagents used in this study. This work was supported by a grant from The Karl and Lionora Fund for Cancer Research, Sackler School of Medicine, Tel Aviv University, Israel. Parts of this work were completed by L.B.-D. in partial fulfillment of the requirements for the Ph.D. degree from the Sackler School of Medicine at Tel Aviv University.
Abbreviations used:
- APC
adenomatous polyposis coli
- Eng
Drosophila Engrailed
- En-1
Engrailed-1
- Fz
Frizzled
- HA
hemagglutinin
- β-Gal
β-galactosidase
- GSK
glycogen synthase kinase
- Gro
Groucho
- LEF/TCF
lymphoid enhancer factor/T-cell factor
- TLE
transducin-like enhancer of split
- Wg
Wingless.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0052) on March 29, 2006.
REFERENCES
- Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T. Wnt/beta-catenin signaling. Cytokine Growth Factor Rev. 2000;11:273–282. doi: 10.1016/s1359-6101(00)00011-3. [DOI] [PubMed] [Google Scholar]
- Araki I., Nakamura H. Engrailed defines the position of dorsal dimesencephalic boundary by repressing diencephalic fate. Development. 1999;126:5127–5135. doi: 10.1242/dev.126.22.5127. [DOI] [PubMed] [Google Scholar]
- Bejsovec A., Martinez Arias A. Roles of wingless in patterning the larval epidermis of Drosophila. Development. 1991;113:471–485. doi: 10.1242/dev.113.2.471. [DOI] [PubMed] [Google Scholar]
- Brabletz T., Jung A., Reu S., Porzner M., Hlubek F., Kunz-Schughart L. A., Knuechel R., Kirchner T. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., Peifer M., Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Jia S. Transcriptional repression: the long and the short of it. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- Davies M. L., Roberts G. T., Spiller D. G., Wakeman J. A. Density-dependent location and interactions of truncated APC and beta-catenin. Oncogene. 2004;23:1412–1419. doi: 10.1038/sj.onc.1207266. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Sher E., Heemskerk-Jongens J., Kassis J. A., O'Farrell P. H. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Gazit A., Yaniv A., Bafico A., Pramila T., Igarashi M., Kitajewski J., Aaronson S. A. Human frizzled 1 interacts with transforming Wnts to transduce a TCF dependent transcriptional response. Oncogene. 1999;18:5959–5966. doi: 10.1038/sj.onc.1202985. [DOI] [PubMed] [Google Scholar]
- Gibert J. M. The evolution of engrailed genes after duplication and speciation events. Dev. Genes Evol. 2002;212:307–318. doi: 10.1007/s00427-002-0243-2. [DOI] [PubMed] [Google Scholar]
- Golan T., Yaniv A., Bafico A., Liu G., Gazit A. The human Frizzled 6 (HFz6) acts as a negative regulator of the canonical Wnt. beta-catenin signaling cascade. J. Biol. Chem. 2004;279:14879–14888. doi: 10.1074/jbc.M306421200. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sancho J. M., Aguilera O., Garcia J. M., Pendas-Franco N., Pena C., Cal S., Garcia de Herreros A., Bonilla F. M. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098–1103. doi: 10.1038/sj.onc.1208303. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose W., Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- Heemskerk J., DiNardo S., Kostriken R., O'Farrell P. H. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991;352:404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J., Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr. Opin. Genet. Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- Hurlstone A., Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P. S., Melton D. A. A molecular mechanism for the effect of lithium on development Proc. Natl. Acad. Sci. USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs F. T., Hu G., Dang C. V., Fearon E. R. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Lecourtois M., Alexandre C., Dubois L., Vincent J. P. Wingless capture by Frizzled and Frizzled2 in Drosophila embryos. Dev. Biol. 2001;235:467–475. doi: 10.1006/dbio.2001.0320. [DOI] [PubMed] [Google Scholar]
- Leung J. Y., Kolligs F. T., Wu R., Zhai Y., Kuick R., Hanash S., Cho K. R., Fearon E. R. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 2002;277:21657–21665. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- Liu J., Stevens J., Rote C. A., Yost H. J., Hu Y., Neufeld K. L., White R. L., Matsunami N. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- Logan C., Hanks M. C., Noble-Topham S., Nallainathan D., Provart N. J., Joyner A. L. Cloning and sequence comparison of the mouse, human, and chicken engrailed genes reveal potential functional domains and regulatory regions. Dev. Genet. 1992;13:348–358. doi: 10.1002/dvg.1020130505. [DOI] [PubMed] [Google Scholar]
- Lustig B., et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias A. Interactions between Wingless and Notch during the assignment of cell fates in Drosophila. Int. J. Dev. Biol. 1998;42:325–333. [PubMed] [Google Scholar]
- Matsuzawa S., Reed J. C. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell. 2001;7:915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K. W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Munemitsu S., Albert I., Souza B., Rubinfeld B., Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc. Natl. Acad. Sci. USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastasi T., et al. Ozz-E3, a muscle-specific ubiquitin ligase, regulates beta-catenin degradation during myogenesis. Dev. Cell. 2004;6:269–282. doi: 10.1016/s1534-5807(04)00020-6. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y., Horikoshi M., Roeder R. G., Despla C. Engrailed, a homeodomain protein, can repress in vitro transcription by competition with the TATA box-binding protein transcription factor IID. Proc. Natl. Acad. Sci. USA. 1990;87:2289–2293. doi: 10.1073/pnas.87.6.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmeda D., Castel S., Vilaro S., Cano A. Beta-catenin regulation during the cell cycle: implications in G2/M and apoptosis. Mol. Biol. Cell. 2003;14:2844–2860. doi: 10.1091/mbc.E03-01-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K., Crockett C., Jensen J. P., Wessman A. M., Byers S. W. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J. Biol. Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- Peifer M., Polakis P. Wnt signaling in oncogenesis and embryogenesis–a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- Polakis P. Casein kinase 1, a Wnt'er of disconnect. Curr. Biol. 2002;23:499–501. doi: 10.1016/s0960-9822(02)00969-7. [DOI] [PubMed] [Google Scholar]
- Roose J., Clevers H. TCF transcription factors: molecular switches in carcinogenesis. Biochim. Biophys. Acta. 1999;1424:23–37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., Van de Wetering M., Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Rousset R., Mack J. A., Wharton K. A., Jr, Axelrod J. D., Cadigan K. M., Fish M. P., Nusse R., Scott M. P. Naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev. 2001;15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano N., Maschat F. Molecular mechanism of polyhomeotic activation by Engrailed. EMBO J. 1998;17:3704–3713. doi: 10.1093/emboj/17.13.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano N., Brock H. W., Maschat F. beta3-tubulin is directly repressed by the engrailed protein in Drosophila. Development. 1997;124:2527–2536. doi: 10.1242/dev.124.13.2527. [DOI] [PubMed] [Google Scholar]
- Sharma C., Pradeep A., Wong L., Rana A., Rana B. J. Peroxisome proliferator-activated receptor gamma activation can regulate beta-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J. Biol. Chem. 2004;279:35583–35594. doi: 10.1074/jbc.M403143200. [DOI] [PubMed] [Google Scholar]
- Shtutman M., Zhurinsky J., Simcha I., Albanese C., D'Amico M., Pestell R., Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano P. J., Mugat B., Martin D., Girard F., Huibant J. M., Ferraz C., Jacq B., Demaille J., Maschat F. Genome-wide identification of in vivo Drosophila Engrailed-binding DNA fragments and related target genes. Development. 2003;130:1243–1254. doi: 10.1242/dev.00348. [DOI] [PubMed] [Google Scholar]
- Spiegelman V. S., Slaga T. J., Pagano M., Minamoto T., Ronai Z., Fuchs S. Y. Wnt/beta-catenin signaling induces the expression and activity of betaTrCP ubiquitin ligase receptor. Mol. Cell. 2000;5:877–882. doi: 10.1016/s1097-2765(00)80327-5. [DOI] [PubMed] [Google Scholar]
- Tolkunova E. N., Fujioka M., Kobayashi M., Deka D., Jaynes J. B. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol. Cell Biol. 1998;18:2804–2814. doi: 10.1128/mcb.18.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol L., Jiang X., Choi H., Garrett-Beal L., Carolan P. J., Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J. Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vider B. Z., Zimber A., Chastre E., Prevot S., Gespach C., Estlein D., Wolloch Y., Tronick S. R., Gazit A., Yaniv A. Evidence for the involvement of the Wnt 2 gene in human colorectal cancer. Oncogene. 1996;4:153–158. [PubMed] [Google Scholar]
- Xiao J. H., Ghosn C., Hinchman F., Forbes C., Wang J., Snider N., Cordrey A., Zhao Y., Chandraratna R.A.S. Adenomatous polyposis coli (APC)-independent regulation of beta-catenin degradation via a retinoid X receptor-mediated pathway. J. Biol. Chem. 2003;278:29954–29962. doi: 10.1074/jbc.M304761200. [DOI] [PubMed] [Google Scholar]